20:
287:
and ChIP-chip is established by the specific site of the protein binding identification. The main difference comes from the efficacy of the two techniques, ChIP-seq produces results with higher sensitivity and spatial resolution because of the wide range of genomic coverage. Even though ChIP-seq has proven to be more efficient than ChIP-chip, ChIP-seq is not always the first choice for scientists. The cost and accessibility of ChIP-seq is a major disadvantage, which has led to the more predominant use of ChIP-chip in laboratories across the world.
402:: This novel method ChIP uses discs of inert, porous polymer functionalized with either Protein A or G in spin columns or microplates. The chromatin-antibody complex is selectively retained by the disc and eluted to obtain enriched DNA for downstream applications such as qPCR and sequencing. The porous environment is specifically designed to maximize capture efficiency and reduce non-specific binding. Due to less manual handling and optimized protocols, ChIP can be performed in 5 hours.
291:
275:, also known as ChIP-chip, is an experimental technique used to isolate and identify genomic sites occupied by specific DNA-binding proteins in living cells. ChIP-on-chip is a relatively newer technique, as it was introduced in 2001 by Peggy Farnham and Michael Zhang. ChIP-on-chip gets its name by combining the methods of
168:, or magnetic beads. Alternatively, chromatin-antibody complexes can be selectively retained and eluted by inert polymer discs. The immunoprecipitated complexes (i.e., the bead–antibody–protein–target DNA sequence complex) are then collected and washed to remove non-specifically bound chromatin, the protein–DNA
216:
fragments of one nucleosome (200bp) to five nucleosomes (1000bp) in length. Thereafter, methods similar to XChIP are used for clearing the cell debris, immunoprecipitating the protein of interest, removing protein from the immunoprecipitated complex, and purifying and analyzing the complex-associated DNA.
433:
reversal and DNA isolation. However, the fast protocol is suitable only for large cell samples (in the range of 10~10). Up to 24 sheared chromatin samples can be processed to yield PCR-ready DNA in 5 hours, allowing multiple chromatin factors be probed simultaneously and/or looking at genomic events
374:
proteins bound to DNA in living bacterial cells. Following lysis of cross-linked cells and immunoprecipitation of bacterial RNA polymerase, DNA associated with enriched RNA polymerase was hybridized to probes corresponding to different regions of known genes to determine the in vivo distribution and
215:
modifiers. Generally, native chromatin is used as starting chromatin. As histones wrap around DNA to form nucleosomes, they are naturally linked. Then the chromatin is sheared by micrococcal nuclease digestion, which cuts DNA at the length of the linker, leaving nucleosomes intact and providing DNA
286:
The two methods seek similar results, as they both strive to find protein binding sites that can help identify elements in the human genome. Those elements in the human genome are important for the advancement of knowledge in human diseases and biological processes. The difference between ChIP-seq
268:
is the primary technique to complete this task, as it has proven to be extremely effective in resolving how proteins and transcription factors influence phenotypical mechanisms. Overall ChIP-seq has risen to be a very efficient method for determining these factors, but there is a rivaling method
468:(PAT-ChIP): This technique allows ChIP from pathology formalin-fixed and paraffin-embedded tissues and thus the use of pathology archives (even those that are several years old) for epigenetic analyses and the identification of candidate epigenetic biomarkers or targets.
462:-based ChIP assay with increased throughput and a simplified procedure. All steps are done in microplate wells without sample transfers, enabling potential for automation. It enables 96 ChIP assays for histone and various DNA-bound proteins in a single day.
1735:
Li, Guoliang; Fullwood, Melissa J; Xu, Han; Mulawadi, Fabianus
Hendriyan; Velkov, Stoyan; Vega, Vinsensius; Ariyaratne, Pramila Nuwantha; Mohamed, Yusoff Bin; Ooi, Hong-Sain; Tennakoon, Chandana; Wei, Chia-Lin; Ruan, Yijun; Sung, Wing-Kin (February 2010).
414:
cells as carrier chromatin to reduce loss and facilitate precipitation of the target chromatin. However, it demands highly specific primers for detection of the target cell chromatin from the foreign carrier chromatin background, and it takes two to three
247:
But XChIP and NChIP have different aims and advantages relative to each other. XChIP is for mapping target sites of transcription factors and other chromatin-associated proteins; NChIP is for mapping target sites of histone modifiers (see Table 1).
442:(QChIP): The assay uses 100,000 cells as starting material and is suitable for up to 1,000 histone ChIPs or 100 transcription factor ChIPs. Thus many chromatin samples can be prepared in parallel and stored, and QChIP can be undertaken in a day.
395:., 1988, and has also been developed and refined quickly. The typical ChIP assay usually takes 4–5 days and requires 10~ 10 cells at least. Now new techniques on ChIP could be achieved as few as 100~1000 cells and completed within one day.
448:(μChIP): chromatin is usually prepared from 1,000 cells and up to 8 ChIPs can be done in parallel without carriers. The assay can also start with 100 cells, but only suit for one ChIP. It can also use small (1 mm) tissue
497:
Large Scale assays using ChIP is challenging using intact model organisms. This is because antibodies have to be generated for each TF, or, alternatively, transgenic model organisms expressing epitope-tagged TFs need to be
149:(bp) in length. Mild formaldehyde crosslinking followed by nuclease digestion has been used to shear the chromatin. Chromatin fragments of 400 - 500bp have proven to be suitable for ChIP assays as they cover two to three
1228:
O'Neill, Laura P; VerMilyea, Matthew D; Turner, Bryan M (July 2006). "Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations".
