31:
393:
region is associated with decreased transcriptional activity and the opposite for hypomethylation. In other words, the more "tightly" held the DNA region then the more stable and "younger" the species. Looking at DNA methylation's properties in tissues, it was found to be almost zero for embryonic tissues, it can be used to determine acceleration of age and the results can be reproduced in chimpanzee tissue.
380:
physiological conditions however these supercentenarians, subjected to single cell profiling of their T-cell receptors, revealed accumulations of cytotoxic CD4 T-cells through clonal expansion. The conversion of helper CD4 T-cells to a cytotoxic variety might be an adaptation to the late stage of aging aiding in the fighting infections and potentially enhancing tumor surveillance.
392:
One of the applications of this finding would allow for identification of the biological age of a person. DNA methylation uses the structure of dna at different stages of life to determine an age. DNA methylation is the methylation of the cysteine in the CG or Cpg region. The hypermethylation of this
182:
In addition to the core histones, H2A, H2B, H3, and H4, there are other versions of the histone proteins that can be significantly different in their sequence and are important for regulating chromatin dynamics. Histone H3.3 is a variant of histone H3 that is incorporated into the genome independent
1454:
Moqri, Mahdi; Herzog, Chiara; Poganik, Jesse R.; Justice, Jamie; Belsky, Daniel W.; Higgins-Chen, Albert; Moskalev, Alexey; Fuellen, Georg; Cohen, Alan A.; Bautmans, Ivan; Widschwendter, Martin; Ding, Jingzhong; Fleming, Alexander; Mannick, Joan; Han, Jing-Dong Jackie; Zhavoronkov, Alex; Barzilai,
335:
is associated with transcriptional repression and hypomethylation of these sites is associated with transcriptional activation. Many studies have shown that there is a loss of DNA methylation during ageing in many species such as, rats, mice, cows, hamsters, and humans. It has also been shown that
190:
There are multiple variants of histone 2, the one most notably implicated in aging is macroH2A. The function of macroH2A has generally been assumed to be transcriptional silencing; most recently, it has been suggested that macroH2A is important in repressing transcription at
Senescence-Associated
375:
Recent data suggests that an increased frequency of senescent CD8+ T cells in the peripheral blood is associated with the development of hyperglycemia from a pre-diabetic state suggestive of senescence playing a role in metabolic aging. Senescent Cd8+ T cells could be utilized as a biomarker to
388:
The main mechanisms identified as potential biomarkers of aging are DNA methylation, loss of histones, and histone modification. The uses for biomarkers of aging are ubiquitous and identifying a physical parameter of biological aging would allow humans to determine our true age, mortality, and
400:) is currently examining the application of these biomarkers to identify longevity interventions and ways to validate them. Moreover, open-source resources, such as the R package methylCIPHER and the Python package pyaging are available to the public as hubs for several biomarkers of aging.
389:
morbidity. The change in the physical biomarker should be proportional to the change in the age of the species. Thus after establishing a biomarker of aging, humans would be able to dive into research on extending life spans and finding timelines for the arise of potential genetic diseases.
379:
Recently, Hashimoto and coworkers profiled thousands of circulating immune cells from supercentenarians at single-cell resolution. They identified a unique increase in cytotoxic CD4 T cells in these supercentenarians. Generally, CD4 T-cells have helper, but not cytotoxic, functions under
68:
and not a predisposition to disease, should cause a minimal amount of trauma to assay in the organism, and should be reproducibly measurable during a short interval compared to the lifespan of the organism. An assemblage of biomarker data for an organism could be termed its "ageotype".
1504:
Moqri, Mahdi; Herzog, Chiara; Poganik, Jesse R.; Ying, Kejun; Justice, Jamie N.; Belsky, Daniel W.; Higgins-Chen, Albert T.; Chen, Brian H.; Cohen, Alan A.; Fuellen, Georg; Hägg, Sara; Marioni, Riccardo E.; Widschwendter, Martin; Fortney, Kristen; Fedichev, Peter O. (February 2024).
233:, the loss of any of the three Trithorax proteins that catalyze the trimethylation of H3K4 such as, WDR-5 and the methyltransferases SET-2 and ASH-2, lowers the levels of H3K4me3 and increases lifespan. Loss of the enzyme that demethylates H3K4me3, RB-2, increases H3K4me3 levels in
142:
MNase-seq (Micrococcal
Nuclease sequencing) showed a loss of nucleosomes of ~50%. Proper histone dosage is important in yeast as shown from the extended lifespans seen in strains that are overexpressing histones. A consequence of histone loss in yeast is the amplification of
112:
on the large datasets from South Korean, Canadian, and
Eastern European populations demonstrated that biomarkers of aging may be population-specific and predictive of mortality. It is also possible to predict the human chronological age using the
1455:
Nir; Kaeberlein, Matt; Cummings, Steven; Kennedy, Brian K.; Ferrucci, Luigi; Horvath, Steve; Verdin, Eric; Maier, Andrea B.; Snyder, Michael P.; Sebastiano, Vittorio; Gladyshev, Vadim N.; Gladyshev, V. N. (2023).
