890:, hypothesizing that specific circRNA would be differentially expressed in AD cases compared to controls and that those effects could be detected early in the disease. They optimized and validated a novel analyses pipeline for circular RNAs (circRNA). They performed a three-stage study design, using the Knight ADRC brain RNA-seq data as discovery (stage 1), using the data from Mount Sinai as replication (stage 2) and a meta-analysis (stage 3) to identify the most significant circRNA differentially expressed in Alzheimer disease. Using his pipeline, they found 3,547 circRNA that passed stringent QC in the Knight ADRC cohort that includes RNA-seq from 13 controls and 83 Alzheimer cases, and 3,924 circRNA passed stringent QC in the MSBB dataset. A meta-analysis of the discovery and replication results revealed a total of 148 circRNAs that were significantly correlated with CDR after FDR correction. In addition, 33 circRNA passed the stringent gene-based, Bonferroni multiple test correction of 5×10-6, including circHOMER1 (P =2.21×10) and circCDR1-AS (P = 2.83 × 10), among others. They also performed additional analyses to demonstrate that the expression of circRNA were independent of the lineal form as well as the cell proportion that can confound the brain RNA-seq analyses in Alzheimer disease studies. They performed co-expression analyses of all the circRNA together with the lineal forms and found that circRNA, including those that were differentially expressed in Alzheimer disease compared to controls co-expressed with known causal Alzheimer genes, such as APP and PSEN1, indicating that some circRNA are also part of the causal pathway. They also demonstrated that circRNA brain expression explained more about Alzheimer clinical manifestations that the number of APOε4 alleles, suggesting that could be used as a potential biomarker for Alzheimer disease. This is an important study for the field, as it is the first time that circRNA are quantified and validated (by real-time PCR) in human brain samples at genome-wide scale and in large and well-characterized cohorts. It also demonstrates that these RNA forms are likely to be implicated on complex traits including Alzheimer disease will help to understand the biological events that leads to disease.
438:
1188:
592:
20:
1022:
single circular RNA. Treatment with circular RNA activates the differentiation and maturation of dendritic cells which then secrete a large number of different cytokines and chemokines by expressing the genes for IL-1β, IL-6 and TNFa. After immunization with circular RNA that encodes the antigen sequence, CD8+ mediator T cell responses to the target antigen are enhanced. Circular RNA has the very advantageous properties of stability and long shelf life, so it is useful for use as biomarkers and
642:
975:
CircRNAs show positive effects such as circ-ITCH which regulates lung cancer associated with oncogenic sponges miR7 and miR214 and overexpression of circ-ITCH inhibits cell proliferation in lung cancer. From different research studies it has been found that F-circM-9, F-circPR and F-circEA, FcircEA-2 are involved in the development of leukemia and cancer. In osteosarcoma cell circ-0016347 induces tumor and downregulation of
494:. Circular introns produced by eukaryotic spliceosomal splicing are circularized intron lariats known as circular intronic RNAs (ciRNAs). Due to circularization, ciRNAs can avoid degradation and are believed to be highly overrepresented. CiRNA function is currently unknown; however, it is speculated they may play a role in enhancing the transcription of genes they are produced from, as they interact with RNA polymerase II.
378:
their linear controls and, according to ribosome profiling, are not translated.< As previously noted, circRNAs have the ability to act as antagonists of miRNA, which is also known as the potential to act as microRNA sponges. Aside from CDR1as, very few circRNAs have the potential to act as microRNA sponges. As a whole, the majority of circular RNAs were found to be inconsequential side-products of imperfect splicing.
172:
950:. Renal chronicity is associated with miR-150, which is negatively regulated by circHLA-C, in patients with lupus nephritis. There is also evidence that circular RNA is involved in acute kidney injury. In these circumstances circular RNA proves to be a novel biomarker and is also used for targeted therapy of kidney disease because its pseudogene can alter DNA composition.
693:
hypothesis by investigating a circular sponge called CDR1as/CiRS-7 in Detail, while other groups found no direct evidence for circular RNAs acting as miRNA sponges by analyzing the potential interaction of circular RNAs with the
Argonaut (AGO) Protein using high-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation (HITS-CLIP) data .