1060:
Solomon, Mark J; Larsen Pamela L; Varshavsky, Alexander. (June 1988). "Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene".
97:
The associated DNA fragments are purified and their sequence is determined. Enrichment of specific DNA sequences represents regions on the genome that the protein of interest is associated with
501:
Researchers studying differential gene expression patterns in small organisms also face problems as genes expressed at low levels, in a small number of cells, in narrow time window.
379:
on fruit fly heat shock genes. These reports are considered the pioneering studies in the field of chromatin immunoprecipitation. XChIP was further modified and developed by
264:. Knowing how the proteins in the human body interact with DNA to regulate gene expression is a key component of our knowledge of human diseases and biological processes.
1597:
Fanelli, Mirco; Amatori, Stefano; Barozzi, Iros; Soncini, Matias; Zuffo, Roberto Dal; Bucci, Gabriele; Capra, Maria; Quarto, Micaela; Dellino, Gaetano Ivan (2010-12-14).
2150:
94:
DNA fragments associated with the protein(s) of interest are selectively immunoprecipitated from the cell debris using an appropriate protein-specific antibody.
1417:"QChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells"
370:
and David
Gilmour, at the time a graduate student in the Lis lab, used UV irradiation, a zero-length protein-nucleic acid crosslinking agent, to covalently
244:
e-amino groups in the N-terminals, disrupting the epitopes. This is likely to explain the consistently low efficiency of XChIP protocols compared to NChIP.
1977:
388:
133:
Cross-linked ChIP is mainly suited for mapping the DNA target of transcription factors or other chromatin-associated proteins, and uses reversibly
2407:
391:
using formaldehyde cross-linking. This technique was extensively developed and refined thereafter. NChIP approach was first described by Hebbes
2143:
156:
Cell debris in the sheared lysate is then cleared by sedimentation and protein–DNA complexes are selectively immunoprecipitated using specific
1599:"Pathology tissue–chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples"
654:
Jackson, Vaughn (November 1978). "Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent".
1797:
66:
modifications are associated with, indicating the target of the histone modifiers. ChIP is crucial for the advancements in the field of
2030:
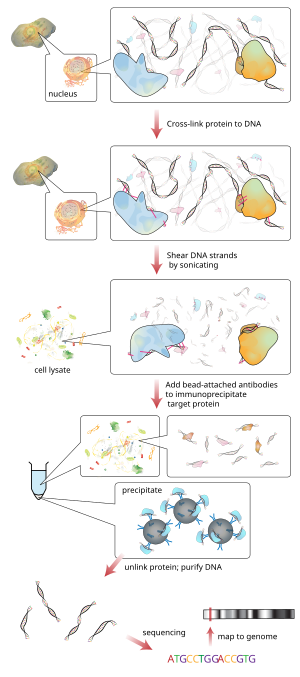
2387:
2136:
1869:
603:
Rosenfeld, John M; Cooke, Tracy; Li, Zirong; Saito, Kan; Taganov, Konstantin; Thyagarajan, Bhaskar; Solache, Alejandra (March 2013).
805:
Beynon, Amy L.; Parkes, Lindsay J.; Turner, Matthew L.; Knight, Steve; Conlan, Steve; Francis, Lewis; Stocks, Ben (September 2014).
2117:
709:
2244:
2112:
1325:
Nelson, Joel D; Denisenko, Oleg; Bomsztyk, Karol (2006). "Protocol for the fast chromatin immunoprecipitation (ChIP) method".
605:"Systematic optimization of parameters involved in preparation of chromatin and chromatin immunoprecipitation (ChIP) workflow"
521:, a technique that adds exonuclease treatment to the ChIP process to obtain up to single base pair resolution of binding sites
375:
density of RNA polymerase at these genes. A year later they used the same methodology to study the distribution of eukaryotic
2402:
1548:"Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events"
1522:
1391:
425:
an ultrasonic bath accelerates the rate of antibody binding to target proteins—and thereby reduces immunoprecipitation time
1843:
Chromatin
Immunoprecipitation (ChIP) of Protein Complexes: Mapping of Genomic Targets of Nuclear Proteins in Cultured Cells
1105:
Orlando V (March 2000). "Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation".
110:
There are mainly two types of ChIP, primarily differing in the starting chromatin preparation. The first uses reversibly
2194:
1681:
1967:
1947:
46:
in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as
1928:
1013:
1838:
Chromatin
Immunopreciptation (ChIP) on Unfixed Chromatin from Cells and Tissues to Analyze Histone Modifications
2167:
1107:
228:
specificity. Most antibodies to modified histones are raised against unfixed, synthetic peptide antigens. The
1862:
276:
1987:
2397:
2229:
2209:
2005:
1506:
1456:
Dahl, John Arne; Collas, Philippe (2008). "A rapid micro chromatin immunoprecipitation assay (microChIP)".
1375:
488:(ChIA-PET), a technique developed for large-scale, de novo analysis of higher-order chromatin structures.
2339:
2104:
2010:
1938:
1822:
1501:
Dahl, John Arne; Collas, Philippe (2009). "μChIP: Chromatin
Immunoprecipitation for Small Cell Numbers".
260:, is an experimental technique used to identify transcription factor binding events throughout an entire
901:"Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes"
2412:
864:
Viens A; et al. (2004). "Use of protein biotinylation in vivo for chromatin immunoprecipitation".