107:
is a promising biomarker of aging and can accurately predict human chronological age. Basic blood biochemistry and cell counts can also be used to accurately predict the chronological age. Further studies of the
159:, and higher occupancy of repressive chromatin factors. In older cells, however, the same genes nucleosome loss at the promoter is more prevalent which leads to higher transcription of these genes.
396:
More recently, biomarkers of aging has been used in multiple clinical trials to measure slowing or reversing of age-related decline or biological aging. The
Biomarkers of Aging Consortium (
1229:
Lee, Yong-ho; Kim, So Ra; Han, Dai Hoon; Yu, Hee Tae; Han, Yoon Dae; Kim, Jin Hee; Kim, Soo Hyun; Lee, Chan Joo; Min, Byoung-Hoon; Kim, Dong-Hyun; Kim, Kyung Hwan (2018-11-02).
1973:
1278:
Hashimoto, Kosuke; Kouno, Tsukasa; Ikawa, Tomokatsu; Hayatsu, Norihito; Miyajima, Yurina; Yabukami, Haruka; Terooatea, Tommy; Sasaki, Takashi; Suzuki, Takahiro (2019-05-20).
526:
Ahadi, Sara; Zhou, Wenyu; Schüssler-Fiorenza Rose, Sophia Miryam; Sailani, M. Reza; Contrepois, Kévin; Avina, Monika; Ashland, Melanie; Brunet, Anne; Snyder, Michael (2020).
1993:
1998:
2060:
2035:
1415:
721:
2100:
612:"Biomarkers of aging: prediction of longevity by using age-sensitive T-cell subset determinations in a middle-aged, genetically heterogeneous mouse population"
114:
2065:
109:
1925:
2105:
1935:
1930:
1660:
2110:
2070:
809:
Mamoshina P, Kochetov K, Putin E, Cortese F, Aliper A, Lee WS, Ahn SM, Uhn L, Skjodt N, Kovalchuk O, Scheibye-Knudsen M, Zhavoronkov A (October 2018).
2153:
2143:
2030:
2050:
2045:
1886:
2085:
2010:
1952:
1947:
183:
of replication. It is the major form of histone H3 seen in the chromatin of senescent human cells, and it appears that excess H3.3 can drive
2090:
2075:
2040:
1983:
1978:
1920:
497:
147:. In younger cells, genes that are most induced with age have specific chromatin structures, such as fuzzy nuclear positioning, lack of a
2148:
2138:
2080:
1988:
811:"Population Specific Biomarkers of Human Aging: A Big Data Study Using South Korean, Canadian, and Eastern European Patient Populations"
2128:
2095:
2015:
2003:
1942:
229:
has been tied to life span regulation in many organisms, specifically H3K4me3, an activating mark, and H4K27me3, a repressing mark. In
191:
Heterochromatin Foci (SAHF). Chromatin that contains macroH2A is impervious to ATP-dependent remodeling proteins and to the binding of
2055:
1821:
60:
would be a means of validating biomarkers of aging, it would not be a practical means for long-lived species such as humans because
2133:
2025:
2020:
1957:
215:
seen due to the loss of histones. There is also a reduction in the levels of H3K56ac during aging and an increase in the levels of
760:
Putin E, Mamoshina P, Aliper A, Korzinkin M, Moskalev A, Kolosov A, Ostrovskiy A, Cantor C, Vijg J, Zhavoronkov A (May 2016).
174:
that activate the DNA damage response. Loss of core histones may be a general epigenetic mark of aging across many organisms.
341:
204:
2188:
1915:
575:
Van Neste D, Tobin DJ (2004). "Hair cycle and hair pigmentation: dynamic interactions and changes associated with aging".
45:
that could predict functional capacity at some later age better than chronological age. Stated another way, biomarkers of
1690:
162:
This phenomenon is not only seen in yeast, but has also been seen in aging worms, during aging of human diploid primary
2261:
2183:
170:
human cells. In human primary fibroblasts, reduced synthesis of new histones was seen to be a consequence of shortened
1037:"Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA"
2322:
1833:
274:
80:
and other common changes seen with aging are not better indicators of future functionality than chronological age.
84:
have continued efforts to find and validate biomarkers of aging, but success thus far has been limited. Levels of
1135:"Stress-associated H3K4 methylation accumulates during postnatal development and aging of rhesus macaque brain"
17:
2266:
2193:
56:, because changes in the biomarkers would be observable throughout the lifespan of the organism. Although
1663:
300:
knockdowns showed an increase in lifespan
Changes in H3K27me3 levels also have affects on aging cells in
1774:
73:
1843:
1816:
1730:
138:. Most of the evidence shows that loss of histones is linked to cell division. In aging and dividing
1592:
858:
Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, et al. (October 2015).