400:
promote circular RNA formation. On the other hand, exon circularization efficiency was determined to be affected by the competition of RNA pairing, such that alternative RNA pairing, and its competition, leads to alternative circularization. Finally, both exon circularization and its regulation were found to be evolutionarily dynamic.
974:
Circular RNA has both positive and negative functions in cancer. For example, ciRS-7 was found to be an oncogene in colorectal cancer tissue that regulates the disease. Overexpression of this ciRS-7 leads to deregulated gene expression leading to malignant phenotypic features. On the other hand, some
535:
A recent study of human circRNAs revealed that these molecules are usually composed of 1–5 exons. Each of these exons can be up to three times longer than the average expressed exon, suggesting that exon length may play a role in deciding which exons to circularize. 85% of circularized exons overlap
377:
data. Most circRNAs were found to be minor splice isoforms and to be expressed in only a few cell types, with 7,112 human circRNAs having circular fractions (the fraction of similarity an isoform has to transcripts the same locus) of at least 10%. CircRNAs were also found to be no more conserved than
1021:
Circular RNA acts as a very active immune agent when it combines with soluble protein antigens and induces adaptive immunity that does not require a specific route of administration. Plasma circular RNA and combined circRNa have higher efficiency in diagnosis than tissue specific treatment and
711:
AGO2 is miR-7's associated
Argonaute protein (see above). Though CDR1as/CiRS-7 can be cleaved by miR-671 and its associated Argonaute protein, it cannot be cleaved by miR-7 and AGO2. MicroRNA cleavage activity depends on complementarity beyond the 12th nucleotide position; none of CiRS-7's binding
929:
has been observed in some patients after cardiac surgery where circRNA_025016 is used as a biomarker. Although the relevance of circular RNA overexpression and downregulation to heart disease has been found from various research studies, it is still unclear. Therefore, further research is needed to
256:
after transcription. RNA editing occurs mainly in Alu elements of protein-coding genes. A-to-I RNA editing in up- and downstream intronic Alu elements flanking the back-splice site (BSS) can reduces the formation of circRNAs in the human heart. In the failing human heart, a predominant reduction in
243:
represent approximately 10% of the human genome. The presence of Alu elements in flanking introns of protein-coding genes adjacent to the first and last exons that form circRNAs, influence the formation of circRNAs. It is important that the flanking intronic Alu elements are complementary, as
1042:
proliferation in a negative effect on the myogenesis process. circLMO7 is involved in overexpression of HDAC4 and downregulates MEF2A expression by upregulating miR-378a-3p leading to myoblast differentiation. CircSVIL a positive regulator induces miR-203 activity that is the inhibitor of myoblast
903:
Recent studies have shown that circRNA is associated with heart failure and heart disease. circFOXO3, Titin genes, circSLC8A1-1 and circAmotl1 play an important role in cardiac function through upregulation or inhibition relevant to heart disease. Overexpression of circFOXO3 and its downregulation
1043:
production and differentiation. circFUT10 is involved in inhibition of myoblast proliferation but enhances differentiation through enhancement of SRF expression. circSNX29 sponges miR-744 and circFGFR2 sponges miR-133a-5p and miR-29b-1-5p that promote myoblast differentiation. circSNX29 activates
862:, which is a brain illness of unknown origin. Perhaps CiRS-7's sponge activity could help in countering miR-7 activity. If circular sponge activity can indeed help in countering harmful miRNA activity, scientists will need to figure out the best way to introduce sponge expression, perhaps via a
187:
is a phenomenon through which one RNA transcript can yield different protein products based on which segments are considered "introns" and "exons" during a splicing event. Although not specific to humans, it is a partial explanation for the fact that humans and other much simpler species (such as
872:
Circular RNA was discovered to play a role in assisting HIV-1 in evading the body's immune defenses. HIV protein known as Vpr induced the production of a circular RNA molecule referred to as ciTRAN. Subsequently, ciTRAN was observed binding to a specific protein called SRSF1, thereby effectively
806:
Usually, intronic lariats (see above) are debranched and rapidly degraded. However, a debranching failure can lead to the formation of circular intronic long non-coding RNAs, also known as ciRNAs. CiRNA formation, rather than being a random process, seems to depend on the presence of specific
692:
that suppress the ability of the miRNA to bind its mRNA targets, thanks to the presence of multiple binding sites that recognize a specific seed region. Certain circular RNAs have many miRNA binding sites, which yielded a clue that they may function in sponging. Two recent papers confirmed this
608:
measured the half-lives of 60 circRNAs and their linear counterparts expressed from the same host gene and revealed that the median half-life of circRNAs of mammary cells (18.8 to 23.7 hours) is at least 2.5 times longer than the median half-life of their linear counterparts (4.0 to 7.4 hours).