1193:
O'Neill, Laura P; Turner, Bryan M (September 2003). "Immunoprecipitation of native chromatin: NChIP".
145:. Then the cross-linked chromatin is usually sheared by sonication, providing fragments of 300 - 1000
2361:
2128:
1962:
1952:
1942:
1904:
1546:
Flanagin, Steve; Nelson, Joel D; Castner, David G; Denisenko, Oleg; Bomsztyk, Karol (February 2008).
188:
1842:
1837:
1832:
1721:
2392:
2324:
2249:
1982:
1855:
1826:
1734:
2274:
1789:
325:
Suitable for transcriptional factors, or any other weakly binding chromatin associated proteins.
2199:
2053:
2015:
1738:"ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing"
1552:
1280:
421:(qChIP): The fast ChIP assay reduced the time by shortening two steps in a typical ChIP assay:
2314:
2204:
1972:
472:
ChIP has also been applied for genome-wide analysis by combining with microarray technology (
84:
The DNA-protein complexes (chromatin-protein) are then sheared into ~500 bp DNA fragments by
1545:
294:
This photo compares the efficacy of the two experimental techniques, ChIP-seq and ChIP-chip.
2334:
2020:
1899:
1664:
Fullwood, Melissa J; Han, Yuyuan; Wei, Chia-Lin; Ruan, Xiaoan; Ruan, Yijun (January 2010).
1610:
1370:
Nelson J, Denisenko O, Bomsztyk K (2009). "The Fast
Chromatin Immunoprecipitation Method".
912:
380:
47:
19:
8:
2329:
2298:
2176:
1933:
1924:
91:
35:
1614:
916:
557:
Collas, Philippe. (January 2010). "The
Current State of Chromatin Immunoprecipitation".
2264:
2259:
1919:
1766:
1737:
1709:
1692:
1663:
1641:
1598:
1574:
1547:
1483:
1352:
1302:
1275:
1273:
1256:
1227:
1161:
1144:"A direct link between core histone acetylation and transcriptionally active chromatin"
1087:
1037:
1008:
782:
755:
681:
639:
604:
582:
118:
called cross-linked ChIP (XChIP). Native ChIP (NChIP) uses native chromatin sheared by
51:
1206:
1170:
1143:
1120:
1075:
984:
959:
935:
900:
731:
700:
2351:
2074:
1771:
1697:
1677:
1646:
1628:
1579:
1528:
1518:
1475:
1438:
1397:
1387:
1344:
1307:
1248:
1210:
1175:
1124:
1079:
1042:
989:
940:
881:
828:
787:
736:
673:
669:
626:
574:
533:, an alternative location mapping technique that does not require specific antibodies
376:
1487:
1356:
1260:
1091:
1059:
685:
586:
184:
biotinylation can be used instead of antibodies to the native protein of interest.
2282:
2254:
2089:
1761:
1751:
1687:
1669:
1636:
1618:
1569:
1561:
1510:
1467:
1458:
1428:
1379:
1336:
1327:
1297:
1289:
1240:
1202:
1165:
1157:
1148:
1116:
1071:
1032:
1022:
979:
971:
930:
920:
873:
818:
777:
769:
726:
718:
665:
634:
616:
566:
459:
55:
773:
348:
May cause false positive result due to fixation of transient proteins to chromatin
335:
Better chromatin and protein revery efficiency due to better antibody specificity
232:
they need to recognize in the XChIP may be disrupted or destroyed by formaldehyde
2346:
2189:
2058:
1882:
1847:
1673:
1231:
960:"In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster"
505:
481:
477:
265:
257:
200:
137:
chromatin as starting material. The agent for reversible cross-linking could be
81:
in living cells or tissues are crosslinked (this step is omitted in Native ChIP).
1514:
1433:
1416:
1383:
621:
346:
Inefficient chromatin recovery due to antibody target protein epitope disruption
1997:
1914:
1742:
1062:
656:
280:
62:. ChIP also aims to determine the specific location in the genome that various
570:
2381:
2286:
2094:
2084:
2025:
1756:
1632:
877:
832:
630:
1623:
1324:
925:
754:
Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T (January 2002).
2319:
2159:
2079:
2048:
1909:
1775:
1701:
1650:
1583:
1532:
1479:
1442:
1401:
1348:
1311:
1252:
1214:
1141:
1128:
1046:
1027:
885:
791:
760:
578:
524:
473:
272:
199:), molecular cloning and sequencing, or direct high-throughput sequencing (
196:
173:
138:
1471:
1340:
1179:
1083:
993:
944:
740:
2291:
2221:
1565:
1293:
975:
849:
722:
677:
367:
192:
142:
119:
67:
823:
806:
290:
2356:
2163:
1891:
756:"Methylation at arginine 17 of histone H3 is linked to gene activation"
430:
429:
a resin-based (Chelex-100) DNA isolation procedure reduces the time of
410:
384:
371:
350:
Wide range of chromatin shearing size due to random cut by sonication.
237:
233:
187:
The DNA associated with the complex is then purified and identified by
169:
157:
150:
134:
115:
111:
85:
2158:
1274:
Nelson, Joel D; Denisenko, Oleg; Sova, Pavel; Bomsztyk, Karol (2006).
160:
to the protein(s) of interest. The antibodies are commonly coupled to
165:
146:
78:
854:
Revolutionary solid state platform for chromatin immunopreciptation.