64:
would take far too much time. Ideally, biomarkers of aging should assay the biological process of
2343:
1683:
262:
212:
144:
2317:
2235:
2218:
2213:
1757:
505:
762:"Deep biomarkers of human aging: Application of deep neural networks to biomarker development"
2286:
1740:
1579:
1280:"Single-cell transcriptomics reveals expansion of cytotoxic CD4 T-cells in supercentenarians"
1786:
1735:
1299:
1291:
1035:
Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, et al. (January 2005).
991:
871:
419:
352:
192:
8:
2358:
2353:
2348:
2307:
2225:
1851:
1828:
254:
250:
246:
226:
220:
167:
152:
49:
would give the true "biological age", which may be different from the chronological age.
1634:
1607:
1539:
1481:
1295:
995:
875:
1856:
1676:
1383:
1348:
1324:
1279:
1185:
1164:
1107:
1082:
1012:
979:
950:
925:
892:
859:
835:
810:
786:
761:
689:
662:
638:
611:
552:
527:
501:
478:
414:
356:
219:. Increased H4K16ac in old yeast cells is associated with the decline in levels of the
61:
1457:"Biomarkers of aging for the identification and evaluation of longevity interventions"
96:
have been used to give good predictions of the expected lifespan of middle-aged mice.
1881:
1793:
1639:
1624:
1544:
1526:
1486:
1407:
1388:
1370:
1329:
1260:
1252:
1211:
1156:
1133:
Han Y, Han D, Yan Z, Boyd-Kirkup JD, Green CD, Khaitovich P, Han JD (December 2012).
1112:
1058:
1017:
955:
897:
840:
791:
713:
694:
643:
592:
557:
470:
466:
242:
1168:
482:
281:
memory of stresses and damages experienced by the organism as it develops and ages.
76:
increases with age, hair graying cannot be called a biomarker of ageing. Similarly,
2276:
2256:
2173:
1769:
1629:
1619:
1567:
1534:
1518:
1476:
1468:
1436:
1428:
1399:
1378:
1360:
1319:
1309:
1242:
1201:
1146:
1102:
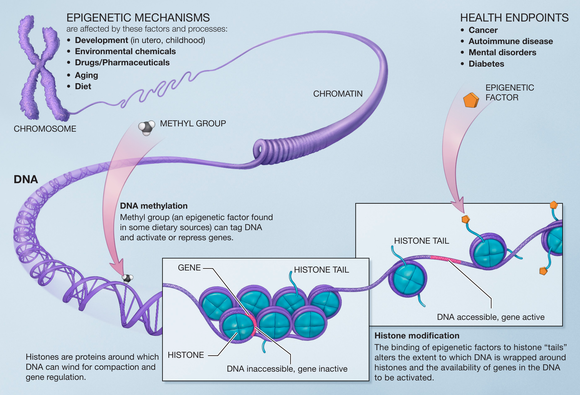
1094:
1048:
1007:
999:
945:
937:
887:
879:
830:
822:
781:
773:
742:
734:
705:
684:
674:
633:
623:
584:
547:
539:
462:
409:
324:
270:
104:
57:
1900:
1781:
1720:
1098:
1053:
1036:
588:
312:
1562:
Thrush, Kyra L.; Higgins-Chen, Albert T.; Liu, Zuyun; Levine, Morgan E. (2022).
2312:
1871:
1838:
1808:
1752:
1522:
1472:
1365:
1284:
Proceedings of the
National Academy of Sciences of the United States of America
941:
815:
The
Journals of Gerontology. Series A, Biological Sciences and Medical Sciences
679:
628:
616:
The
Journals of Gerontology. Series A, Biological Sciences and Medical Sciences
351:
Hypomethylation of DNA can lower genomic stability, induce the reactivation of
238:
81:
53:
1571:
1403:
1206:
1189:
709:
543:
528:"Personal aging markers and ageotypes revealed by deep longitudinal profiling"
296:
expression correlates with a decrease in H3K27me3 and a decrease in lifespan.
2337:
2271:
2230:
2178:
1530:
1456:
1374:
1256:
89:
1563:
1506:
1314:
826:
1876:
1866:
1861:
1643:
1548:
1490:
1411:
1392:
1333:
1264:
1215:
1160:
1116:
1062:
1021:
1003:
959:
901:
844:
795:
717:
698:
647:
596:
561:
364:
93:
777:
474:
103:
analysis allowed for the new types of "aging clocks" to be developed. The
2205:
1764:
1747:
278:
208:
131:
1425:
has been checked and does not affect the cited material, please replace
731:
has been checked and does not affect the cited material, please replace
30:
1725:
1151:
1134:
883:
525:
424:
184:
163:
148:
52:
Validated biomarkers of aging would allow for testing interventions to
1247:
1231:"Senescent T Cells Predict the Development of Hyperglycemia in Humans"
1230:
2302:
2281:
1699:
266:
42:
450:
1304:
1190:"DNA methylation, ageing and the influence of early life nutrition"
1083:"Histone methylation and aging: lessons learned from model systems"
332:
328:
320:
285:
171:
156:
100:
1608:"pyaging: a Python-based compendium of GPU-optimized aging clocks"
397:
1715:
1422:
728:
216:
135:
77:
1087:
Biochimica et
Biophysica Acta (BBA) - Gene Regulatory Mechanisms
860:"The transcriptional landscape of age in human peripheral blood"
376:
signal the transition from pre-diabetes to overt hyperglycemia.