1064:
are mostly plant pathogens, which consist of short stretches (a few hundred nucleobases) of highly complementary, circular, single-stranded, and non-coding RNAs without a protein coat. Compared with other infectious plant pathogens, viroids are extremely small in size, ranging from 246 to 467
992:
can encode circular RNAs such as circEBNA_W1_C1 (EBV) and circE7 (HPV) that play a role in oncogenesis in infected individuals. As circRNAs involved in cancer development or regulation process so that it has the potential to use as a biomarker in cancer surveillance and identification process.
916:
regulator miR-133 and leads to heart failure. Apart from circRNA-mediated cardiac disease, some circRNAs have played a role in cardiac damage repair. For example, circAmotl1 overexpression increases cardiomyocyte longevity through binding and translocation of AKT that regulates cardiac repair.
399:
densities that can form inverted repeated Alu pairs (IRAlus). IRAlus, either convergent or divergent, are juxtaposed across flanking introns of circRNAs in a parallel way with similar distances to adjacent exons. IRAlus, and other non-repetitive, but complementary, sequences were also found to
394:
genes, suggesting that the circular RNA formation is generally coupled to RNA splicing. It was determined that most circular RNAs contain multiple, most commonly, two to three, exons. Exons from circRNAs with only one circularized exon were found to be much longer than those from circRNAs with
634:. After digesting total RNA with RNase R, they were able to identify circular species, indicating that circRNAs are not specific to eukaryotes. However, these archaeal circular species are probably not made via splicing, suggesting that other mechanisms to generate circular RNA likely exist.
340:
RNA was treated with RNase R to enrich for circular RNAs, followed by the categorization of circular transcripts based on their abundance (low, medium, high). Approximately 1 in 8 expressed genes were found to produce detectable levels of circRNAs, including those of low abundance, which was
543:
Introns surrounding exons that are selected to be circularized are, on average, up to three times longer than those not flanking pre-circle exons, although it is not yet clear why this is the case. Compared to regions not resulting in circles, these introns are much more likely to contain
357:, and extensively validating them. The expression of circRNAs was often found to be tissue/developmental stage specific. Additionally, circRNAs were found to have the ability to act as antagonists of miRNAs, microRNAs which interfere with translation of mRNAs, as exemplified by the circRNA
429:, indicating that some circRNAs are also part of the causal pathway. Altogether, circRNA brain expression was found to explain more about Alzheimer's clinical manifestations than the number of APOε4 alleles, suggesting that circRNAs could be used as a potential biomarker for Alzheimer's.
719:, which do not have the CDR1 locus in their genome, provides evidence for CiRS-7's sponge activity. During development, miR-7 is strongly expressed in the zebrafish brain. To silence miR-7 expression in zebrafish, Memczak and colleagues took advantage of a tool called
1017:
Circular RNA plays a significant role in immune regulation and induction of T cell responses. circRNA100783 is involved in immunity and senescence of CD8+ T cells. circRNA-003780 and circRNA-010056 also have major roles for macrophage differentiation and polarization.
637:
CircRNAs were found to be largely conserved between human and sheep. By analyzing total RNA sequencing data from sheep's parietal lobe cortex and peripheral blood mononuclear cells it was shown that 63% of the detected circRNAs are homologous to known human circRNAs.
