20:
141:
a part to the community sign the
Contributor Agreement, agreeing not to assert against Users Contributor-held intellectual property rights that might limit the use of the contributed materials. Signers of the User Agreement may freely use the whole collection of parts given by contributors. There is no requirement for users to contribute to the community in order to use the parts, and users may assert intellectual property rights to inventions developed by using the parts. The User Agreement allows users to establish invention of uses of parts, to disclose patents on parts combinations, and to freely build on the contributions of other users.
413:
resistance genes. This method reduces noise from uncut plasmids by amplifying a desired insert using PCR prior to digestion and treating the mixture with the restriction enzyme DpnI, which digests methylated DNA like plasmids. Eliminating the template plasmids with DpnI leaves only the insert to be amplified by PCR. To decrease the possibility of creating plasmids with unwanted combinations of insert and backbone, the backbone can be treated with phosphatase to prevent its religation.
73:
170:
404:
a correctly assembled plasmid. The destination plasmids also have different antibiotic resistance genes than the plasmids carrying the BioBrick parts. All three plasmids are digested with an appropriate restriction enzyme and then allowed to ligate. Only the correctly assembled part will produce a viable composite part contained in the destination plasmid. This allows a good selection as only the correctly assembled BioBrick parts survive.
464:
components. One example of a professional parts registry is the USA-based publicly funded facility, The
International Open Facility Advancing Biotechnology (BIOFAB), which contains detailed descriptions of each biological part. It is also an open-source registry, and is available commercially. BIOFAB aims to catalogue high-quality BioBrick parts to accommodate the needs of professional synthetic biology community.
323:
132:. BioBricks Foundation's activities include hosting SBx.0 Conferences, technical and educational programs. The SBx.0 conferences are international conferences on synthetic biology hosted across the world. Technical programs are aimed at the production of a series of standard biological parts, and their education expansion is creating acts which help create open, standardized sources of biological parts.
111:. This team introduced a cloning strategy for the assembly of short DNA fragments. However, this early attempt was not widely recognised by the scientific research community at the time. In 1999, Arkin and Endy realized that the heterogeneous elements that made up a genetic circuit were lacking standards, so they proposed a list of standard biological parts. BioBricks were described and introduced by
459:
Every BioBrick part has its unique identification code which makes the search for the desired BioBrick part easier (for example, BBa_J23100, a constitutive promoter). The registry is open access, whereby anyone can submit a BioBrick part. Most of the BioBrick submission is from students participating
403:
The 3A assembly method is the most commonly used, as its compatible with assembly
Standard 10, Silver standard as well as the Freiburg standard. This assembly method involves two BioBrick parts and a destination plasmid. The destination plasmid contains a toxic (lethal) gene, to ease the selection of
282:
Tom Knight later developed the BB-2 assembly standard in 2008 to address problems with joining the scars of protein domains and that the scars consist of eight bases, which will yield an altered reading frame when joining protein domains. The enzymes used for digestion of the initial parts are almost
140:
As an alternative to traditional biotechnology patent systems and in an effort to allow BioBricks to be utilized as an open-source community standard, the BioBricks
Foundation created the BioBrick Public Agreement, which consists of a Contributor Agreement and a User Agreement. Those who want to give
438:
The 4R/2M assembly method was designed to combine parts (BioBrick
Assembly Standard 10 or Silver Standard) within existing plasmids (i.e. without PCR or subcloning). The plasmids are reacted in vivo with sequence-specific DNA methyltransferases, so that each is modified and protected from one of two
467:
The BioBrick
Foundation (BBF) is a public-benefit organization established to promote the use of standardized BioBrick parts on a scale beyond the iGEM competition. The BBF is currently working on the derivation of standard framework to promote the production high quality BioBrick parts which would
447:
The MIT group led by Tom Knight that developed BioBricks and
International Genetically Engineered Machines (iGEM) competition are also the pioneers of The Registry of Standard Biological Parts (Registry). Registry being one of the foundations of synthetic biology, provides web-based information and