1707:
360:
345:
316:
258:
46:
857:
2251:
808:
759:
337:
139:
65:
1668:
1184:
1561:
223:
Sir2, which can increase the life span when overexpressed.
1277:
1453:
1034:
261:. These increases reflect progressively more active and
85:
1503:
1349:"DNA methylation age of human tissues and cell types"
663:"DNA methylation age of human tissues and cell types"
1132:
924:
Sen P, Shah PP, Nativio R, Berger SL (August 2016).
923:
211:
state as an organism ages, similar to the increased
134:
mark found in studies of aging cells is the loss of
383:
288:demethylase, plays a critical role in the aging of
1188:, Hoile SP, Grenfell L, Burdge GC (August 2014).
2335:
155:, weak chromatin phasing, a higher frequency of
1128:
1126:
496:Harrison, Ph.D., David E. (November 11, 2011).
495:
926:"Epigenetic Mechanisms of Longevity and Aging"
574:
1684:
1123:
1080:
1228:
802:
753:
654:
448:
1691:
1677:
269:structures that are often associated with
1633:
1623:
1538:
1480:
1382:
1364:
1323:
1313:
1303:
1246:
1205:
1150:
1106:
1052:
1011:
949:
891:
834:
785:
688:
678:
637:
627:
551:
444:
442:
440:
1194:The Proceedings of the Nutrition Society
977:
237:and decreases their life spans. In the
198:
29:
1605:
1346:
660:
207:contributes to chromatin taking a more
14:
2336:
1439:|...|checked=yes}}
745:|...|checked=yes}}
609:
437:
1672:
1606:Camillo, Lucas Paulo de Lima (2024).
1180:
1178:
1081:McCauley BS, Dang W (December 2014).
1076:
1074:
1072:
973:
971:
969:
919:
917:
915:
913:
911:
277:response. These changes may form an
370:
1507:"Validation of biomarkers of aging"
177:
125:
120:
24:
1175:
1069:
966:
908:
307:
25:
2370:
1661:Biomarkers of Aging News Advisory
1654:
359:, all of which can contribute to
257:during postnatal development and
1834:Biodemography of human longevity
384:Applications of aging biomarkers
1599:
1555:
1497:
1447:
1340:
1271:
1222:
1028:
398:https://www.agingconsortium.org
323:base is methylated and becomes
1625:10.1093/bioinformatics/btae200
851:
603:
568:
519:
489:
34:Chromosomes during methylation
13:
1:
1698:
978:Pal S, Tyler JK (July 2016).
498:"V. Life span as a biomarker"
430:
331:context. Hypermethylation of
151:depleted region (NDR) at the
1099:10.1016/j.bbagrm.2014.05.008
1054:10.1016/j.devcel.2004.10.019
589:10.1016/j.micron.2003.11.006
467:10.1016/0531-5565(88)90025-3
449:Baker GT, Sprott RL (1988).
315:is a common modification in
7:
1664:National Institute on Aging
403:
10:
2375:
1775:research into centenarians
1523:10.1038/s41591-023-02784-9
1473:10.1016/j.cell.2023.08.003
1366:10.1186/gb-2013-14-10-r115
942:10.1016/j.cell.2016.07.050
680:10.1186/gb-2013-14-10-r115
2295:
2244:
2204:
2166:
2121:
1966:
1908:
1897:
1844:Longevity escape velocity
1807:
1731:aging-associated diseases
1706:
1572:10.1101/2022.07.13.499978
1404:10.1186/s13059-015-0649-6
1207:10.1017/S0029665114000081
710:10.1186/s13059-015-0649-6
544:10.1038/s41591-019-0719-5
327:, most often when in the
2323:Longest-living organisms
629:10.1093/gerona/56.4.b180
610:Miller RA (April 2001).
455:Experimental Gerontology
355:, and cause the loss of
344:decrease with aging and
1347:Horvath, Steve (2013).
1315:10.1073/pnas.1907883116
980:"Epigenetics and aging"
227:Methylation of histones
205:acetylation of histones
2318:Transhumanist politics
2236:Immortality in fiction
2214:Biological immortality
1758:cognitive epidemiology
1587:Cite journal requires
1004:10.1126/sciadv.1600584
273:responses such as the
35:
2122:regions by continents
1741:negligible senescence
1736:degenerative diseases
1431:|...}}
864:Nature Communications
827:10.1093/gerona/gly005
778:10.18632/aging.100968
737:|...}}
451:"Biomarkers of aging"
353:transposable elements
265:accessible (or open)
199:Histone modifications
193:transcription factors
33:
1967:regions of countries
1787:LGBT life expectancy
1618:(btae200): btae200.