307:
In 2012, in an effort to initially identify cancer-specific exon scrambling events, scrambled exons were discovered in large numbers in both normal and cancer cells. It was found that scrambled exon isoforms comprised about 10% of the total transcript isoforms in
818:. In addition, these molecules contain few (if any) miRNA binding sites. Instead of acting as sponges, ciRNAs seem to function in regulating the expression of their parent genes. For example, a relatively abundant ciRNA called ci-ankrd52 positively regulates
822:
transcription. Many ciRNAs remain at their "sites of synthesis" in the nucleus. However, ciRNA may have roles other than simply regulating their parent genes, as ciRNAs do localize to additional sites in the nucleus other than their "sites of synthesis".
502:
These are first spliced from precursors as linear molecules and then circularized with a ligase. They are essential in allowing for the rearrangement in RNA sequence order and vital in the biogenesis of permuted tRNA genes in certain algae and archaea.
408:
The
Cruchaga lab performed the first large scale analyses of circRNA in Alzheimer disease (AD) and demonstrated the role of circRNAs in health and disease. A total of 148 circRNAs were found to be significantly associated in multiple datasets with
835:, it is important to consider how circular RNA can be used as a tool to help mankind. Given its abundance, evolutionary conservation, and potential regulatory role, it is worthwhile to look into how circular RNA can be used to study
421:(FDR) correction. The expression of circRNAs was independent of the lineal form and that circRNA expression was also corrected by cell proportion. CircRNAs were also found to be co-expressed with known causal Alzheimer genes, such as
778:, whereas that without an IRES did not. Although the tested circRNA was a purely artificial construct, Chen and Sarnow stated in their paper that they would be interested to see whether circles naturally contain IRES elements.
548:
repeats, Alu being the most common transposon in the genome. By the Alu repeats base pairing to one another, it has been proposed that this may enable the splice sites to find each other, thus facilitating circularization.
523:
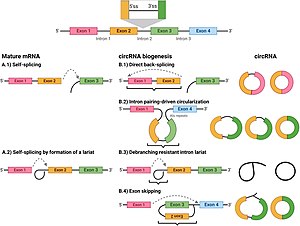
Circular RNAs produced by back-splicing (a form of exon scrambling) occur when a 5′ splice site is joined to an upstream 3′ splice site. Currently, more than 25,000 different circRNAs have been identified in humans.
983:
when it overexpresses and down-regulates COL1A1 protein function. In contrast, silencing of hsa_circRNA_002178 reduced IL-6 and TNF-α production, which inhibited tumor growth and inflammation. Some viruses such as
742:(IRs) over 15.5 kilobases (kb) in length. When one or both of the IRs are deleted, circularization does not occur. It was this finding that introduced the idea of inverted repeats enabling circularization.
328:, which are not present in circular molecules. Overall, 98% of scrambled isoforms were found to represent circRNAs, circRNAs were found to be located in the cytoplasm, and circRNAs were found to be abundant.
768:(RBPs) and RNAs besides miRNAs to form RNA-protein complexes. These complexes could regulate RBP & RNA interactions with, for example, the canonical linear transcript of the gene or viral infection.
221:, in which two RNA transcripts fuse, resulting in a linear transcript containing exons that, for example, may be derived from genes encoded on two different chromosomes. Trans-splicing is very common in
665:
kinases which produce a large amount of circRNA from one particular exon in both species. Evolutionary conservation reinforces the likelihood of a relevant and significant role for RNA circularization.
745:
Because circular RNA sponges are characterized by high expression levels, stability, and a large number of miRNA binding sites, they are likely to be more effective sponges than those that are linear.
1065:
nucleobases; they thus consist of fewer than 10,000 atoms. In comparison, the genome of the smallest known viruses capable of causing an infection by themselves are around 2,000 nucleobases long.
2270:
Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (November 1982). "Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of
Tetrahymena".
