24:
352:. It is also a necessary cofactor in the metabolic pathway of alkene-oxidizing bacteria. CoM helps eliminate the toxic epoxides formed from the oxidation of alkenes such as propylene. The structure of this coenzyme was discovered by CD Taylor and RS Wolfe in 1974 while they were studying methanogenesis, the process by which carbon dioxide is transformed into methane in some archaea. The coenzyme is an
516:
and propylene or ethylene in aerobic bacteria. Specifically, in bacteria that oxidize alkenes into epoxides. After the propylene (or other alkene) undergoes epoxidation and becomes epoxypropane it becomes electrophilic and toxic. These epoxides react with DNA and proteins, affecting cell function.
677:
Partovi, Sarah E.; Mus, Florence; Gutknecht, Andrew E.; Martinez, Hunter A.; Tripet, Brian P.; Lange, Bernd Markus; DuBois, Jennifer L.; Peters, John W. (2018-04-06).
523:
use a metabolic pathway in which CoM is conjugated with an aliphatic epoxide. This step creates a nucleophilic compound which can react with CO
679:"Coenzyme M biosynthesis in bacteria involves phosphate elimination by a functionally distinct member of the aspartase/fumarase superfamily"
240:
1115:
978:
974:
907:
60:
2-mercaptoethylsulfonate; 2-mercaptoethanesulfonate; coenzyme M anion; H-S-CoM; AC1L1HCY; 2-sulfanylethane-1-sulfonate; CTK8A8912
741:
Krishnakumar, Arathi M.; Sliwa, Darius; Endrizzi, James A.; Boyd, Eric S.; Ensign, Scott A.; Peters, John W. (September 2008).
819:
970:
966:
215:
115:
318:
1034:
900:
1092:
46:
191:
956:
799:
493:
842:"Biochemistry of methanogenesis: a tribute to Marjory Stephenson:1998 Marjory Stephenson Prize Lecture"
519:
893:
1187:
1200:
919:
1100:
942:
1110:
1105:
952:
36:
1018:
798:
Parry, Ronald J. (1999-01-01), Barton, Sir Derek; Nakanishi, Koji; Meth-Cohn, Otto (eds.),
200:
90:
155:
82:
8:
1317:
1192:
998:
72:
135:
1307:
1214:
811:
775:
742:
713:
678:
559:"Specificity and biological distribution of coenzyme M (2-mercaptoethanesulfonic acid)"
654:
607:
583:
558:
1285:
1044:
1026:
871:
863:
815:
780:
762:
718:
700:
659:
588:
853:
807:
770:
754:
708:
690:
649:
578:
570:
501:
473:, 7-thioheptanoylthreoninephosphate, to give a heterodisulfide, releasing methane:
263:
574:
1312:
1048:
527:. The eventual carboxylation produces acetoacetate, breaking down the propylene.
180:
858:
841:
1064:
420:
312:
1301:
1177:
1130:
1082:
867:
766:
704:
695:
497:
1182:
1172:
1140:
784:
722:
875:
758:
663:
1167:
1120:
592:
1247:
1125:
988:
470:
345:
338:
297:
146:
1242:
1237:
1072:
1058:
424:
403:
311:
Except where otherwise noted, data are given for materials in their
885:
349:
334:
23:
114:
1278:
1222:
934:
743:"Getting a Handle on the Role of Coenzyme M in Alkene Metabolism"
466:
342:
167:
1252:
1227:
1161:
1157:
1153:
1149:
1008:
916:
490:
391:
740:
536:
399:
395:
353:
126:
104:
398:
is most available. Mercaptoethanesulfonate contains both a
1257:
1232:
1145:
800:"1.29 - Biosynthesis of Sulfur-containing Natural Products"
676:
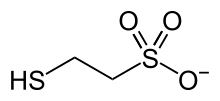
386:. It is named 2-mercaptoethanesulfonate and abbreviated
539:– a cancer chemotherapy adjuvant with the same structure
224:
InChI=1S/C2H6O3S2/c3-7(4,5)2-1-6/h6H,1-2H2,(H,3,4,5)/p-1
423:. It is converted to methyl-coenzyme M thioether, the
512:
Coenzyme M is also used to make acetoacetate from CO
406:group, which confers solubility in aqueous media.