420:Biomarker (medicine)
115:transcriptomic clock
62:longitudinal studies
2308:Longevity insurance
2226:Digital immortality
1852:calorie restriction
1839:Indefinite lifespan
1829:Anti-aging movement
1799:Biomarkers of aging
1296:2019PNAS..11624242H
1290:(48): 24242–24251.
996:2016SciA....2E0584P
876:2015NatCo...6.8570.
110:hematological clock
39:Biomarkers of aging
1958:World macroregions
1857:Diet and longevity
1152:10.1111/acel.12007
1041:Developmental Cell
884:10.1038/ncomms9570
661:Horvath S (2013).
502:Jackson Laboratory
415:Hallmarks of aging
313:Methylation of DNA
36:
27:Type of biomarkers
2331:
2330:
2162:
2161:
1882:stem-cell therapy
1794:Maximum life span
1467:(18): 3758–3775.
1248:10.2337/db17-1218
821:(11): 1482–1490.
508:on April 26, 2012
371:Immune biomarkers
263:transcriptionally
243:prefrontal cortex
82:Biogerontologists
16:(Redirected from
2366:
2174:Longevity claims
1906:
1905:
1770:supercentenarian
1693:
1686:
1679:
1670:
1669:
1648:
1647:
1637:
1627:
1603:
1597:
1596:
1590:
1585:
1583:
1575:
1559:
1553:
1552:
1542:
1501:
1495:
1494:
1484:
1451:
1445:
1444:
1442:
1440:
1432:
1417:Retraction Watch
1396:
1386:
1368:
1344:
1338:
1337:
1327:
1317:
1307:
1275:
1269:
1268:
1250:
1226:
1220:
1219:
1209:
1182:
1173:
1172:
1154:
1130:
1121:
1120:
1110:
1078:
1067:
1066:
1056:
1032:
1026:
1025:
1015:
984:Science Advances
975:
964:
963:
953:
921:
906:
905:
895:
855:
849:
848:
838:
806:
800:
799:
789:
757:
751:
750:
748:
746:
738:
723:Retraction Watch
702:
692:
682:
658:
652:
651:
641:
631:
607:
601:
600:
572:
566:
565:
555:
523:
517:
516:
514:
513:
504:. Archived from
493:
487:
486:
446:
410:Epigenetic clock
363:progression and
325:5-methylcytosine
178:Histone variants
126:Loss of histones
121:Epigenetic marks
105:epigenetic clock
58:maximum lifespan
21:
2374:
2373:
2369:
2368:
2367:
2365:
2364:
2363:
2334:
2333:
2332:
2327:
2291:
2245:Longevity genes
2240:
2200:
2158:
2117:
1962:
1901:life expectancy
1899:
1893:
1803:
1782:Life expectancy
1721:Longevity myths
1702:
1697:
1657:
1652:
1651:
1604:
1600:
1588:
1586:
1577:
1576:
1560:
1556:
1511:Nature Medicine
1502:
1498:
1452:
1448:
1434:
1426:
1420:
1398:(Erratum:
1397:
1345:
1341:
1276:
1272:
1227:
1223:
1183:
1176:
1131:
1124:
1093:(12): 1454–62.
1079:
1070:
1033:
1029:
990:(7): e1600584.
976:
967:
922:
909:
856:
852:
807:
803:
758:
754:
740:
732:
726:
704:(Erratum:
703:
659:
655:
608:
604:
573:
569:
532:Nature Medicine
524:
520:
511:
509:
494:
490:
461:(4–5): 223–39.
447:
438:
433:
406:
386:
373:
310:
308:DNA methylation
201:
180:
128:
123:
74:graying of hair
54:extend lifespan
28:
23:
22:
15:
12:
11:
5:
2372:
2362:
2361:
2356:
2351:
2346:
2344:Biogerontology
2329:
2328:
2326:
2325:
2320:
2315:
2313:Longevity risk
2310:
2305:
2299:
2297:
2293:
2292:
2290:
2289:
2284:
2279:
2274:
2269:
2264:
2259:
2254:
2248:
2246:
2242:
2241:
2239:
2238:
2233:
2228:
2223:
2222:
2221:
2210:
2208:
2202:
2201:
2199:
2198:
2197:
2196:
2191:
2186:
2176:
2170:
2168:
2164:
2163:
2160:
2159:
2157:
2156:
2151:
2146:
2141:
2136:
2131:
2125:
2123:
2119:
2118:
2116:
2115:
2114:
2113:
2108:
2098:
2093:
2088:
2083:
2078:
2073:
2068:
2063:
2058:
2053:
2048:
2043:
2038:
2033:
2028:
2023:
2018:
2013:
2008:
2007:
2006:
1996:
1991:
1986:
1981:
1976:
1970:
1968:
1964:
1963:
1961:
1960:
1955:
1950:
1945:
1940:
1939:
1938:
1933:
1923:
1918:
1912:
1910:
1903:
1895:
1894:
1892:
1891:
1890:
1889:
1884:
1879:
1874:
1872:organ printing
1869:
1864:
1859:
1854:
1846:
1841:
1836:
1831:
1826:
1825:
1824:
1813:
1811:
1809:Life extension
1805:
1804:
1802:
1801:
1796:
1791:
1790:
1789:
1779:
1778:
1777:
1772:
1762:
1761:
1760:
1755:
1753:biogerontology
1745:
1744:
1743:
1738:
1733:
1723:
1718:
1712:
1710:
1704:
1703:
1696:
1695:
1688:
1681:
1673:
1667:
1666:
1656:
1655:External links
1653:
1650:
1649:
1612:Bioinformatics
1598:
1589:|journal=
1554:
1517:(2): 360–372.