723:, which can base pair and sequester target molecules. Morpholino treatment had the same severe effect on midbrain development as ectopically expressing CiRS-7 in zebrafish brains using injected
511:
Certain archaeal species have circRNAs that are produced from excised circularized tRNA introns. Circularization of functional noncoding RNAs is thought to work as a protective mechanism against
231:
The notion that circularized transcripts are byproducts from imperfect splicing is supported by the low abundance and the lack of sequence conservation of most circRNAs, but has been challenged.
320:
being identified. Additionally, about 1 in 50 expressed genes produced scrambled transcript isoforms at least 10% of the time. Tests used to recognize circularity included treating samples with
753:
Though recent attention has been focused on circRNA's "sponge" functions, scientists are considering several other functional possibilities as well. For example, some areas of the mouse adult
866:, which is a synthetic gene that is transferred between organisms. It is also important to consider how transgenes can be expressed only in specific tissues, or expressed only when induced.
208:
Exon scrambling, also called exon shuffling, describes an event in which exons are spliced in a "non-canonical" (atypical) order. There are three ways in which exon scrambling can occur:
993:
Circular RNA has the advantage of stability, tissue specificity and it can be found in the blood, saliva, urine, cerebrospinal fluid, and human body fluid secretion that has abundance in
466:
In viroids and HDV, single-stranded circRNAs are vital in RNA replication. Circularity allows for one initiation event to lead to multiple genomic copies in a process otherwise known as
921:. Cardiac dysfunction occurs post myocardial infection due to CircNfix downregulation. Since various types of circular RNA are related to heart disease, it can be used as a potential
257:
A-to-I RNA editing leads to an increased formation of circRNAs, which is presumably mediated by better complementary pairing of RNA of the Alu elements flanking the back-splice site.
785:
2013: Tested natural circRNAs that contained a translation "start codon". However, none of these molecules bound to ribosomes, suggesting that many circRNAs may not be translated
677:(miRNAs) are small (~21nt) non-coding RNAs that repress translation of messenger RNAs involved in a large, diverse set of biological processes. They directly base-pair to target
794:
Transporting miRNAs inside the cell. The fact that CiRS-7 can be sliced by miR-671 might indicate the existence of a system to release a "load" of miRNAs at the appropriate time.
395:
multiple circularized exons, indicating that processing may prefer a certain length to maximize exon(s) circularization. The introns of circularized exons generally contain high
688:
proteins are the "effector proteins" which help miRNAs carry out their job, while microRNA sponges are RNAs that "sponge up" miRNAs of a particular family, thereby serving as
153:, which is flanked by specific sequences at its 5' and 3' ends, known as a donor splice site (or 5' splice site) and an acceptor splice site (or 3' splice site), respectively.
2224:
Grabowski PJ, Zaug AJ, Cech TR (February 1981). "The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of
Tetrahymena".
79:
normally present in an RNA molecule have been joined together. This feature confers numerous properties to circular RNA, many of which have only recently been identified.
609:
Generally, the lifetime of RNA molecules defines their response time. Accordingly, it was reported that mammary circRNAs respond slowly to stimulation by growth factors.
270:
Early discoveries of circular RNAs led to the belief that they lacked significance due to their rarity. These early discoveries included the analysis of genes like the
851:/ARF expression, which, in turn, increases risk for atherosclerosis. Further study of cANRIL expression could potentially be used to prevent or treat atherosclerosis.
797:
Regulating mRNA in the cell through limited base pairing. It is formally possible that miR-7 moderates CiRS-7's regulatory activity instead of the other way around!
1038:
mechanisms such as circRBFOX2, circLMO7 acts as a negative regulator and CircSVIL acts as a positive regulator. circRBFOX2 regulates miR-206 expression and induces
2017:
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. (March 2013). "Circular RNAs are a large class of animal RNAs with regulatory potency".
564:, where the number of circular RNA transcripts derived from a gene can be up to ten times greater than the number of associated linear RNAs generated from that
930:
trace disease progression in different stages of cardiac dysfunction using circular RNA as a biomarker and can be used for gene delivery purposes in cells.
478:
Circular molecules are produced by introns produced from spliceosomal splicing, tRNA splicing, and group I and group II (self-splicing ribozymes) introns.
2316:
Zaug AJ, Grabowski PJ, Cech TR (17–23 February 1983). "Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction".
