248:
173:
512:
35:
648:
409:
535:(T) as it is identical to adenine (A) but has an amine group at position 2 forming 3 intermolecular hydrogen bonds, eliminating the major difference between the two types of basepairs (weak:A-T and strong:C-G). This improved stability affects protein-binding interactions that rely on those differences.
1482:
Trzaska, C; Amand, S; Bailly, C; Leroy, C; Marchand, V; Duvernois-Berthet, E; Saliou, JM; Benhabiles, H; Werkmeister, E; Chassat, T; Guilbert, R; Hannebique, D; Mouray, A; Copin, MC; Moreau, PA; Adriaenssens, E; Kulozik, A; Westhof, E; Tulasne, D; Motorin, Y; Rebuffat, S; Lejeune, F (20 March 2020).
1089:
Pezo, Valerie; Jaziri, Faten; Bourguignon, Pierre-Yves; Louis, Dominique; Jacobs-Sera, Deborah; Rozenski, Jef; Pochet, Sylvie; Herdewijn, Piet; Hatfull, Graham F.; Kaminski, Pierre-Alexandre; Marliere, Philippe (30 April 2021). "Noncanonical DNA polymerization by aminoadenine-based siphoviruses".
1446:
Zouharova, D; Lipenska, I; Fojtikova, M; Kulich, P; Neca, J; Slany, M; Kovarcik, K; Turanek-Knotigova, P; Hubatka, F; Celechovska, H; Masek, J; Koudelka, S; Prochazka, L; Eyer, L; Plockova, J; Bartheldyova, E; Miller, AD; Ruzek, D; Raska, M; Janeba, Z; Turanek, J (29 February 2016). "Antiviral
1037:
Sleiman, Dona; Garcia, Pierre Simon; Lagune, Marion; Loc’h, Jerome; Haouz, Ahmed; Taib, Najwa; Röthlisberger, Pascal; Gribaldo, Simonetta; Marlière, Philippe; Kaminski, Pierre
Alexandre (30 April 2021). "A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes".
1141:
Zhou, Yan; Su, Xuexia; Wei, Yifeng; Cheng, Yu; Guo, Yu; Khudyakov, Ivan; Liu, Fuli; He, Ping; Song, Zhangyue; Li, Zhi; Gao, Yan; Ang, Ee Lui; Zhao, Huimin; Zhang, Yan; Zhao, Suwen (30 April 2021).
1540:
Kamiya, Y; Donoshita, Y; Kamimoto, H; Murayama, K; Ariyoshi, J; Asanuma, H (5 October 2017). "Introduction of 2,6-Diaminopurines into
Serinol Nucleic Acid Improves Anti-miRNA Performance".
903:
Kirnos, MD; Khudyakov, IY; Alexandrushkina, NI; Vanyushin, BF (November 1977). "2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA".
1447:
activities of 2,6-diaminopurine-based acyclic nucleoside phosphonates against herpesviruses: In vitro study results with pseudorabies virus (PrV, SuHV-1)".
576:; expression of this system is toxic to the cell. The structures of MazZ (subtype 2) and PurZ are also determined, showing a possible link between PurZ and
416:
520:
947:
294:
1325:
Weckbecker, G; Cory, JG (1989). "Metabolic activation of 2,6-diaminopurine and 2,6-diaminopurine-2'-deoxyriboside to antitumor agents".
1276:
Burchenal, JH; Karnofsky, DA; Kingsley-Pillers, EM; Southam, CM; Myers, WP; Escher, GC; Craver, LF; Dargeon, HW; Rhoads, CP (May 1951).
570:
In August 2021, it was shown that DatZ, MazZ and PurZ are sufficient to replace some occurrence of A by Z in the bacterial genome of
1294:
1277:
967:
Czernecki, Dariusz; Legrand, Pierre; Tekpinar, Mustafa; Rosario, Sandrine; Kaminski, Pierre-Alexandre; Delarue, Marc (2021-04-23).
1585:"The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA"
1278:"The effects of the folic acid antagonists and 2,6-diaminopurine on neoplastic disease, with special reference to acute leukemia"
790:
Callahan, M.P.; Smith, K.E.; Cleaves, H.J.; Ruzica, J.; Stern, J.C.; Glavin, D.P.; House, C.H.; Dworkin, J.P. (11 August 2011).