338:
Pam Silver's lab created the Silver assembly standard to overcome the issue surrounding the formation of fusion protein. This assembly standard is also known as
Biofusion standard, and is an improvement of the BioBrick assembly standard 10. Silver's standard involves deletion of one nucleotide from
463:
Professional parts registries have also been developed. Since most of the BioBrick parts are submitted by undergraduates as part of the iGEM competition, the parts may lack important characterisation data and metadata which would be essential when it comes to designing and modelling the functional
308:
GATCCaaaCTCGAG), and lacking in these same restriction sites internally. The upstream part in the pairwise assembly is purified from an EcoRI/BamHI digest, and the downstream part+vector is purified from an EcoRI/BglII digest. Ligation of these two fragments creates a composite part reforming the
291:
The BglBrick assembly standard was proposed by J. Christopher
Anderson, John E. Dueber, Mariana Leguia, Gabriel C. Wu, Jonathan C. Goler, Adam P. Arkin, and Jay D. Keasling in September 2009 as a standard very similar in concept to BioBrick, but enabling the generation of fusion proteins without
412:
The amplified insert assembly method does not depend on prefix and suffix sequences, allowing to be used in combination with a majority of assembly standards. It also has a higher transformation rate than 3A assembly, and it does not require the involved plasmids to have different antibiotic
270:
process: multiple applications do not change the end product, and maintain the prefix and suffix. Although the BioBrick standard assembly allows for the formation of functional modules, there is a limitation to this standard 10 approach. The 8 bp scar site does not allow the creation of a
153:
methods. The BioBrick assembly standard is a more reliable approach for combining parts to form larger composites. The assembly standard enables two groups of synthetic biologists in different parts of the world to re-use a BioBrick part without going through the whole cycle of design and
160:
cloning method, the assembly standard process is faster and promotes automation. The BioBrick assembly standard 10 was the first assembly standard to be introduced. Over the years, several other assembly standards, such as the
Biofusion standard and Freiburg standard have been developed.
394:
Different methods are used when it comes to assembling BioBricks. This is because some standards require different materials and methods (use of different restriction enzymes), while others are due to preferences in protocol because some methods of assembly have higher efficiency and is
385:
respectively. This scar sequence results in a much more stable protein as the glycine forms a stable N-terminal, unlike the arginine, which signals for N-terminal degradation. The assembly technique proposed by the Freiburg team diminishes the limitations of the Biofusion standard.
127:
The BioBricks Foundation was formed in 2006 by engineers and scientists alike as a not-for-profit organization to standardize biological parts across the field. The Foundation focuses on improving in areas of Technology, Law, Education and the Global Community as they apply to
217:(P) sites. The prefix and the suffix are not considered part of the BioBrick part. To facilitate the assembly process, the BioBrick part itself must not contain any of these restriction sites. During the assembly of two different parts, one of the plasmids is digested with
371:
The 2007 Freiburg iGEM team introduced a new assembly standard to overcome the disadvantages of the existing Biofusion standard technique. The Freiburg team created a new set of prefix and suffix sequences by introducing additional restriction enzyme sites,
429:
ends of sequences, creating single-stranded DNA in the ends of all sequences where the different components are designed to anneal. DNA polymerase then adds DNA parts to gaps in the anneal components, and a Taq ligase can seal the final strands.
98:
The development of standardized biological parts allows for the rapid assembly of sequences. The ability to test individual parts and devices to be independently tested and characterized also improves the reliability of higher-order systems.
380:
to the existing prefix and suffix respectively. These newly introduced restriction enzyme sites are BioBrick standard compatible. The Freiburg standard still forms a 6 bp scar site, but the scar sequence (ACCGGC) now codes for threonine and
253:
sites will also ligate as the digestion produces compatible ends. Now, both the DNA parts are in one plasmid. The ligation produces an 8 base pair "scar" site between the two BioBrick parts. Since the scar site is a hybrid of the
313:
scar sequence at the junction of the parts, a scar that encodes the amino acids glycine and serine when fusing CDS parts together in-frame, convenient due to the GlySer dipeptide being a popular linker of protein domains.