1299:
179:
348:, and in the metabolism of other substrates in
89:
81:
402:, which is the main site of reactivity, and a
337:required for methyl-transfer reactions in the
901:
605:
908:
894:
747:Microbiology and Molecular Biology Reviews
556:
154:
857:
774:
712:
694:
653:
608:"Structure and methylation of coenzyme M(
582:
469:formation. Methyl-coenzyme M reacts with
199:
804:Comprehensive Natural Products Chemistry
1300:
839:
806:, Oxford: Pergamon, pp. 825–863,
606:Taylor CD, Wolfe RS (10 August 1974).
889:
797:
736:
734:
732:
227:Key: ZNEWHQLOPFWXOF-UHFFFAOYSA-M
134:
915:
507:
683:The Journal of Biological Chemistry
489:This induction is catalyzed by the
409:
170:
13:
812:10.1016/b978-0-08-091283-7.00031-x
729:
14:
1329:
414:
840:Thauer, Rudolf K. (1998-09-01).
419:The coenzyme is the C1 donor in
275:
22:
517:Alkene-oxidizing bacteria like
315:(at 25 °C , 100 kPa).
833:
791:
670:
599:
550:
287:
281:
269:
1:
655:10.1016/S0021-9258(19)42403-4
575:10.1128/JB.137.1.256-263.1979
543:
465:, in the penultimate step to
7:
859:10.1099/00221287-144-9-2377
557:Balch WE, Wolfe RS (1979).
530:
494:methyl-coenzyme M reductase
10:
1334:
520:Xanthobacter autotrophicus
204: (sulfonic acid form)
86: (sulfonic acid form)
1270:
1213:
1091:
933:
926:
309:
256:
236:
211:
65:
57:
51:2-Sulfanylethanesulfonate
45:
40:2-Sulfanylethanesulfonate
35:
30:
21:
696:10.1074/jbc.RA117.001234
394:is unimportant, but the
94: (sulfonate form)
759:10.1128/MMBR.00005-08
47:Systematic IUPAC name
481:–S–CoM + HS–CoB → CH
305: g·mol
18:
496:, which restricts
319:Infobox references
16:
1295:
1294:
1266:
1265:
821:978-0-08-091283-7
689:(14): 5236–5246.
508:Alkene metabolism
356:with the formula
327:Chemical compound
325:
324:
116:Interactive image
1325:
931:
930:
910:
903:
896:
887:
886:
880:
879:
861:
852:(9): 2377–2406.
837:
831:
830:
829:
828:
795:
789:
788:
778:
738:
727:
726:
716:
698:
674:
668:
667:
657:
637:
636:
635:
627:
626:
618:
617:
603:
597:
596:
586:
554:
502:prosthetic group
464:
463:
462:
454:
453:
445:
444:
436:
435:
410:Biochemical role
385:
384:
383:
375:
374:
366:
365:
304:
289:
283:
277:
271:
264:Chemical formula
203:
183:
172:
158:
138:
118:
93:
85:
26:
19:
15:
1333:
1332:
1328:
1327:
1326:
1324:
1323:
1322:
1298:
1297:
1296:
1291:
1262:
1209:
1204:
1196:
1087:
1078:
1070:
1054:
1040:
1030:
1022:
1014:
1004:
994:
984:
962:
948:
922:
914:
884:
883:
838:
834:
826:
824:
822:
796:
792:
739:
730:
675:
671:
648:(15): 4879–85.
634:
631:
630:
629:
625:
622:
621:
620:
616:
613:
612:
611:
609:
604:
600:
555:
551:
546:
533:
526:
515:
510:
484:
480:
461:
458:
457:
456:
452:
449:
448:
447:
443:
440:
439:
438:
434:
431:
430:
429:
427:
417:
412:
382:
379:
378:
377:
373:
370:
369:
368:
364:
361:
360:
359:
357:
328:
321:
316:
302:
292:
286:
280:
274:
266:
252:
249:
244:
243:
232:
229:
228:
225:
219:
218:
207:
186:
173:
161:
141:
121:
108:
97:
75:
61:
53:
52:
41:
12:
11:
5:
1331:
1321:
1320:
1315:
1310:
1293:
1292:
1290:
1289:
1274:
1272:
1268:
1267:
1264:
1263:
1261:
1260:
1255:
1250:
1245:
1240:
1235:
1230:
1225:
1219:
1217:
1211:
1210:
1208:
1207:
1202:
1198:
1194:
1190:
1185:
1180:
1175:
1170:
1165:
1143:
1138:
1133:
1128:
1123:
1118:
1113:
1108:
1103:
1097:
1095:
1089:
1088:
1086:
1085:
1080:
1076:
1068:
1062:
1056:
1052:
1042:
1038:
1028:
1020:
1016:
1012:
1006:
1002:
996:
992:
986:
982:
964:
960:
950:
946:
939:
937:
928:
924:
923:
913:
912:
905:
898:
890:
882:
881:
832:
820:
790:
753:(3): 445–456.