776:
1201:"Characterization of a triad of genes in cyanophage S-2L sufficient to replace adenine by 2-aminoadenine in bacterial DNA"
762:
691:
262:
17:
423:
745:
DAP is used similarly to other nuclear acid analogues in the investigation of enzyme structures and mechanisms.
538:
Four papers published April 2021 further describes the use and production of the Z-base. It is now known that:
109:
969:"How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA"
205:
168:
735:
226:
129:
555:
responsible for handling the Z base occurs in the same gene cluster as the three aforementioned enzymes;
714:
virus, a economically important livestock disease. This base, in its free form, is able to correct UGA
528:
854:
545:
The Z base is produced by a pathway involving DUF550 (MazZ) and PurZ in S-2L and Vibrio phage PhiVC8;
690:
in mouse leukemia cells by 1989. Cancer cells are known to become resistant to DAP by losing their
243:
879:
1199:
Czernecki, Dariusz; Bonhomme, Frédéric; Kaminski, Pierre-Alexandre; Delarue, Marc (2021-08-05).
662:
506:
47:
542:
The S-2L phage avoids incorporating A bases in the genome by hydrolyzing dATP (DatZ enzyme);
1643:
1496:
1373:
1220:
1212:
1154:
1099:
1047:
980:
912:
807:
214:
150:
549:
85:
8:
247:
172:
75:
1500:
1377:
1360:
Shao, C; Deng, L; Henegariu, O; Liang, L; Stambrook, PJ; Tischfield, JA (20 June 2000).
1216:
1158:
1103:
1051:
984:
916:
811:
661:
Please expand the article to include this information. Further details may exist on the
1565:
1517:
1484:
1307:
1253:
1200:
1178:
1123:
1071:
1011:
968:
830:
791:
1609:
1584:
1614:
1557:
1522:
1464:
1428:
1401:
1396:
1361:
1342:
1338:
1299:
1258:
1240:
1182:
1170:
1142:
1127:
1115:
1075:
1063:
1016:
998:
928:
835:
715:
474:
1569:
1311:
1143:"A widespread pathway for substitution of adenine by diaminopurine in phage genomes"
742:
nucleic acid type) incorporating base Z instead of A show enhanced binding to RNA.
1638:
1604:
1596:
1549:
1512:
1504:
1456:
1391:
1381:
1334:
1289:
1248:
1230:
1162:
1107:
1055:
1006:
988:
920:
825:
815:
631:
387:
317:
1362:"Chromosome instability contributes to loss of heterozygosity in mice lacking p53"
194:
1460:
1508:
1366:
Proceedings of the
National Academy of Sciences of the United States of America
1235:
993:
552:
400:
1225:
948:"Some Viruses Have a Completely Different Genome to The Rest of Life on Earth"
792:"Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases"
588:
2-aminoadenine is produced in two steps. The enzyme MazZ (homologous to MazG,
1632:
1600:
1244:
1002:
455:
376:
161:
1386:
1166:
1111:
1059:
820:
511:
1561:
1553:
1526:
1468:
1405:
1303:
1295:
10.1002/1097-0142(195105)4:3<549::aid-cncr2820040308>3.0.co;2-j
1262:
1174:
1119:
1067:
1020:
952:
883:
839:
711:
559:
524:
1618:
1485:"2,6-Diaminopurine as a highly potent corrector of UGA nonsense mutations"
1346:
1432:
1275:
932:
563:
494:
902:
777:"Some viruses thwart bacterial defenses with a unique genetic alphabet"
719:
687:
612:
589:
466:
345:
141:
924:
707:
399:
Except where otherwise noted, data are given for materials in their
271:
InChI=1S/C5H6N6/c6-3-2-4(9-1-8-2)11-5(7)10-3/h1H,(H5,6,7,8,9,10,11)=
34:
683:
577:
447:
281:
InChI=1S/C5H6N6/c6-3-2-4(9-1-8-2)11-5(7)10-3/h1H,(H5,6,7,8,9,10,11)
880:"DNA Building Blocks Can Be Made in Space, NASA Evidence Suggests"
108:
739:
696:
572:
532:
490:
486:
451:
366:
181:
1445:
1198:
647:
1539:
966:
630:
The resulting dSMP is processed by host enzymes analogously to
120:
1418:
723:
686:
since as early as 1951. It is known to arrest progression of
470:
98:
855:"NASA Researchers: DNA Building Blocks Can Be Made in Space"
659:
about results of the altered H-bond strength in DNA and RNA.