262:
sites, it is not recognized by either restriction enzyme. The prefix and suffix sequences remain unchanged by this digestion and ligation process, which allows for subsequent assembly steps with more BioBrick parts.
189:. The vector acts as a transport system to carry the BioBrick parts. The first approach towards a BioBrick standard was the introduction of standard sequences, the prefix and suffix sequences, which flank the 5
460:
in the annual iGEM competition hosted every summer. The Registry allows exchange of data and materials online which allows rapid re-use and modifications of parts by the participating community.
347:
site, which shortens the scar site by 2 nucleotides, which now forms a 6 bp scar sequence. The 6 bp sequence allows the reading frame to be maintained. The scar sequence codes for the amino acid
292:
altering the reading frame or introducing stop codons and while creating a relatively neutral amino acid linker scar (GlySer). A BglBrick part is as a DNA sequence flanked by 5
421:
The Gibson scarless assembly method allows the joining of multiple BioBricks simultaneously. This method requires the desired sequences to have an overlap of 20 to 150
80:
The BioBrick parts are used by applying engineering principles of abstraction and modularization. BioBrick parts form the base of the hierarchical system on which
623:
425:. Because BioBricks do not have this overlap, this method requires PCR primers to create overhangs between adjacent BioBricks. T5 exonuclease attacks the 5
326:
Biofusion assembly of two BioBrick parts.The schematic diagram shows the 6 base pair scar site made due to the deletion and insertion of nucleotide in the
154:
manipulation. This means the newly designed part can be used by other teams of researchers more easily. Besides that, when compared to the old-fashioned
821:
Mark, Fischer; Lee, Crews; Jennifer, Lynch; Jason, Schultz; David, Grewal; Drew, Endy (2009-10-18). "The BioBrick Public Agreement v1 (draft)".
1094:
487:
91:
Device: Collection set of parts with defined function. In simple terms, a set of complementary BioBrick parts put together forms a device.
119:
in 2003. Since then, various research groups have utilized the BioBrick standard parts to engineer novel biological devices and systems.
359:
is a disadvantage to the Biofusion assembly technique: these properties of arginine result in the destabilisation of the protein by the
42:
from individual parts and combinations of parts with defined functions, which would then be incorporated into living cells such as
197:
ends of the DNA part respectively. These standard sequences encode specific restriction enzyme sites. The prefix sequence encodes
355:(AGA). This minor improvement allows for the formation of in-frame fusion protein. However, arginine's being a large, charged
173:
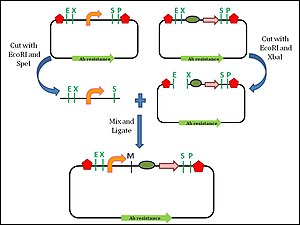
Standard assembly of two BioBrick parts (promoter and coding sequence) by digestion and ligation which forms a "scar" site(M).
1048:
1073:
177:
Assembly standard 10 was developed by Tom Knight, and is the most widely used assembly standard. It involves the use of
1165:
719:
1002:
630:
1205:
743:
1113:
19:
39:
772:
1237:
149:
The BioBrick assembly standard was introduced to overcome the lack of standardization posed by traditional
279:
which prevents the continuous reading of codons, which is required for the formation of fusion protein.
1242:
186:
1140:
Matsumura I. 2020. Methylase-assisted subcloning for high throughput BioBrick assembly. PeerJ 8:e9841
1031:
Røkke, G.; Korvald, E.; Pahr, J.; Oyås, O.; Lale, R. (2014-01-01). Valla, Svein; Lale, Rahmi (eds.).
847:
439:
restriction endonucleases that are later used to linearize undesired circular ligation products.