807:"Chromatrap 96: a new solid-state platform for high-throughput ChIP"
504:
ChIP experiments cannot discriminate between different TF isoforms (
327:
Applicable to any organisms where native protein is hard to prepare
1244:
1192:
536:
518:
485:
225:
122:
59:
38:
experimental technique used to investigate the interaction between
2239:
1878:
229:
212:
177:
161:
63:
39:
2366:
1500:
1455:
1414:
449:
408:(CChIP): This approach could use as few as 100 cells by adding
261:
241:
1790:"ChIA-PET: Novel Method For 3-D Whole Genome Mapping Research"
1666:
Chromatin interaction analysis using paired-end tag sequencing
486:
Chromatin
Interaction Analysis using Paired End Tag sequencing
332:
Better antibody specificity as target protein naturally intact
2234:
2040:
753:
530:
1596:
1142:
Hebbes, Tim R; Thorne, Alan W; Crane-Robinson C (May 1988).
211:
Native ChIP is mainly suited for mapping the DNA target of
1009:"Discovery of cellular regulation by protein degradation"
701:"In vivo interactions of RNA polymerase II with genes of
539:, a similar technique to analyze RNA-protein interactions
43:
804:
256:
1369:
299:
Table 1 Advantages and disadvantages of NChIP and XChIP
251:
172:
is reversed and proteins are removed by digestion with
1782:
602:
1877:
1668:. Vol. Chapter 21. pp. Unit 21.15.1–25.
957:
898:
476:) or second-generation DNA-sequencing technology (
556:
383:and co-workers, who examined the distribution of
2379:
1415:Dahl, John Arne; Collas, Philippe (April 2007).
219:
73:Briefly, the conventional method is as follows:
1603:Proceedings of the National Academy of Sciences
653:
361:
180:-tagged version of the protein of interest, or
70:and learning more about epigenetic phenomena.
2144:
1863:
353:Usually not suitable for non-histone proteins
1728:
1657:
1539:
1494:
1449:
1408:
1363:
1318:
1267:
1221:
1186:
1098:
1006:
1000:
128:
1798:Agency for Science, Technology and Research
1135:
1053:
747:
550:
355:Nucleosomes may rearrange during digestion
2151:
2137:
1870:
1856:
1276:"Fast chromatin immunoprecipitation assay"
698:
692:
527:, combines ChIP with microarray technology
1825:at the U.S. National Library of Medicine
1765:
1755:
1691:
1640:
1622:
1573:
1432:
1301:
1169:
1104:
1036:
1026:
983:
934:
924:
822:
781:
730:
647:
638:
620:
452:and microChIP can be done within one day.
289:
18:
2245:Enzyme multiplied immunoassay technique
863:
2380:
2113:Photoactivated localization microscopy
2031:Protein–protein interaction prediction
206:
2132:
1851:
1503:Chromatin Immunoprecipitation Assays
1372:Chromatin Immunoprecipitation Assays
844:
842:
598:
596:
252:Comparison of ChIP-seq and ChIP-chip
1988:Freeze-fracture electron microscopy
13:
2408:Protein–protein interaction assays
2195:Ouchterlony double immunodiffusion
1162:10.1002/j.1460-2075.1988.tb02956.x
958:Gilmour DS, Lis JT (August 1985).
699:Gilmour DS, Lis JT (August 1985).
14:
2424:
1816:
1509:. Vol. 567. pp. 59–74.
1378:. Vol. 567. pp. 45–57.
839:
798:
615:(S1): P122, 1756–8935–6-S1-P122.
593:
2388:Biochemical separation processes
1968:Isothermal titration calorimetry
1948:Dual-polarization interferometry
224:The major advantage of NChIP is
1800:(A*STAR), Singapore. 2009-11-08
1590:
1014:Journal of Biological Chemistry
951:
480:). ChIP can also combine with
283:, thus creating ChIP-on-chip.
105:
77:DNA and associated proteins on
1108:Trends in Biochemical Sciences
1007:Varshavsky A (December 2008).
892:
857:
710:Molecular and Cellular Biology
491:
1:
2185:Chromatin immunoprecipitation
1958:Chromatin immunoprecipitation
1823:Chromatin+immunoprecipitation
1207:10.1016/S1046-2023(03)00090-2
1121:10.1016/S0968-0004(99)01535-2
1076:10.1016/S0092-8674(88)90469-2
543:
330:Testable antibody specificity
277:Chromatin Immunoprecipitation
220:Comparison of XChIP and NChIP
28:Chromatin immunoprecipitation
2403:Molecular biology techniques
2230:Chemiluminescent immunoassay
2210:Counterimmunoelectrophoresis
2021:Protein structural alignment
2006:Protein structure prediction
1674:10.1002/0471142727.mb2115s89
1507:Methods in Molecular Biology
1376:Methods in Molecular Biology
670:10.1016/0092-8674(78)90278-7
362:History and New ChIP methods
7:
2340:Direct fluorescent antibody
2105:Super-resolution microscopy
2011:Protein function prediction
1939:Peptide mass fingerprinting
1934:Protein immunoprecipitation
1515:10.1007/978-1-60327-414-2_4
1434:10.1634/stemcells.2006-0430
1384:10.1007/978-1-60327-414-2_3
899:Gilmour DS, Lis JT (1984).