1496:
1446:
1353:Genome Biology
1339:
1305:10.1101/643528
1270:
1241:(1): 156–162.
1221:
1174:
1145:(6): 1055–64.
1122:
1068:
1027:
965:
936:(4): 822–839.
907:
850:
801:
772:(5): 1021–33.
752:
667:Genome Biology
653:
602:
583:(3): 193–200.
567:
518:
488:
435:
434:
432:
429:
428:
427:
422:
417:
412:
405:
402:
385:
382:
372:
369:
309:
306:
239:rhesus macaque
200:
197:
179:
176:
127:
124:
122:
119:
90:memory T cells
26:
18:Biological age
9:
6:
4:
3:
2:
2371:
2360:
2357:
2355:
2352:
2350:
2347:
2345:
2342:
2341:
2339:
2324:
2321:
2319:
2316:
2314:
2311:
2309:
2306:
2304:
2301:
2300:
2298:
2294:
2288:
2285:
2283:
2280:
2278:
2275:
2273:
2270:
2268:
2265:
2263:
2260:
2258:
2255:
2253:
2250:
2249:
2247:
2243:
2237:
2234:
2232:
2231:Eternal youth
2229:
2227:
2224:
2220:
2217:
2216:
2215:
2212:
2211:
2209:
2207:
2203:
2195:
2192:
2190:
2187:
2185:
2182:
2181:
2180:
2179:Oldest people
2177:
2175:
2172:
2171:
2169:
2165:
2155:
2154:South America
2152:
2150:
2147:
2145:
2144:North America
2142:
2140:
2137:
2135:
2132:
2130:
2127:
2126:
2124:
2120:
2112:
2109:
2107:
2104:
2103:
2102:
2099:
2097:
2094:
2092:
2089:
2087:
2084:
2082:
2079:
2077:
2074:
2072:
2069:
2067:
2064:
2062:
2059:
2057:
2054:
2052:
2049:
2047:
2044:
2042:
2039:
2037:
2034:
2032:
2029:
2027:
2024:
2022:
2019:
2017:
2014:
2012:
2009:
2005:
2002:
2001:
2000:
1997:
1995:
1992:
1990:
1987:
1985:
1982:
1980:
1977:
1975:
1972:
1971:
1969:
1965:
1959:
1956:
1954:
1951:
1949:
1946:
1944:
1941:
1937:
1936:South America
1934:
1932:
1931:North America
1929:
1928:
1927:
1924:
1922:
1919:
1917:
1914:
1913:
1911:
1907:
1904:
1902:
1896:
1888:
1885:
1883:
1880:
1878:
1875:
1873:
1870:
1868:
1865:
1863:
1860:
1858:
1855:
1853:
1850:
1849:
1847:
1845:
1842:
1840:
1837:
1835:
1832:
1830:
1827:
1823:
1820:
1819:
1818:
1817:Topic outline
1815:
1814:
1812:
1810:
1806:
1800:
1797:
1795:
1792:
1788:
1785:
1784:
1783:
1780:
1776:
1773:
1771:
1768:
1767:
1766:
1763:
1759:
1756:
1754:
1751:
1750:
1749:
1746:
1742:
1739:
1737:
1734:
1732:
1729:
1728:
1727:
1724:
1722:
1719:
1717:
1714:
1713:
1711:
1709:
1705:
1701:
1694:
1689:
1687:
1682:
1680:
1675:
1674:
1671:
1665:
1662:
1659:
1658:
1645:
1641:
1636:
1631:
1626:
1621:
1617:
1613:
1609:
1602:
1594:
1581:
1573:
1569:
1565:
1558:
1550:
1546:
1541:
1536:
1532:
1528:
1524:
1520:
1516:
1512:
1508:
1500:
1492:
1488:
1483:
1478:
1474:
1470:
1466:
1462:
1458:
1450:
1438:
1430:
1424:
1419:
1418:
1413:
1409:
1405:
1401:
1394:
1390:
1385:
1380:
1376:
1372:
1367:
1362:
1358:
1354:
1350:
1343:
1335:
1331:
1326:
1321:
1316:
1311:
1306:
1301:
1297:
1293:
1289:
1285:
1281:
1274:
1266:
1262:
1258:
1254:
1249:
1244:
1240:
1236:
1232:
1225:
1217:
1213:
1208:
1203:
1200:(3): 413–21.