1098:
112:
53:
861:
Smolke, Christina D (2009). "Building outside of the box: iGEM and the BioBricks Foundation".
23:
Synthetic Biology Open Language (SBOL) standard visual symbols for use with BioBricks Standard
834:
61:
672:
8:
178:
49:
676:
986:
951:
926:
886:
597:
562:
35:
455:
Includes a catalogue which describes the function, performance and design of each part
1161:
1054:
1044:
1008:
998:
956:
878:
700:
695:
656:
602:
584:
482:
477:
150:
129:
81:
1036:
990:
982:
946:
938:
890:
870:
822:
690:
680:
592:
574:
532:
44:
88:
Parts: Pieces of DNA that form a functional unit (for example promoter, RBS, etc.)
57:
1040:
924:
107:
The first attempt to create a list of standard biological parts was in 1996, by
38:
assembly standard. These building blocks are used to design and assemble larger
925:
Sleight, S. C.; Bartley, B. A.; Lieviant, J. A.; Sauro, H. M. (12 April 2010).
272:
979:
Assembly of BioBrick standard biological parts using three antibiotic assembly
48:
cells to construct new biological systems. Examples of BioBrick parts include
1231:
1035:. Methods in Molecular Biology. Vol. 1116. Humana Press. pp. 1–24.
904:
685:
588:
528:
1058:
1012:
960:
882:
606:
1114:"dspace.mit.edu/bitstream/handle/1721.1/45140/BBF_RFC%2025.pdf?sequence=1"
1074:"A New Biobrick Assembly Strategy Designed for Facile Protein Engineering"
704:
579:
1209:
942:
874:
751:
276:
1033:
BioBrick Assembly Standards and Techniques and Associated Software Tools
994:
826:
536:
72:
360:
356:
267:
169:
94:
System: Combination of a set of devices that performs high-level tasks.
452:
Information and characterisation data for all parts, device and system
309:
original flanking sites required in the part definition and leaving a
1141:
422:
348:
322:
654:
233:. This leaves both plasmids with 4 base pair (bp) overhangs at the 5
655:
Rebatchouk, Dmitri; Daraselia, N.; Narita, J. O. (1 October 1996).
504:
352:
245:
sites will ligate since they are complementary to each other. The
801:
780:
382:
182:
661:
DNA manipulation applied to promoter analysis and vector design"
225:. The plasmid carrying the other BioBrick part is digested with
156:
977:
Shetty, R.; Lizarazo, M.; Rettberg, R.; Knight, T. F. (2011).
561:
Knight, Thomas F; Reshma P Shetty; Drew Endy (14 April 2008).
560:
181:. Every BioBrick part is a DNA sequence which is carried by a
529:"Idempotent Vector Design for Standard Assembly of Biobricks"
199:
1181:
976:
448:
data on over 20,000 BioBrick parts. The Registry contains:
213:
981:. Methods in Enzymology. Vol. 498. pp. 311–26.
116:
76:
Abstraction hierarchy allows the breakdown of complexity.
31:
499:
433:
1072:
Silver, Pamela A.; Ira E. Phillips (April 18, 2006).
1071:
897:
1030:
820:
802:"Frequently Asked Questions | BioBricks Foundation"
283:the same, but with modified prefixes and suffixes.