231:
1419:
Kocharian, ShM; Chukanova, TI; Sukhodolets, VV (1977). "".
858:
803:
462:
381:
117 to 122 °C (243 to 252 °F; 390 to 395 K)
1088:
789:
482:
478:
473:, was published suggesting 2,6-diaminopurine and related
1036:
1481:
1359:
566:, based on the occurrence of the said gene cluster.
877:
1630:
193:
796:Proceedings of the National Academy of Sciences
531:). Diaminopurine base (Z) pairs perfectly with
84:
1324:
1140:
446:) is a compound once used in the treatment of
527:), diaminopurine is used instead of adenine (
493:, may have been formed extraterrestrially in
852:
846:
246:
171:
149:
1608:
1516:
1395:
1385:
1293:
1252:
1234:
1224:
1032:
1030:
1010:
992:
829:
819:
213:
510:
450:. As the Z base, it is found instead of
783:
694:(APRT) function, a process shared with
558:The Z base is quite widespread in both
242:
14:
1631:
1582:
1027:
945:
274:Key: MSSXOMSJDRHRMC-UHFFFAOYSA-N=
162:
1194:
1192:
623:+ L-aspartate = (d)ADP + phosphate +
611:The enzyme PurZ (homologous to PurA,
128:
1134:
1082:
878:ScienceDaily Staff (9 August 2011).
755:
641:
454:(A) in the genetic material of some
853:Steigerwald, John (8 August 2011).
637:
625:2-aminodeoxyadenylosuccinate (dSMP)
461:In August 2011, a report, based on
361:White to yellow crystalline powder
184:
24:
1189:
871:
25:
1655:
729:
692:adenine phosphoribosyltransferase
646:
407:
335:
329:
33:
1576:
1533:
1475:
1439:
1412:
1353:
1318:
1269:
583:
403:(at 25 °C , 100 kPa).
960:
946:Bowler, Jacinta (4 May 2021).
939:
896:
769:
323:
13:
1:
1327:Advances in Enzyme Regulation
748:
500:
1583:Bailly, C (1 October 1998).
1461:10.1016/j.vetmic.2016.01.010
1339:10.1016/0065-2571(89)90068-x
722:, through the inhibition of
7:
736:anti-miRNA oligonucleotides
10:
1660:
1509:10.1038/s41467-020-15140-z
1236:10.1038/s41467-021-25064-x
994:10.1038/s41467-021-22626-x
682:2,6-DAP was used to treat
504:
1226:10.1101/2021.04.30.442174
720:translational readthrough
515:DiampurineT DNA base pair
397:
310:
290:
258:
68:
60:
46:
41:
32:
1449:Veterinary Microbiology
1387:10.1073/pnas.97.13.7405
1167:10.1126/science.abe4882
1112:10.1126/science.abe6542
1060:10.1126/science.abe6494
821:10.1073/pnas.1106493108
63:2-aminoadenine; 2,6-DAP
1601:10.1093/nar/26.19.4309
1589:Nucleic Acids Research
1554:10.1002/cbic.201700272
657:is missing information
516:
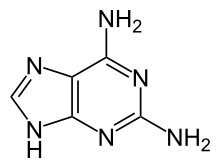
507:nucleic acid analogues
1489:Nature Communications
1205:Nature Communications
973:Nature Communications
763:"George H. Hitchings"
514:
703:DAP derivatives are
1501:2020NatCo..11.1509T
1378:2000PNAS...97.7405S
1217:2021NatCo..12.4710C
1159:2021Sci...372..512Z
1104:2021Sci...372..520P
1052:2021Sci...372..516S
985:2021NatCo..12.2420C
917:1977Natur.270..369K
812:2011PNAS..10813995C
738:(specifically, the
734:In bioengineering,
388:Solubility in water
353: g·mol
55:-purine-2,6-diamine
29:
716:nonsense mutations
592:) first performs:
580:versions of PurA.
517:
424:Infobox references
393:2.38 g/L at 20 °C
28:2,6-Diaminopurine
27:
1595:(19): 4309–4314.
1548:(19): 1917–1922.
1153:(6541): 512–516.
1098:(6541): 520–524.
1046:(6541): 516–520.
765:. nobelprize.org.
680:
679:
634:to produce dZTP.