390:
data, was developed. The vast majority of identified highly expressed exonic circular RNAs were found to be processed from exons located in the middle of
2378:
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (March 2013). "Natural RNA circles function as efficient microRNA sponges".
869:
Circular RNAs were found to be regulated by hypoxia, especially the circRNA cZNF292 was found to have proangiogenic activities in endothelial cells.
3444:
Zhang, Pengpeng; Chao, Zhe; Zhang, Rui; Ding, Ruoqi; Wang, Yaling; Wu, Wei; Han, Qiu; Li, Cencen; Xu, Haixia; Wang, Lei; Xu, Yongjie (2019-08-13).
540:, although the circular RNAs themselves do not appear to be translated. During circRNA formation, exon 2 is often the upstream "acceptor" exon.
188:
nematodes) have similar numbers of genes (in the range of 20 - 25 thousand). One of the most striking examples of alternative splicing is in the
1354:
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, et al. (April 2002). "A draft sequence of the rice genome (Oryza sativa L. ssp. indica)".
353:
At the same time, a computational method to detect circRNAs was developed, leading to de novo detection of circRNAs in humans, mice, and
1009:
progression acting as a miRNA sponge which regulates DNA methylation, adaptive immune activation, and costimulatory molecule secretion.
843:
Circular ANRIL (cANRIL) is the circular form of ANRIL, a long non-coding RNA (ncRNA). Expression of cANRIL is correlated with risk for
2813:
Dubin RA, Kazmi MA, Ostrer H (December 1995). "Inverted repeats are necessary for circularization of the mouse testis Sry transcript".
893:
774:
Chen and Sarnow 1995 showed that a synthetic circRNA that contained an IRES (internal ribosome entry site) produced a protein product
227:
A splice donor site being joined to a splice acceptor site further upstream in the primary transcript, yielding a circular transcript.
938:
Various studies have demonstrated that circular RNA acts as a prognostic agent and biomarker in kidney diseases including renal cell
757:
show expression of CiRS-7 but not miR-7, suggesting that CiRS-7 may have roles that are independent of interacting with the miRNA.
552:
Introns within the circRNAs are retained at a relatively high frequency (~25%), thus adding extra sequence to the mature circRNAs.
604:
CircRNAs lack a polyadenylated tail and, therefore, are predicted to be less prone to degradation by exonucleases. In 2015, Enuka
588:. However, certain circRNAs, such as CiRS-7/CDR1as, are expressed in neuronal tissues, where mitotic division is not prevalent.
2604:
Ding XC, Weiler J, Grosshans H (January 2009). "Regulating the regulators: mechanisms controlling the maturation of microRNAs".
2883:
Chen CY, Sarnow P (April 1995). "Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs".
2073:"An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations"
3024:"Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk"
2490:
2137:
3189:
Jin, Juan; Sun, Haolu; Shi, Chao; Yang, Hui; Wu, Yiwan; Li, Wanhai; Dong, Yu-hang; Cai, Liang; Meng, Xiao-ming (June 2020).
3372:
Amaya, Laura; Grigoryan, Lilit; Li, Zhijian; Lee, Audrey; Wender, Paul A.; Pulendran, Bali; Chang, Howard Y. (2023-05-16).
3505:"Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures"
2172:"Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies"
886:., demonstrated for the first time that brain circular RNAs (circRNA) are part of the pathogenic events that lead to
2737:
Summerton J (December 1999). "Morpholino antisense oligomers: the case for an RNase H-independent structural type".
894:
Circular RNA pathogenesis in Heart disease, Renal disease, Liver disease, autoimmune disease, and Cancer development
105:
and are presumably more stable than most linear RNA in cells. Circular RNA has been linked to some diseases such as
908:
and protein ID1, FAK, causing cardiomyopathy induced by DOX. Titin gene derived circRNA induces cardiotoxicity in
2124:. Advances in Experimental Medicine and Biology. Vol. 1087. Singapore: Springer Singapore. pp. 41–52.