84:is based. There are three levels to the hierarchy:
920:
918:
618:
616:
563:"Engineering BioBrick vectors from BioBrick parts"
1105:
1229:
1095:"Freiburg07/report fusion parts - 2007.igem.org"
927:"In-Fusion BioBrick assembly and re-engineering"
720:"A Standard Parts List for Biological Circuitry"
164:
915:
665:Proceedings of the National Academy of Sciences
613:
317:
286:
972:
970:
398:
207:(X) sites, while the suffix sequence encodes
144:
135:
16:Standard for components used in DNA synthesis
407:
1151:
1149:
416:
967:
950:
694:
684:
648:
596:
578:
1146:
556:
554:
552:
550:
548:
546:
522:
520:
321:
168:
71:
18:
1155:
122:
1230:
860:
526:
1026:
1024:
1022:
543:
517:
505:Registry of Standard Biological Parts
711:
366:
1198:
434:Methylase-assisted (4R/2M) Assembly
13:
1142:https://doi.org/10.7717/peerj.9841
1111:
1019:
987:10.1016/B978-0-12-385120-8.00013-9
389:
14:
1254:
1097:. August 29, 2021. Archived from
773:"Programs - BioBricks Foundation"
717:
657:"NOMAD: a versatile strategy for
567:Journal of Biological Engineering
493:
468:be freely available to everyone.
442:
296:EcoRI and BglII sites (GAATTCaaaA
1174:
1160:. London: Imperial College Pr.
1134:
1087:
1065:
854:
814:
794:
765:
744:"About - BioBricks Foundation"
736:
1:
510:
165:BioBrick assembly standard 10
54:ribosomal binding sites (RBS)
40:synthetic biological circuits
34:sequences which conform to a
1206:"About BioBricks Foundation"
624:"SynBio Standards -BioBrick"
7:
1041:10.1007/978-1-62703-764-8_1
471:
318:Silver (Biofusion) standard
287:BglBricks assembly standard
67:
10:
1259:
1182:"Main Page - ung.igem.org"
1158:Synthetic Biology A Primer
399:3 Antibiotic (3A) Assembly
145:BioBrick Assembly standard
136:BioBricks Public Agreement
102:
408:Amplified Insert Assembly
275:. The scar site causes a
686:10.1073/pnas.93.20.10891
417:Gibson Scarless Assembly
1156:Baldwin, Geoff (2012).
905:"The BioBrick approach"
527:Knight, Thomas (2003).
1081:Harvard Medical School
931:Nucleic Acids Research
842:Cite journal requires
335:
304:BamHI and XhoI sites (
174:
77:
24:
580:10.1186/1754-1611-2-5
325:
172:
75:
22:
1112:Muller, Kristian M.
875:10.1038/nbt1209-1099
863:Nature Biotechnology
777:BioBricks Foundation
748:BioBricks Foundation
500:BioBricks Foundation
266:This assembly is an
123:BioBricks Foundation
1238:Genetics techniques
1212:on 13 November 2015
677:1996PNAS...9310891R
671:(20): 10891–10896.
179:restriction enzymes
943:10.1093/nar/gkq179
336:
185:, which acts as a
175:
78:
36:restriction-enzyme
25:
1243:Synthetic biology
1050:978-1-62703-763-1
869:(12): 1099–1102.
478:Synthetic biology
367:Freiburg standard
151:molecular cloning
130:synthetic biology
82:synthetic biology
1250:
1222:
1221:
1219:
1217:
1208:. Archived from
1202:
1196:
1195:
1193:
1192:
1178:
1172:
1171:
1153:
1144:
1138:
1132:
1131:
1129:
1127:
1118:
1109:
1103:
1102:
1091:
1085:
1084:
1078:
1069:
1063:
1062:
1028:
1017:
1016:
974:
965:
964:
954:
937:(8): 2624–2636.