615:) then performs:
475:organic molecules
436:2,6-diaminopurine
432:Chemical compound
430:
429:
227:CompTox Dashboard
110:Interactive image
18:2,6-diaminopurine
16:(Redirected from
1651:
1623:
1622:
1612:
1580:
1574:
1573:
1537:
1531:
1530:
1520:
1479:
1473:
1472:
1443:
1437:
1436:
1416:
1410:
1409:
1399:
1389:
1357:
1351:
1350:
1322:
1316:
1315:
1297:
1273:
1267:
1266:
1256:
1238:
1228:
1196:
1187:
1186:
1138:
1132:
1131:
1086:
1080:
1079:
1034:
1025:
1024:
1014:
996:
964:
958:
957:
943:
937:
936:
925:10.1038/270369a0
911:(5635): 369–70.
900:
894:
893:
891:
890:
875:
869:
868:
866:
865:
850:
844:
843:
833:
823:
787:
781:
780:
773:
767:
766:
759:
675:
672:
666:
650:
642:
638:In cellular life
632:adenylosuccinate
477:, including the
442:, also known as
414:
411:
410:
352:
337:
331:
325:
318:Chemical formula
302:Nc1nc(N)nc2c1nc2
251:
250:
235:
233:
217:
197:
186:
175:
164:
153:
132:
112:
88:
37:
30:
26:
21:
1659:
1658:
1654:
1653:
1652:
1650:
1649:
1648:
1629:
1628:
1627:
1626:
1581:
1577:
1538:
1534:
1480:
1476:
1444:
1440:
1427:(10): 1821–30.
1417:
1413:
1372:(13): 7405–10.
1358:
1354:
1323:
1319:
1274:
1270:
1197:
1190:
1139:
1135:
1087:
1083:
1035:
1028:
965:
961:
944:
940:
901:
897:
888:
886:
876:
872:
863:
861:
851:
847:
806:: 13995–13998.
788:
784:
775:
774:
770:
761:
760:
756:
751:
732:
718:by encouraging
710:useful against
676:
670:
667:
660:
651:
640:
602:
586:
521:cyanophage S-2L
509:
503:
433:
426:
421:
420:
419: ?)
412:
408:
404:
390:
350:
340:
334:
328:
320:
306:
303:
298:
297:
286:
283:
282:
276:
275:
272:
266:
265:
254:
236:
229:
220:
200:
187:
156:
136:
115:
102:
91:
78:
64:
56:
23:
22:
15:
12:
11:
5:
1657:
1647:
1646:
1641:
1625:
1624:
1575:
1532:
1474:
1438:
1411:
1352:
1317:
1268:
1188:
1133:
1081:
1026:
959:
938:
895:
870:
845:
782:
768:
753:
752:
750:
747:
731:
730:Bioengineering
728:
678:
677:
654:
652:
645:
639:
636:
628:
627:
609:
608:
600:
585:
582:
568:
567:
556:
553:DNA polymerase
546:
543:
505:Main article:
502:
499:
444:2-aminoadenine
431:
428:
427:
422:
406:
405:
401:standard state
398:
395:
394:
391:
386:
383:
382:
379:
373:
372:
369:
363:
362:
359:
355:
354:
348:
342:
341:
338:
332:
326:
321:
316:
313:
312:
308:
307:
305:
304:
301:
293:
292:
291:
288:
287:
285:
284:
280:
279:
277:
273:
270:
269:
261:
260:
259:
256:
255:
253:
252:
239:
237:
225:
222:
221:
219:
218:
210:
208:
202:
201:
199:
198:
190:
188:
180:
177:
176:
166:
158:
157:
155:
154:
146:
144:
138:
137:
135:
134:
125:
123:
117:
116:
114:
113:
105:
103:
96:
93:
92:
90:
89:
81:
79:
74:
71:
70:
66:
65:
62:
58:
57:
50:
44:
43:
39:
38:
9:
6:
4:
3:
2:
1656:
1645:
1642:
1640:
1637:
1636:
1634:
1620:
1616:
1611:
1606:
1602:
1598:
1594:
1590:
1586:
1579:
1571:
1567:
1563:
1559:
1555:
1551:
1547:
1543:
1536:
1528:
1524:
1519:
1514:
1510:
1506:
1502:
1498:
1494:
1490:
1486:
1478:
1470:
1466:
1462:
1458:
1454:
1450:
1442:
1434:
1430:
1426:
1422:
1415:
1407:
1403:
1398:
1393:
1388:
1383:
1379:
1375:
1371:
1367:
1363:
1356:
1348:
1344:
1340:
1336:
1332:
1328:
1321:
1313:
1309:
1305:
1301:
1296:
1291:
1288:(3): 549–69.