774:10.1093/embo-reports/kvf013
622:10.1186/1756-8935-6-S1-P122
609:Epigenetics & Chromatin
512:
440:Quick and quantitative ChIP
10:
2429:
2362:Total complement activity
2307:
2273:
2220:
2175:
2103:
2067:
2039:
1996:
1963:Surface plasmon resonance
1953:Microscale thermophoresis
1943:Protein mass spectrometry
1905:Green fluorescent protein
1890:
571:10.1007/s12033-009-9239-8
434:over several time points.
189:polymerase chain reaction
129:Cross-linked ChIP (XChIP)
2325:Complement fixation test
1983:Cryo-electron microscopy
1827:Medical Subject Headings
1757:10.1186/gb-2010-11-2-r22
905:Proc Natl Acad Sci U S A
878:10.1016/j.ab.2003.10.015
269:known as ChIP-on-chip.
23:ChIP-sequencing workflow
2016:Protein–protein docking
1929:Protein electrophoresis
1624:10.1073/pnas.1007647107
926:10.1073/pnas.81.14.4275
866:Analytical Biochemistry
703:Drosophila melanogaster
559:Molecular Biotechnology
2200:Radial immunodiffusion
1915:Protein immunostaining
1553:Nucleic Acids Research
1281:Nucleic Acids Research
1028:10.1074/jbc.X800009200
295:
240:are likely to involve
236:, particularly as the
88:or nuclease digestion.
58:, and possibly define
24:
2315:Diagnostic immunology
2205:Immunoelectrophoresis
1973:X-ray crystallography
1472:10.1038/nprot.2008.68
1341:10.1038/nprot.2006.27
293:
114:chromatin sheared by
48:transcription factors
22:
2335:Immunohistochemistry
1900:Protein purification
976:10.1128/mcb.5.8.2009
723:10.1128/mcb.5.8.2009
381:Alexander Varshavsky
2398:Genomics techniques
2330:Immunocytochemistry
2299:Latex fixation test
2177:Immunoprecipitation
1925:Gel electrophoresis
1615:2010PNAS..10721535F
1609:(50): 21535–21540.
917:1984PNAS...81.4275G
824:10.1038/nmeth.f.372
207:Native ChIP (NChIP)
36:immunoprecipitation
2265:Immunofluorescence
2260:Radiobinding assay
2068:Display techniques
1920:Protein sequencing
1566:10.1093/nar/gkn001
1294:10.1093/nar/gnj004
296:
25:
2413:Immunologic tests
2375:
2374:
2352:Skin allergy test
2126:
2125:
2075:Bacterial display
1524:978-1-60327-413-5
1393:978-1-60327-413-5
377:RNA polymerase II
359:
358:
56:DNA binding sites
16:Genomic technique
2420:
2283:Hemagglutination
2255:Radioimmunoassay
2153:
2146:
2139:
2130:
2129:
2090:Ribosome display
2026:Protein ontology
1872:
1865:
1858:
1849:
1848:
1833:EpigenomeNOE.com
1810:
1809:
1807:
1805:
1786:
1780:
1779:
1769:
1759:
1732:
1726:
1725:
1719:
1715:
1713:
1705:
1695:
1661:
1655:
1654:
1644:
1626:
1594:
1588:
1587:
1577:
1543:
1537:
1536:
1498:
1492:
1491:
1459:Nature Protocols
1453:
1447:
1446:
1436:
1412:
1406:
1405:
1367:
1361:
1360:
1328:Nature Protocols
1322:
1316:
1315:
1305:
1271:
1265:
1264:
1225:
1219:
1218:
1190:
1184:
1183:
1173:
1149:The EMBO Journal
1139:
1133:
1132:
1102:
1096:
1095:
1057:
1051:
1050:
1040:
1030:
1021:(50): 34469–89.
1004:
998:
997:
987:
955:
949:
948:
938:
928:
896:
890:
889:
861:
855:
853:
846:
837:
836:
826:
802:
796:
795:
785:
751:
745:
744:
734:
696:
690:
689:
651:
645:
644:
642:
624:
600:
591:
590:
554:
389:heat shock genes
303:
302:
2428:
2427:
2423:
2422:
2421:
2419:
2418:
2417:
2393:Protein methods
2378:
2377:
2376:
2371:
2347:Epitope mapping
2303:
2269:
2216:
2190:Immunodiffusion
2171:
2157:
2127:
2122:
2099:
2063:
2059:Secretion assay
2035:
1992:
1886:
1876:
1819:
1814:
1813:
1803:
1801:
1788:
1787:
1783:
1733:
1729:
1717:
1716:
1707:
1706:
1684:
1662:
1658:
1595:
1591:
1544:
1540:
1525:
1499:
1495:
1454:
1450:
1413:
1409:
1394:
1368:
1364:
1323:
1319:
1272:
1268:
1232:Nature Genetics
1226:
1222:
1191:
1187:
1156:(5): 1395–402.