989:
649:
In a closer evolutionary connection, a comparison of RNA from mouse testes vs. RNA from a human cell found 69
163:
The free 5' exon then attacks the 3' splice site, joining the two exons and releasing a structure known as an
91:
291:
271:
160:
attack by a downstream sequence called the branch point, resulting in a circular structure called a lariat.
684:
MicroRNAs are grouped in "seed families". Family members share nucleotides 2–7, known as the seed region.
1469:"Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types"
2434:"Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor"
386:
In the same year, CIRCexplorer, a tool used to identify thousands of circRNAs in humans without RNase R
467:
2120:
Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN (2018). "Circular RNA Splicing". In Xiao J (ed.).
731:
697:
422:
275:
1187:
479:
414:
1407:"Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated"
1145:
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. (April 2017).
437:
3073:
Bhardwaj V, Singh A, Choudhary A, Dalavi R, Ralte L, Chawngthu RL, et al. (September 2023).
2848:
Bhardwaj V, Singh A, Dalavi R, Ralte L, Chawngthu RL, Senthil Kumar N, et al. (2022-11-04).
1897:"Reduction of A-to-I RNA editing in the failing human heart regulates formation of circular RNAs"
212:
195:
gene, which can give rise to approximately 30 thousand distinct alternatively spliced isoforms.
985:
887:
859:
689:
410:
2849:
137:
in the mature mRNA, which can subsequently be translated to produce the protein product. The
847:, a disease in which the arteries become hard. It has been proposed that cANRIL can modify
141:, a protein-RNA complex located in the nucleus, catalyzes splicing in the following manner:
3516:
3385:
3086:
2892:
2387:
2325:
2026:
1699:
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. (January 2015).
1604:
1480:
1363:
943:
418:
184:
2557:"Novel circRNA discovery in sheep shows evidence of high backsplice junction conservation"
681:(mRNAs), and can trigger cleavage of the mRNA depending on the degree of complementarity.
8:
2696:
Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, et al. (October 2015).
1895:
Kokot KE, Kneuer JM, John D, Rebs S, Möbius-Winkler MN, Erbe S, et al. (June 2022).
926:
913:
765:
623:
317:
102:
3520:
3414:
3389:
3373:
3107:
3090:
3074:
2896:
2391:
2329:
2030:
1608:
1484:
1367:
1228:
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. (February 2013).
324:, an enzyme that degrades linear but not circular RNAs, and testing for the presence of
3480:
3445:
3349:
3316:
3287:
3252:
3251:
Zeng, Xianhui; Yuan, Xianglei; Cai, Qiuyu; Tang, Chengwei; Gao, Jinhang (August 2021).
3223:
3190:
3166:
3131:
3050:
3023:
2955:
2930:
2865:
2670:
2645:
2581:
2556:
2532:
2507:
2458:
2433:
2411:
2349:
2295:
2249:
2198:
2171:
2097:
2072:
2050:
1975:
1948:
1921:
1896:
1872:
1815:
1790:
1766:
1741:
1676:
1649:
1625:
1592:
1561:
1534:
1503:
1468:
1431:
1406:
1387:
1305:
1278:
1254:
1229:
1171:
1146:
1127:
1114:
1087:
1047:
by enhancing Wnt5a and CaMKIId expression which are involved in myogenesis regulation.
1039:
1006:
662:
650:
3539:
3504:
2750:
1701:"Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals"
617:
3544:
3485:
3467:
3419:
3401:
3354:
3336:
3292:
3274:
3228:
3210:
3171:
3153:
3112:
3055:
3004:
2960:
2908:
2869:
2830:
2826:
2795:
2790:
2773:
2772:
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. (June 1993).
2754:
2719:
2675:
2621:
2586:
2537:
2486:
2463:
2415:
2403:
2341:
2287:
2283:
2241:
2237:
2203:
2170:
Nisar S, Bhat AA, Singh M, Karedath T, Rizwan A, Hashem S, et al. (2021-02-05).
2143:
2133:
2102:
2042:
1980:
1926:
1864:
1820:
1771:
1722:
1681:
1630:
1566:
1508:
1436:
1391:
1379:
1337:
1310:
1259:
1176:
1131:
1119:
994:
832:
708:). It has over 60 miR-7 binding sites, far more than any known linear miRNA sponge.