1287:
1283:
1279:
1272:
1264:
1260:
1255:
1250:
1246:
1242:
1237:
1232:
1227:
1222:
1218:
1214:
1210:
1206:
1202:
1195:
1193:
1184:
1180:
1176:
1172:
1168:
1164:
1160:
1156:
1152:
1148:
1144:
1137:
1129:
1125:
1121:
1117:
1113:
1109:
1105:
1101:
1097:
1093:
1085:
1077:
1073:
1069:
1065:
1061:
1057:
1053:
1049:
1045:
1041:
1033:
1031:
1022:
1018:
1013:
1008:
1004:
1000:
995:
990:
986:
982:
978:
974:
970:
963:
955:
954:
949:
942:
934:
930:
926:
922:
918:
914:
910:
906:
899:
885:
881:
874:
860:
856:
849:
841:
837:
832:
827:
822:
817:
813:
809:
805:
801:
797:
793:
786:
779:. 5 May 2021.
778:
772:
764:
758:
754:
746:
743:
741:
737:
727:
725:
721:
717:
713:
709:
706:
701:
699:
698:
693:
689:
685:
674:
664:
658:
655:This article
653:
649:
644:
643:
635:
633:
626:
622:
618:
617:
616:
614:
607:+ diphosphate
606:
598:
595:
594:
593:
591:
581:
579:
575:
574:
565:
561:
557:
554:
551:
547:
544:
541:
540:
539:
536:
534:
530:
526:
522:
513:
508:
498:
496:
492:
488:
484:
480:
476:
472:
468:
465:studies with
464:
459:
457:
456:bacteriophage
453:
449:
445:
441:
437:
425:
418:
402:
396:
392:
389:
385:
384:
380:
378:
377:Melting point
375:
374:
370:
368:
365:
364:
360:
357:
356:
349:
347:
344:
343:
322:
319:
315:
314:
309:
300:
299:
296:
289:
278:
268:
267:
264:
257:
249:
245:
244:DTXSID0062052
241:
240:
238:
228:
224:
223:
216:
212:
211:
209:
207:
204:
203:
196:
192:
191:
189:
183:
179:
178:
174:
170:
167:
165:
163:ECHA InfoCard
160:
159:
152:
148:
147:
145:
143:
140:
139:
131:
127:
126:
124:
122:
119:
118:
111:
107:
106:
104:
100:
95:
94:
87:
83:
82:
80:
77:
73:
72:
67:
59:
54:
49:
45:
40:
36:
31:
19:
1592:
1588:
1578:
1545:
1541:
1535:
1492:
1488:
1477:
1452:
1448:
1441:
1424:
1420:
1414:
1369:
1365:
1355:
1330:
1326:
1320:
1285:
1281:
1271:
1208:
1204:
1150:
1146:
1136:
1095:
1091:
1084:
1043:
1039:
976:
972:
962:
953:ScienceAlert
951:
941:
908:
904:
898:
887:. Retrieved
884:ScienceDaily
873:
862:. Retrieved
848:
799:
795:
785:
771:
757:
744:
733:
712:pseudorabies
704:
702:
695:
681:
671:October 2021
668:
656:
629:
624:
620:
610:
604:
596:
587:
584:Biosynthesis
571:
569:
560:Siphoviridae
537:
529:host evasion
525:Siphoviridae
518:
460:
443:
439:
435:
434:
130:ChEMBL388596
69:Identifiers
61:Other names
52:
1644:Nucleobases
1542:ChemBioChem
1495:(1): 1509.
1211:(1): 4710.
979:(1): 2420.
564:Podoviridae
550:PrimPol/AEP
495:outer space
485:components
371:1.743 g/cm
358:Appearance
311:Properties
169:100.016.006
1633:Categories
1333:: 125–44.
889:2011-08-09
864:2011-08-10
749:References
708:antivirals
688:cell cycle
613:EC 6.3.4.4
590:EC 3.6.1.8
501:In viruses
467:meteorites
346:Molar mass
215:49P95BAU4Z
142:ChemSpider
97:3D model (
76:CAS Number
48:IUPAC name
1455:: 84–93.