1140:
1136:
1103:
1099:
1058:
1054:
1005:
1001:
964:Mol. Cell. Biol
956:
952:
897:
893:
862:
858:
848:
847:
840:
803:
799:
752:
748:
697:
693:
652:
648:
601:
594:
555:
551:
546:
515:
506:Protein isoform
494:
482:paired-end tags
478:Chip-Sequencing
364:
354:
349:
347:
333:
331:
326:
254:
222:
209:
131:
108:
34:) is a type of
17:
12:
11:
5:
2426:
2416:
2415:
2410:
2405:
2400:
2395:
2390:
2373:
2372:
2370:
2369:
2364:
2359:
2354:
2349:
2344:
2343:
2342:
2332:
2327:
2322:
2317:
2311:
2309:
2305:
2304:
2302:
2301:
2296:
2295:
2294:
2279:
2277:
2271:
2270:
2268:
2267:
2262:
2257:
2252:
2247:
2242:
2237:
2232:
2226:
2224:
2218:
2217:
2215:
2214:
2213:
2212:
2207:
2202:
2197:
2187:
2181:
2179:
2173:
2172:
2156:
2155:
2148:
2141:
2133:
2124:
2123:
2121:
2120:
2115:
2109:
2107:
2101:
2100:
2098:
2097:
2092:
2087:
2082:
2077:
2071:
2069:
2065:
2064:
2062:
2061:
2056:
2051:
2045:
2043:
2037:
2036:
2034:
2033:
2028:
2023:
2018:
2013:
2008:
2002:
2000:
1998:Bioinformatics
1994:
1993:
1991:
1990:
1985:
1980:
1975:
1970:
1965:
1960:
1955:
1950:
1945:
1936:
1931:
1922:
1917:
1912:
1907:
1902:
1896:
1894:
1888:
1887:
1875:
1874:
1867:
1860:
1852:
1846:
1845:
1840:
1835:
1830:
1818:
1817:External links
1815:
1812:
1811:
1781:
1743:Genome Biology
1727:
1718:|journal=
1683:978-0471142720
1682:
1656:
1589:
1538:
1523:
1493:
1466:(6): 1032–45.
1448:
1427:(4): 1037–46.
1407:
1392:
1362:
1317:
1266:
1245:10.1038/ng1820
1220:
1185:
1134:
1097:
1052:
999:
970:(8): 2009–18.
950:
911:(14): 4275–9.
891:
856:
838:
811:Nature Methods
797:
746:
717:(8): 2009–18.
691:
646:
592:
548:
547:
545:
542:
541:
540:
534:
528:
522:
514:
511:
510:
509:
502:
499:
493:
490:
484:sequencing in
470:
469:
466:Pathology-ChIP
463:
453:
443:
436:
435:
416:
403:
400:Bead-free ChIP
363:
360:
357:
356:
351:
344:
338:
337:
328:
323:
317:
316:
311:
306:
281:DNA microarray
253:
250:
221:
218:
208:
205:
130:
127:
107:
104:
103:
102:
95:
89:
82:
15:
9:
6:
4:
3:
2:
2425:
2414:
2411:
2409:
2406:
2404:
2401:
2399:
2396:
2394:
2391:
2389:
2386:
2385:
2383:
2368:
2365:
2363:
2360:
2358:
2355:
2353:
2350:
2348:
2345:
2341:
2338:
2337:
2336:
2333:
2331:
2328:
2326:
2323:
2321:
2318:
2316:
2313:
2312:
2310:
2306:
2300:
2297:
2293:
2290:
2289:
2288:
2287:Hemagglutinin
2284:
2281:
2280:
2278:
2276:
2275:Agglutination
2272:
2266:
2263:
2261:
2258:
2256:
2253:
2251:
2248:
2246:
2243:
2241:
2238:
2236:
2233:
2231:
2228:
2227:
2225:
2223:
2219:
2211:
2208:
2206:
2203:
2201:
2198:
2196:
2193:
2192:
2191:
2188:
2186:
2183:
2182:
2180:
2178:
2174:
2169:
2165:
2161:
2160:Medical tests
2154:
2149:
2147:
2142:
2140:
2135:
2134:
2131:
2119:
2116:
2114:
2111:
2110:
2108:
2106:
2102:
2096:
2095:Yeast display
2093:
2091:
2088:
2086:
2085:Phage display
2083:
2081:
2078:
2076:
2073:
2072:
2070:
2066:
2060:
2057:
2055:
2054:Protein assay
2052:
2050:
2047:
2046:
2044:
2042:
2038:
2032:
2029:
2027:
2024:
2022:
2019:
2017:
2014:
2012:
2009:
2007:
2004:
2003:
2001:
1999:
1995:
1989:
1986:
1984:
1981:
1979:
1976:
1974:
1971:
1969:
1966:
1964:
1961:
1959:
1956:
1954:
1951:
1949:
1946:
1944:
1940:
1937:
1935:
1932:
1930:
1926:
1923:
1921:
1918:
1916:
1913:
1911:
1908:
1906:
1903:
1901:
1898:
1897:
1895:
1893:
1889:
1884:
1880:
1873:
1868:
1866:
1861:
1859:
1854:
1853:
1850:
1844:
1841:
1839:
1836:
1834:
1831:
1828:
1824:
1821:
1820:
1799:
1795:
1791:
1785:
1777:
1773:
1768:
1763:
1758:
1753:
1749:
1745:
1744:
1739:
1731:
1723:
1711:
1703:
1699:
1694:
1689:
1685:
1679:
1675:
1671:
1667:
1660:
1652:
1648:
1643:
1638:
1634:
1630:
1625:
1620:
1616:
1612:
1608:
1604:
1600:
1593:
1585:
1581:
1576:
1571:
1567:
1563:
1559:
1555:
1554:
1549:
1542:
1534:
1530:
1526:
1520:
1516:
1512:
1508:
1504:
1497:
1489:
1485:
1481:
1477:
1473:
1469:
1465:
1461:
1460:
1452:
1444:
1440:
1435:
1430:
1426:
1422:
1418:
1411:
1403:
1399:
1395:
1389:
1385:
1381:
1377:
1373:
1366:
1358:
1354:
1350:
1346:
1342:
1338:
1335:(1): 179–85.