1199:
1195:
1191:
1187:
1181:
1179:
1170:
1166:
1162:
1158:
1153:
1148:
1144:
1140:
1136:
1129:
1127:
1118:
1114:
1109:
1104:
1100:
1096:
1092:
1088:
1084:
1077:
1075:
1073:
1064:
1060:
1055:
1050:
1046:
1042:
1038:
1031:
1023:
1019:
1014:
1009:
1005:
1001:
997:
993:
989:
985:
981:
974:
972:
970:
961:
957:
952:
947:
943:
939:
935:
931:
927:
920:
918:
916:
914:
912:
903:
899:
894:
889:
885:
881:
877:
873:
869:
865:
861:
854:
846:
842:
837:
832:
828:
824:
820:
816:
812:
805:
797:
793:
788:
783:
779:
775:
771:
767:
763:
756:
744:
736:
730:
725:
724:
719:
715:
711:
707:
700:
696:
691:
686:
681:
676:
672:
668:
664:
657:
649:
645:
640:
635:
630:
625:
622:(4): B180-6.
621:
617:
613:
606:
598:
594:
590:
586:
582:
578:
571:
563:
559:
554:
549:
545:
541:
537:
533:
529:
522:
507:
503:
499:
492:
484:
480:
476:
472:
468:
464:
460:
456:
452:
445:
443:
441:
436:
426:
423:
421:
418:
416:
413:
411:
408:
407:
401:
399:
394:
390:
381:
377:
368:
366:
362:
358:
354:
349:
347:
343:
339:
334:
330:
326:
322:
318:
314:
305:
303:
299:
295:
291:
287:
282:
280:
276:
272:
268:
264:
260:
256:
252:
249:increases at
248:
244:
240:
236:
232:
228:
224:
222:
218:
214:
213:transcription
210:
206:
196:
194:
188:
186:
175:
173:
169:
165:
160:
158:
157:TATA elements
154:
150:
146:
145:transcription
141:
137:
133:
118:
116:
111:
106:
102:
97:
95:
94:naive T cells
91:
87:
83:
79:
78:skin wrinkles
75:
70:
67:
63:
59:
55:
50:
48:
44:
40:
32:
19:
2219:Regeneration
2071:South Africa
1877:rejuvenation
1867:nanomedicine
1862:gene therapy
1798:
1615:
1611:
1601:
1580:cite journal
1557:
1514:
1510:
1499:
1464:
1460:
1449:
1435:{{
1427:{{
1416:
1414:,
1359:(10): R115.
1356:
1352:
1342:
1287:
1283:
1273:
1238:
1234:
1224:
1197:
1193:
1186:Lillycrop KA
1142:
1138:
1090:
1086:
1047:(1): 19–30.
1044:
1040:
1030:
987:
983:
933:
929:
867:
863:
853:
818:
814:
804:
769:
765:
755:
741:{{
733:{{
722:
720:,
673:(10): R115.
670:
666:
656:
619:
615:
605:
580:
576:
570:
538:(1): 83–90.
535:
531:
521:
510:. Retrieved
506:the original
491:
458:
454:
395:
391:
387:
378:
374:
365:pathogenesis
350:
311:
304:and humans.
301:
297:
293:
292:: increased
289:
283:
234:
230:
225:
202:
189:
181:
161:
129:
99:Advances in
98:
71:
51:
38:
37:
2206:Immortality
1765:Centenarian
1748:Gerontology
348:increases.
333:CpG islands
319:cells. The
209:euchromatic
164:fibroblasts
2359:Biomarkers
2354:Senescence
2349:Physiology
2338:Categories
2189:by country
2086:Tajikistan
2051:Kazakhstan
1909:by country
1726:Senescence
1139:Aging Cell
512:2011-12-03
431:References
425:Senescence
357:imprinting
302:Drosophila
279:epigenetic
275:DNA damage
235:C. elegans
231:C. elegans
203:Increased
185:senescence
149:nucleosome
132:epigenetic
43:biomarkers
2303:Blue zone
2061:Palestine
1979:Australia
1974:Argentina
1700:Longevity
1564:"bioRxiv"
1531:1546-170X
1421:. If the
1375:1465-6906
1257:0012-1797
727:. If the
317:mammalian
290:C.elegans
284:UTX-1, a
267:chromatin
255:enhancers
251:promoters
172:telomeres
168:senescent
166:, and in
72:Although
2184:verified
2106:shortest
1926:Americas
1898:Lists of
1848:Methods
1644:38603598
1635:11058068
1549:38355974
1540:11090477
1491:37657418
1482:11088934
1412:25968125
1393:24138928
1334:31719197
1265:30389747
1235:Diabetes
1216:25027290
1169:17523080
1161:22978322
1117:24859460
1063:15621527
1022:27482540
960:27518561
902:26490707
870:: 8570.