1245:2041-1723
1183:233448821
1128:233448788
1076:233448787
1003:2041-1723
663:talk page
619:(d)ATP +
469:found on
458:viruses.
86:1904-98-9
1570:35619213
1562:28748559
1527:32198346
1469:26854349
1421:Genetika
1406:10861008
1312:31262125
1304:14839611
1263:34354070
1175:33926954
1120:33926956
1068:33926955
1021:33893297
840:21836052
705:in vitro
684:leukemia
578:archaeal
448:leukemia
1639:Purines
1619:9742229
1518:7083880
1497:Bibcode
1374:Bibcode
1347:2624171
1254:8342488
1221:bioRxiv
1213:Bibcode
1155:Bibcode
1147:Science
1100:Bibcode
1092:Science
1048:Bibcode
1040:Science
1012:8065100
981:Bibcode
913:Bibcode
831:3161613
808:Bibcode
740:serinol
697:E. coli
573:E. coli
533:thymine
491:guanine
487:adenine
452:adenine
440:2,6-DAP
417:what is
415: (
367:Density
351:150.145
182:PubChem
1617:
1610:147870
1607:
1568:
1560:
1525:
1515:
1467:
1433:348574
1431:
1404:
1394:
1345:
1310:
1302:
1282:Cancer
1261:
1251:
1243:
1223:
1181:
1173:
1126:
1118:
1074:
1066:
1019:
1009:
1001:
933:413053
931:
905:Nature
838:
828:
802:(34).
295:SMILES
121:ChEMBL
42:Names
1566:S2CID
1397:16558
1308:S2CID
1179:S2CID
1124:S2CID
1072:S2CID
724:FTSJ1
471:Earth
263:InChI
195:30976
151:28738
99:JSmol
1615:PMID
1558:PMID
1523:PMID
1465:PMID
1429:PMID
1402:PMID
1343:PMID
1300:PMID
1259:PMID
1241:ISSN
1171:PMID
1116:PMID
1064:PMID
1017:PMID
999:ISSN
929:PMID
859:NASA
836:PMID
804:PNAS
621:dGMP
605:dGMP
603:O =
597:dGTP
562:and
548:The
489:and
481:and
463:NASA
206:UNII
1605:PMC
1597:doi
1550:doi
1513:PMC
1505:doi
1457:doi
1453:184
1392:PMC
1382:doi
1335:doi
1290:doi
1249:PMC
1231:doi
1163:doi
1151:372
1108:doi
1096:372
1056:doi
1044:372
1007:PMC
989:doi
921:doi
909:270
826:PMC
816:doi
800:108
599:+ H
519:In
483:RNA
479:DNA
232:EPA
185:CID
1635::
1613:.
1603:.
1593:26
1591:.
1587:.
1564:.
1556:.
1546:18
1544:.
1521:.
1511:.
1503:.
1493:11
1491:.
1487:.
1463:.
1451:.
1425:13
1423:.
1400:.
1390:.
1380:.
1370:97
1368:.
1364:.
1341:.
1331:28
1329:.
1306:.
1298:.
1284:.
1280:.
1257:.
1247:.
1239:.
1229:.
1219:.
1209:12
1207:.
1203:.
1191:^
1177:.
1169:.
1161:.
1149:.
1145:.
1122:.
1114:.
1106:.
1094:.
1070:.
1062:.
1054:.
1042:.
1029:^
1015:.
1005:.
997:.
987:.
977:12
975:.
971:.
950:.
927:.
919:.
907:.
882:.
857:.
834:.
824:.
814:.
798:.
794:.
726:.
700:.
497:.
1621:.
1599::
1572:.
1552::
1529:.
1507::
1499::
1471:.
1459::
1435:.
1408:.
1384::
1376::
1349:.
1337::
1314:.
1292::
1286:4
1265:.
1233::
1215::
1185:.
1165::
1157::
1130:.
1110::
1102::
1078:.
1058::
1050::
1023:.
991::
983::
956:.
935:.
923::
915::
892:.
867:.
842:.
818::
810::
673:)
669:(
665:.
601:2
523:(
438:(
413:N
339:6
336:N
333:6
330:H
327:5
324:C
234:)
230:(
133:=
101:)
53:H
51:7
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.