1334:
1330:
1329:
1321:
1313:
1309:
1304:
1299:
1295:
1291:
1287:
1283:
1282:
1277:
1270:
1262:
1258:
1254:
1250:
1246:
1242:
1239:(7): 835–41.
1238:
1234:
1233:
1224:
1216:
1212:
1208:
1204:
1200:
1196:
1189:
1181:
1177:
1172:
1167:
1163:
1159:
1155:
1151:
1150:
1145:
1138:
1130:
1126:
1122:
1118:
1115:(3): 99–104.
1114:
1110:
1109:
1101:
1093:
1089:
1085:
1081:
1077:
1073:
1070:(6): 937–47.
1069:
1065:
1064:
1056:
1048:
1044:
1039:
1034:
1029:
1024:
1020:
1016:
1015:
1010:
1003:
995:
991:
986:
981:
977:
973:
969:
965:
961:
954:
946:
942:
937:
932:
927:
922:
918:
914:
910:
906:
902:
895:
887:
883:
879:
875:
871:
867:
860:
851:
845:
843:
834:
830:
825:
820:
816:
812:
808:
801:
793:
789:
784:
779:
775:
771:
767:
763:
762:
757:
750:
742:
738:
733:
728:
724:
720:
716:
712:
711:
706:
704:
695:
687:
683:
679:
675:
671:
667:
664:(3): 945–54.
663:
659:
658:
650:
641:
636:
632:
628:
623:
618:
614:
610:
606:
599:
597:
588:
584:
580:
576:
572:
568:
565:(1): 87–100.
564:
560:
553:
549:
538:
535:
532:
529:
526:
523:
520:
517:
516:
507:
503:
500:
496:
495:
489:
487:
483:
479:
475:
467:
464:
461:
457:
454:
451:
447:
444:
441:
438:
437:
432:
428:
424:
420:
417:
413:
412:
407:
404:
401:
398:
397:
396:
394:
390:
386:
382:
378:
373:
369:
352:
345:
343:
342:Disadvantages
340:
339:
336:
329:
324:
322:
319:
318:
315:
312:
310:
307:
305:
304:
301:
300:
292:
288:
284:
282:
278:
274:
270:
267:
263:
259:
249:
245:
243:
239:
235:
234:cross-linking
231:
227:
217:
214:
204:
202:
198:
194:
190:
185:
183:
179:
175:
171:
167:
163:
159:
154:
152:
148:
144:
140:
136:
126:
124:
121:
117:
113:
100:
96:
93:
90:
87:
83:
80:
76:
75:
74:
71:
69:
65:
61:
57:
53:
49:
45:
41:
37:
33:
29:
21:
2320:Nephelometry
2184:
2170:86000–86849)
2080:mRNA display
2049:Enzyme assay
1957:
1910:Western blot
1892:Experimental
1802:. Retrieved
1794:ScienceDaily
1793:
1784:
1747:
1741:
1730:
1665:
1659:
1606:
1602:
1592:
1557:
1551:
1541:
1502:
1496:
1463:
1457:
1451:
1424:
1420:
1410:
1371:
1365:
1332:
1326:
1320:
1285:
1279:
1269:
1236:
1230:
1223:
1201:(1): 76–82.
1198:
1194:
1188:
1153:
1147:
1137:
1112:
1106:
1100:
1067:
1061:
1055:
1018:
1012:
1002:
967:
963:
953:
908:
904:
894:
872:(1): 68–76.
869:
865:
859:
850:"Chromatrap"
814:
810:
800:
768:(1): 39–44.
765:
761:EMBO Reports
759:
749:
714:
708:
702:
694:
661:
655:
649:
612:
608:
562:
558:
552:
525:ChIP-on-chip
474:ChIP-on-chip
471:
465:
458:: This is a
455:
445:
439:
426:
422:
418:
409:
406:Carrier ChIP
405:
399:
392:
365:
341:
334:
320:
313:
308:
298:
297:
285:
273:ChIP-on-chip
271:
255:
246:
223:
210:
197:ChIP-on-chip
186:
181:
174:proteinase K
155:
139:formaldehyde
135:cross-linked
132:
112:cross-linked
109:
106:Typical ChIP
98:
92:Cross-linked
72:
31:
27:
26:
2292:Coombs test
2222:Immunoassay
2118:Vertico SMI
1978:Protein NMR
817:(9): i–ii.
492:Limitations
456:Matrix ChIP
368:John T. Lis
238:cross-links
193:microarrays
151:nucleosomes
125:digestion.
120:micrococcal
68:epigenomics
2382:Categories
2357:Patch test
2164:immunology
1750:(2): R22.
1560:(3): e17.
1421:Stem Cells
544:References
460:microplate
431:cross-link
411:Drosophila
385:histone H4
372:cross-link
321:Advantages
170:cross-link
158:antibodies
147:base pairs
116:sonication
86:sonication
2250:RAST test
1720:ignored (
1710:cite book
1633:0027-8424
1288:(1): e2.
833:1548-7091
631:1756-8935
498:produced.