922:
913:
912:
901:
895:
894:
858:
852:
851:
845:
840:
838:
830:
818:
812:
811:
809:
808:
798:
792:
791:
789:
788:
779:. Archived from
769:
763:
762:
760:
759:
750:. Archived from
740:
734:
733:
731:
729:
724:
715:
709:
708:
698:
688:
652:
646:
645:
643:
641:
636:on 27 March 2014
635:
629:. Archived from
628:
620:
611:
610:
600:
582:
558:
541:
540:
524:
488:iGEM Competition
428:
303:
295:
240:
236:
196:
192:
183:circular plasmid
109:Rebatchouk et al
58:coding sequences
45:Escherichia coli
1258:
1257:
1253:
1252:
1251:
1249:
1248:
1247:
1228:
1227:
1226:
1225:
1215:
1213:
1204:
1203:
1199:
1190:
1188:
1180:
1179:
1175:
1168:
1154:
1147:
1139:
1135:
1125:
1123:
1116:
1110:
1106:
1093:
1092:
1088:
1076:
1070:
1066:
1051:
1029:
1020:
1005:
975:
968:
923:
916:
903:
902:
898:
859:
855:
843:
841:
832:
831:
819:
815:
806:
804:
800:
799:
795:
786:
784:
771:
770:
766:
757:
755:
742:
741:
737:
727:
725:
722:
716:
712:
653:
649:
639:
637:
633:
626:
622:
621:
614:
559:
544:
525:
518:
513:
496:
474:
445:
436:
426:
419:
410:
401:
395:user-friendly.
392:
390:Assembly method
369:
320:
301:
293:
289:
238:
234:
194:
190:
167:
147:
138:
125:
105:
70:
17:
12:
11:
5:
1256:
1246:
1245:
1240:
1224:
1223:
1197:
1173:
1167:978-1848168633
1166:
1145:
1133:
1104:
1101:on 2021-08-29.
1086:
1064:
1049:
1018:
1003:
966:
914:
896:
853:
844:|journal=
813:
793:
764:
735:
710:
647:
612:
542:
515:
514:
512:
509:
508:
507:
502:
495:
494:External links
492:
491:
490:
485:
480:
473:
470:
457:
456:
453:
444:
443:Parts Registry
441:
435:
432:
418:
415:
409:
406:
400:
397:
391:
388:
368:
365:
319:
316:
288:
285:
273:fusion protein
166:
163:
146:
143:
137:
134:
124:
121:
104:
101:
96:
95:
92:
89:
69:
66:
15:
9:
6:
4:
3:
2:
1255:
1244:
1241:
1239:
1236:
1235:
1233:
1211:
1207:
1201:
1187:
1183:
1177:
1169:
1163:
1159:
1152:
1150:
1143:
1137:
1122:
1115:
1108:
1100:
1096:
1090:
1082:
1075:
1068:
1060:
1056:
1052:
1046:
1042:
1038:
1034:
1027:
1025:
1023:
1014:
1010:
1006:
1004:9780123851208
1000:
996:
992:
988:
984:
980:
973:
971:
962:
958:
953:
948:
944:
940:
936:
932:
928:
921:
919:
910:
906:
900:
892:
888:
884:
880:
876:
872:
868:
864:
857:
849:
836:
828:
824:
817:
803:
797:
783:on 2015-09-17
782:
778:
774:
768:
754:on 2015-11-13
753:
749:
745:
739:
721:
718:Arkin, Adam.