728:
669:
632:
623:
614:
598:
548:
547:
545:
542:
541:
540:
532:
529:
524:
513:
509:
506:
487:
486:
482:
478:
459:
450:
441:
432:
421:methanogenesis
416:
415:Methanogenesis
413:
411:
408:
380:
371:
362:
326:
323:
322:
317:
313:standard state
310:
307:
306:
300:
294:
293:
290:
284:
278:
272:
267:
262:
259:
258:
254:
253:
251:
250:
247:
239:
238:
237:
234:
233:
231:
230:
226:
223:
222:
214:
213:
212:
209:
208:
206:
205:
196:
194:
188:
187:
185:
184:
176:
174:
166:
163:
162:
160:
159:
151:
149:
143:
142:
140:
139:
131:
129:
123:
122:
120:
119:
111:
109:
102:
99:
98:
96:
95:
87:
78:
76:
71:
68:
67:
63:
62:
59:
55:
54:
50:
49:
43:
42:
39:
33:
32:
28:
27:
9:
6:
4:
3:
2:
1330:
1319:
1316:
1314:
1311:
1309:
1306:
1305:
1303:
1288:
1287:
1281:
1280:
1276:
1275:
1273:
1269:
1259:
1256:
1254:
1251:
1249:
1246:
1244:
1241:
1239:
1236:
1234:
1231:
1229:
1226:
1224:
1221:
1220:
1218:
1216:
1212:
1206:
1199:
1197:
1191:
1189:
1186:
1184:
1181:
1179:
1178:Molybdopterin
1176:
1174:
1171:
1169:
1166:
1163:
1159:
1155:
1151:
1147:
1144:
1142:
1139:
1137:
1134:
1132:
1131:Cofactor F430
1129:
1127:
1124:
1122:
1119:
1117:
1114:
1112:
1109:
1107:
1104:
1102:
1099:
1098:
1096:
1094:
1090:
1084:
1083:Coenzyme F420
1081:
1074:
1066:
1065:Phylloquinone
1063:
1060:
1059:Ascorbic acid
1057:
1050:
1046:
1043:
1036:
1032:
1024:
1017:
1010:
1007:
1000:
997:
990:
987:
980:
976:
972:
968:
965:
958:
954:
951:
944:
941:
940:
938:
936:
932:
929:
925:
921:
918:
911:
906:
904:
899:
897:
892:
891:
888:
877:
873:
869:
865:
860:
855:
851:
847:
843:
836:
823:
817:
813:
809:
805:
801:
794:
786:
782:
777:
772:
768:
764:
760:
756:
752:
748:
744:
737:
735:
733:
724:
720:
715:
710:
706:
702:
697:
692:
688:
684:
680:
673:
665:
661:
656:
651:
647:
643:
642:J. Biol. Chem
639:
602:
594:
590:
585:
580:
576:
572:
569:(1): 256–63.