446:MicroChIP
419:Fast ChIP
166:sepharose
79:chromatin
60:cistromes
54:or other
52:promoters
2162:used in
1885:of study
1879:Proteins
1804:14 March
1776:20181287
1702:20069536
1651:21106756
1584:18203739
1533:19588085
1488:29529307
1480:18536650
1443:17272500
1402:19588084
1357:20577722
1349:17406230
1312:16397291
1261:28311996
1253:16767102
1215:12893176
1129:10694875
1092:11169130
1047:18708349
886:14715286
792:11751582
686:25169609
587:24225210
579:20077036
537:RIP-Chip
519:ChIP-exo
513:See also
450:biopsies
366:In 1984
266:ChIP-seq
258:ChIP-seq
230:epitopes
226:antibody
201:ChIP-Seq
143:UV light
123:nuclease
40:proteins
2240:ELISpot
1883:methods
1767:2872882
1693:6924956
1642:3003125
1611:Bibcode
1575:2241906
1303:1325209
1195:Methods
1180:3409869
1084:2454748
1038:3259866
994:3018544
945:6379641
913:Bibcode
783:1083932
741:3018544
640:3620580
213:histone
191:(PCR),
182:in vivo
178:epitope
162:agarose
99:in vivo
64:histone
2367:MELISA
1881:: key
1829:(MeSH)
1774:
1764:
1700:
1690:
1680:
1649:
1639:
1631:
1582:
1572:
1531:
1521:
1486:
1478:
1441:
1400:
1390:
1355:
1347:
1310:
1300:
1259:
1251:
1213:
1178:
1171:458389
1168:
1127:
1090:
1082:
1045:
1035:
992:
985:366919
982:
943:
936:345570
933:
884:
831:
790:
780:
739:
732:366919
729:
684:
678:569554
676:
637:
629:
585:
577:
262:genome
242:lysine
2308:Other
2235:ELISA
2041:Assay
1484:S2CID
1353:S2CID
1257:S2CID
1088:S2CID
682:S2CID
583:S2CID
531:DamID
415:days.
393:et al
314:NChIP
309:XChIP
176:. An
1806:2010
1772:PMID
1722:help
1698:PMID
1678:ISBN
1647:PMID
1629:ISSN
1580:PMID
1529:PMID
1519:ISBN
1476:PMID
1439:PMID
1398:PMID
1388:ISBN
1345:PMID
1308:PMID
1249:PMID
1211:PMID
1176:PMID
1125:PMID
1080:PMID
1063:Cell
1043:PMID
990:PMID
941:PMID
882:PMID
829:ISSN
788:PMID
737:PMID
674:PMID
657:Cell
627:ISSN
575:PMID
427:(ii)
279:and
42:and
32:ChIP
2168:CPT
1762:PMC
1752:doi
1688:PMC
1670:doi
1637:PMC
1619:doi
1607:107
1570:PMC
1562:doi
1511:doi
1468:doi
1429:doi
1380:doi
1337:doi
1298:PMC
1290:doi
1241:doi
1203:doi
1166:PMC
1158:doi
1117:doi
1072:doi
1033:PMC
1023:doi
1019:283
980:PMC
972:doi
931:PMC
921:doi
874:doi
870:325
819:doi
778:PMC
770:doi
727:PMC
719:doi
666:doi
635:PMC
617:doi
567:doi
423:(i)
387:on
203:).
141:or
50:on
44:DNA
2384::
1796:.
1792:.
1770:.
1760:.
1748:11
1746:.
1740:.
1714::
1712:}}
1708:{{
1696:.
1686:.
1676:.
1645:.
1635:.
1627:.
1617:.
1605:.
1601:.
1578:.
1568:.
1558:36
1556:.
1550:.
1527:.
1517:.
1505:.
1482:.
1474:.
1462:.
1437:.
1425:25
1423:.
1419:.
1396:.
1386:.
1374:.
1351:.
1343:.
1331:.
1306:.
1296:.
1286:34
1284:.
1278:.
1255:.
1247:.
1237:38
1235:.
1209:.
1199:31
1197:.
1174:.
1164:.
1152:.
1146:.
1123:.
1113:25
1111:.
1086:.
1078:.
1068:53
1066:.
1041:.
1031:.
1017:.
1011:.
988:.
978:.
966:.
962:.
939:.
929:.
919:.
909:81
907:.
903:.
880:.
868:.
841:^
827:.
815:11
813:.
809:.
786:.
776:.
764:.
758:.
735:.
725:.
713:.
707:.
680:.
672:.
662:15
660:.
633:.
625:.
611:.
607:.
595:^
581:.
573:.
563:45
561:.
508:).
164:,
153:.
2285:/
2166:(
2152:e
2145:t
2138:v
1941:/
1927:/
1871:e
1864:t
1857:v
1808:.
1778:.
1754::
1724:)
1704:.
1672::
1653:.
1621::
1613::
1586:.
1564::
1535:.
1513::
1490:.
1470::
1464:3
1445:.
1431::
1404:.
1382::
1359:.
1339::
1333:1
1314:.
1292::
1263:.
1243::
1217:.
1205::
1182:.
1160::
1154:7
1131:.
1119::
1094:.
1074::
1049:.
1025::
996:.
974::
968:5
947:.
923::
915::
888:.
876::
852:.
835:.
821::
794:.
772::
766:3
743:.
721::
715:5
705:"
688:.
668::
643:.
619::
613:6
589:.
569::
195:(
101:.
30:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.