714:
706:
702:
697:
692:
687:
682:
678:
674:
670:
666:
662:
660:
651:
632:
625:
619:
617:
608:
604:
599:
594:
590:
586:
581:
576:
572:
568:
564:
557:
555:
553:
551:
549:
547:
538:
534:
530:
523:
521:
516:
506:
503:
501:
498:
497:
489:
486:
484:
481:
479:
476:
475:
469:
465:
461:
454:
451:
450:
449:
440:
431:
424:
414:
405:
396:
387:
384:
379:
375:
364:
362:
358:
354:
350:
346:
342:
333:
329:
324:
315:
312:
307:
299:
284:
280:
278:
274:
269:
264:
261:
257:
252:
248:
244:
232:
228:
224:
220:
216:
215:
210:
206:
202:
201:
188:
184:
180:
171:
162:
159:
158:
152:
142:
133:
131:
120:
118:
114:
110:
100:
93:
90:
87:
86:
85:
83:
74:
65:
63:
59:
55:
51:
47:
46:
41:
37:
33:
29:
21:
1214:. Retrieved
1210:the original
1200:
1189:. Retrieved
1185:
1176:
1157:
1136:
1124:. Retrieved
1120:
1107:
1099:the original
1089:
1080:
1067:
1032:
995:1721.1/65066
978:
934:
930:
908:
899:
866:
862:
856:
835:cite journal
827:1721.1/49434
816:
805:. Retrieved
796:
785:. Retrieved
781:the original
776:
767:
756:. Retrieved
752:the original
747:
738:
726:. Retrieved
713:
668:
664:
658:
650:
638:. Retrieved
631:the original
570:
566:
537:1721.1/21168
466:
462:
458:
446:
437:
420:
411:
402:
393:
377:
373:
370:
344:
340:
337:
331:
327:
310:
305:
297:
290:
281:
265:
259:
255:
250:
246:
242:
230:
226:
222:
218:
212:
208:
204:
198:
176:
155:
148:
139:
126:
108:
106:
97:
79:
43:
27:
26:
909:j5.jbei.org
277:frame shift
62:terminators
1232:Categories
1191:2015-11-10
807:2021-10-19
787:2015-11-04
758:2015-11-04
511:References
361:N-end rule
357:amino acid
351:(ACT) and
268:idempotent
241:ends. The
113:Tom Knight
30:parts are
589:1754-1611
349:threonine
50:promoters
1216:27 March
1186:igem.org
1126:27 March
1059:24395353
1013:21601683
961:20385581
883:20010584
728:27 March
659:in vitro
640:27 March
607:18410688
573:(5): 5.
472:See also
353:arginine
211:(S) and
203:(E) and
68:Overview
28:BioBrick
952:2860134
891:5486814
705:8855278
673:Bibcode
598:2373286
483:Wetware
383:glycine
300:) and 3
103:History
1164:
1083:: 1–6.
1057:
1047:
1011:
1001:
959:
949:
889:
881:
703:
693:
605:
595:
587:
378:NgoMIV
334:sites.
311:GGATCT
187:vector
157:ad hoc
1117:(PDF)
1077:(PDF)
887:S2CID
723:(PDF)
696:38253
634:(PDF)
627:(PDF)
298:GATCT
243:EcoRI
237:and 3
227:EcoRI
219:EcoRI
200:EcoRI
193:and 3
1218:2014
1162:ISBN
1128:2014
1055:PMID
1045:ISBN
1009:PMID
999:ISBN
957:PMID
879:PMID
848:help
730:2014
701:PMID
642:2014
603:PMID
585:ISSN
376:and
374:AgeI
345:SpeI
343:and
341:Xbal
339:the
332:SpeI
330:and
328:XbaI
260:SpeI
258:and
256:Xbal
251:SpeI
249:and
247:Xbal
231:Xbal
229:and
223:SpeI
221:and
214:PstI
209:SpeI
205:Xbal
60:and
1121:MIT
1037:doi
991:hdl
983:doi
947:PMC
939:doi
871:doi
823:hdl
691:PMC
681:doi
593:PMC
575:doi
533:hdl
423:bps
117:MIT
115:at
32:DNA
1234::
1184:.
1148:^
1119:.
1079:.
1053:.
1043:.
1021:^
1007:.
997:.
989:.
969:^
955:.
945:.
935:38
933:.
929:.
917:^
907:.
885:.
877:.
867:27
865:.
839::
837:}}
833:{{
775:.
746:.
699:.
689:.
679:.
669:93
667:.
663:.
615:^
601:.
591:.
583:.
569:.
565:.
545:^
531:.
519:^
363:.
64:.
56:,
52:,
1220:.
1194:.
1170:.
1130:.
1061:.
1039::
1015:.
993::
985::
963:.
941::
911:.
893:.
873::
850:)
846:(
829:.
825::
810:.
790:.
761:.
732:.
707:.
683::
675::
644:.
609:.
577::
571:2
539:.
535::
427:′
306:G
302:′
294:′
239:′
235:′
195:′
191:′
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.