568:
564:
560:
553:
549:
538:
535:
534:
528:
522:
521:
505:
503:
499:
498:cofactor F430
495:
492:
485:+ CoB–S–S–CoM
476:
475:
474:
472:
468:
426:
422:
407:
405:
401:
397:
393:
389:
355:
351:
347:
344:
340:
336:
332:
320:
314:
308:
301:
299:
296:
295:
268:
265:
261:
260:
255:
246:
245:
242:
235:
221:
220:
217:
210:
202:
198:
197:
195:
193:
190:
189:
182:
178:
177:
175:
169:
165:
164:
157:
153:
152:
150:
148:
145:
144:
137:
133:
132:
130:
128:
125:
124:
117:
113:
112:
110:
106:
101:
100:
92:
88:
84:
80:
79:
77:
74:
70:
69:
64:
56:
48:
44:
38:
34:
29:
25:
20:
1283:
1277:
1183:Mycofactocin
1173:Methanofuran
1135:
1093:non-vitamins
927:Active forms
849:
846:Microbiology
845:
835:
825:, retrieved
803:
793:
750:
746:
686:
682:
672:
645:
641:
601:
566:
563:J. Bacteriol
562:
552:
518:
511:
488:
418:
387:
330:
329:
248:S(=O)(=O)CCS
66:Identifiers
58:Other names
1168:Lipoic Acid
1146:Heme / Haem
1073:Menaquinone
396:sodium salt
346:methanogens
257:Properties
136:CHEBI:58319
17:Coenzyme M
1318:Sulfonates
1302:Categories
1271:Base forms
1215:metal ions
1141:Coenzyme Q
1136:Coenzyme M
1126:Coenzyme B
989:Coenzyme A
943:TPP / ThDP
827:2022-05-10
544:References
471:coenzyme B
339:metabolism
331:Coenzyme M
298:Molar mass
201:VHD28S0H7F
147:ChemSpider
103:3D model (
91:40292-31-7
73:CAS Number
37:IUPAC name
1308:Coenzymes
1201:THMPT / H
999:PLP / P5P
920:cofactors
868:1350-0872
767:1092-2172
705:1083-351X
425:thioether
404:sulfonate
83:3375-50-6
1286:vitamins
1279:vitamins
1193:THB / BH
1027:DHFA / H
1019:THFA / H
935:vitamins
785:18772284
723:29414784
531:See also
350:bacteria
343:archaeal
335:coenzyme
876:9782487
776:2546864
714:5892593
664:4367810
500:as the
467:methane
390:. The
168:PubChem
1313:Thiols
1233:Fe, Fe
1045:AdoCbl
1009:Biotin
917:Enzyme
874:
866:
818:
783:
773:
765:
721:
711:
703:
662:
593:104960
591:
584:218444
581:
491:enzyme
392:cation
388:HS–CoM
303:141.18
241:SMILES
31:Names
1049:MeCbl
979:NADPH
537:Mesna
400:thiol
354:anion
333:is a
216:InChI
127:ChEBI
105:JSmol
1284:see
1116:PAPS
1111:SAMe
1035:MTHF
975:NADP
971:NADH
872:PMID
864:ISSN
816:ISBN
781:PMID
763:ISSN
719:PMID
701:ISSN
660:PMID
610:HSCH
589:PMID
358:HSCH
192:UNII
181:4077
156:3935
1205:MPT
1188:PQQ
1121:GSH
1106:CTP
1101:ATP
1071:),
1061:(C)
967:NAD
957:FAD
953:FMN
854:doi
850:144
808:doi
771:PMC
755:doi
709:PMC
691:doi
687:293
650:doi
646:249
579:PMC
571:doi
567:137
437:SCH
341:of
171:CID
1304::
1282::
1258:Zn
1253:Ni
1248:Mo
1243:Mn
1238:Mg
1228:Cu
1223:Ca
1160:,
1156:,
1152:,
1075:(K
1067:(K
1053:12
1051:(B
1047:,
1037:(B
1033:,
1031:FA
1025:,
1023:FA
1011:(B
1001:(B
991:(B
981:(B
977:,
973:,
969:,
959:(B
955:,
945:(B
870:.
862:.
848:.
844:.
814:,
802:,
779:.
769:.
761:.
751:72
749:.
745:.
731:^
717:.
707:.
699:.
685:.
681:.
658:.
644:.
640:.
638:)"
628:SO
619:CH
587:.
577:.
565:.
561:.
504:.
477:CH
455:SO
446:CH
428:CH
376:SO
367:CH
1203:4
1195:4
1164:)
1162:O
1158:C
1154:B
1150:A
1148:(
1079:)
1077:2
1069:1
1055:)
1041:)
1039:9
1029:2
1021:4
1015:)
1013:7
1005:)
1003:6
995:)
993:5
985:)
983:3
963:)
961:2
949:)
947:1
909:e
902:t
895:v
878:.
856::
810::
787:.
757::
725:.
693::
666:.
652::
633:3
624:2
615:2
595:.
573::
525:2
514:2
483:4
479:3
460:3
451:2
442:2
433:3
381:3
372:2
363:2
291:2
288:S
285:3
282:O
279:5
276:H
273:2
270:C
107:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.