845:29340580
796:27191382
718:25968125
699:24138928
648:11283189
597:15036274
562:31932806
483:31039588
404:See also
321:cytosine
286:H3K27me3
153:promoter
136:histones
101:big data
88:and CD8
2296:Related
2167:Records
2149:Oceania
2111:longest
2021:Germany
2016:Finland
1984:Belgium
1953:Oceania
1716:Old age
1437:erratum
1429:erratum
1423:erratum
1384:4015143
1325:6883788
1300:bioRxiv
1292:Bibcode
1108:4240748
1013:4966880
992:Bibcode
951:5821249
893:4639797
872:Bibcode
836:6175034
787:4931851
743:erratum
735:erratum
729:erratum
690:4015143
639:7537444
553:7301912
475:3058488
247:H3K4me2
217:H4K16ac
2277:Klotho
2194:living
2139:Europe
2129:Africa
2091:Turkey
2081:Sweden
2066:Russia
2056:Mexico
2036:Israel
2026:Greece
2011:France
2004:Cities
1994:Canada
1989:Brazil
1948:Europe
1921:Africa
1708:Ageing
1642:
1632:
1547:
1537:
1529:
1489:
1479:
1410:
1406:,
1391:
1381:
1373:
1332:
1322:
1302:
1263:
1255:
1214:
1167:
1159:
1115:
1105:
1061:
1020:
1010:
958:
948:
900:
890:
843:
833:
794:
784:
716:
712:,
697:
687:
646:
636:
595:
577:Micron
560:
550:
481:
473:
361:cancer
346:DNMT3b
342:DNMT3a
271:stress
241:brain
130:A new
2282:SIRT1
2252:FOXO3
2076:Spain
2046:Japan
2041:Italy
2031:India
1999:China
1916:World
1822:index
1433:with
1165:S2CID
766:Aging
739:with
479:S2CID
338:DNMT1
298:Utx-1
294:utx-1
259:aging
140:yeast
66:aging
47:aging
2262:CETP
2257:APOE
2134:Asia
1943:Asia
1887:SENS
1640:PMID
1593:help
1545:PMID
1527:ISSN
1487:PMID
1461:Cell
1408:PMID
1389:PMID
1371:ISSN
1330:PMID
1261:PMID
1253:ISSN
1212:PMID
1157:PMID
1113:PMID
1091:1839
1059:PMID
1018:PMID
956:PMID
930:Cell
898:PMID
841:PMID
792:PMID
714:PMID
695:PMID
644:PMID
593:PMID
558:PMID
471:PMID
340:and
253:and
221:HDAC
92:and
41:are
2287:TNF
2272:IL6
2267:ACE
2101:USA
1630:PMC
1620:doi
1568:doi
1535:PMC
1519:doi
1477:PMC
1469:doi
1465:186
1400:doi
1379:PMC
1361:doi
1320:PMC
1310:doi
1288:116
1243:doi
1202:doi
1147:doi
1103:PMC
1095:doi
1049:doi
1008:PMC
1000:doi
946:PMC
938:doi
934:166
888:PMC
880:doi
831:PMC
823:doi
782:PMC
774:doi
706:doi
685:PMC
675:doi
634:PMC
624:doi
585:doi
548:PMC
540:doi
463:doi
329:CpG
86:CD4
2340::
2096:UK
1638:.
1628:.
1616:40
1614:.
1610:.
1584::
1582:}}
1578:{{
1566:.
1543:.
1533:.
1525:.
1515:30
1513:.
1509:.
1485:.
1475:.
1463:.
1459:.
1387:.
1377:.
1369:.
1357:14
1355:.
1351:.
1328:.
1318:.
1308:.
1298:.
1286:.
1282:.
1259:.
1251:.
1239:68
1237:.
1233:.
1210:.
1198:73
1196:.
1192:.
1177:^
1163:.
1155:.
1143:11
1141:.
1137:.
1125:^
1111:.
1101:.
1089:.
1085:.
1071:^
1057:.
1043:.
1039:.
1016:.
1006:.
998:.
986:.
982:.
968:^
954:.
944:.
932:.
928:.
910:^
896:.
886:.
878:.
866:.
862:.
839:.
829:.
819:73
817:.
813:.
790:.
780:.
768:.
764:.
693:.
683:.
671:14
669:.
665:.
642:.
632:.
620:56
618:.
614:.
591:.
581:35
579:.
556:.
546:.
536:26
534:.
530:.
500:.
477:.
469:.
459:23
457:.
453:.
439:^
367:.
245:,
195:.
187:.
117:.
1692:e
1685:t
1678:v
1646:.
1622::
1595:)
1591:(
1574:.
1570::
1551:.
1521::
1493:.
1471::
1443:)
1441:.
1402::
1395:.
1363::
1336:.
1312::
1294::
1267:.
1245::
1218:.
1204::
1171:.
1149::
1119:.
1097::
1065:.
1051::
1045:8
1024:.
1002::
994::
988:2
962:.
940::
904:.
882::
874::
868:6
847:.
825::
798:.
776::
770:8
749:)
747:.
708::
701:.
677::
650:.
626::
599:.
587::
564:.
542::
515:.
485:.
465::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.