482:
form circRNAs through autocatalytic ribozymal action, and while they can be detected
56:
2981:
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. (September 2013).
2299:
2253:
2071:
Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, et al. (November 2019).
1876:
3534:
3524:
3475:
3457:
3409:
3393:
3344:
3328:
3282:
3264:
3218:
3202:
3161:
3143:
3102:
3094:
3045:
3035:
2994:
2950:
2942:
2900:
2857:
2822:
2785:
2746:
2709:
2698:"Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA"
2665:
2657:
2613:
2576:
2568:
2527:
2519:
2453:
2445:
2395:
2353:
2333:
2279:
2233:
2193:
2183:
2125:
2092:
2084:
2054:
2034:
1970:
1960:
1916:
1908:
1854:
1810:
1802:
1761:
1753:
1712:
1671:
1661:
1620:
1612:
1556:
1546:
1498:
1488:
1426:
1418:
1371:
1300:
1290:
1249:
1241:
1166:
1158:
1109:
1099:
577:
565:
309:
282:, all of which expressed circular isoforms. CircRNA producing genes like the human
2714:
2697:
2617:
2432:
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y (February 2016).
1757:
1086:
Dawoud A, Ihab
Zakaria Z, Hisham Rashwan H, Braoudaki M, Youness RA (March 2023).
3040:
2999:
2982:
1717:
1700:
1493:
1422:
1162:
947:
844:
739:
584:, one hypothesis is that the molecules exit the nucleus during this phase of the
487:
342:
325:
287:
3075:"HIV-1 Vpr induces ciTRAN to prevent transcriptional repression of the provirus"
2129:
1104:
3509:
Proceedings of the
National Academy of Sciences of the United States of America
2850:"HIV-1 Vpr induces ciTRAN to prevent transcriptional silencing of the provirus"
2774:"Circular transcripts of the testis-determining gene Sry in adult mouse testis"
2572:
1912:
1859:
1842:
1650:"Alu elements shape the primate transcriptome by cis-regulation of RNA editing"
909:
218:
83:
2861:
2188:
2088:
1551:
1279:"Closing the circle: current state and perspectives of circular RNA databases"
877:
727:. This indicates a significant interaction between CiRS-7 and miR-7 in vivo.
341:
significantly higher than previously suspected, and was attributed to greater
3561:
3471:
3405:
3340:
3278:
3214:
3157:
1666:
980:
678:
591:
130:
48:
24:
3529:
3397:
2904:
1616:
1375:
1088:"Circular RNAs: New layer of complexity evading breast cancer heterogeneity"
810:
CiRNAs are distinct from circRNAs in that they are prominently found in the
403:
3567:
3489:
3423:
3358:
3296:
3232:
3175:
3116:
3098:
3059:
3008:
2964:
2758:
2723:
2679:
2625:
2590:
2555:
Varela-Martínez E, Corsi GI, Anthon C, Gorodkin J, Jugo BM (January 2021).
2541:
2467:
2407:
2207:
2147:
2106:
2046:
1984:
1930:
1868:
1824:
1806:
1775:
1726:
1685:
1634:
1570:
1512:
1440:
1383:
1314:
1263:
1245:
1180:
1123:
958:
Evidence found that circular RNA plays a role in chronic liver disease and
836:
811:
573:
569:
545:
157:
3548:
3503:
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (November 1976).
3315:
Li, Weizhen; Liu, Jia-Qiang; Chen, Ming; Xu, Jiang; Zhu, Di (March 2022).
3269:
3022:
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE (December 2010).
2946:
2912:
2834:
2799:
2523:
2449:
2345:
2291:
2245:
704:
locus (hence the name CDR1as), and targets miR-7 (hence the name CiRS-7 –
19:
3462:
3148:
1535:"Expanded identification and characterization of mammalian circular RNAs"
1295:
1085:
1044:
963:
959:
754:
701:
512:
396:
253:
240:
146:
138:
98:
2661:
2399:
2038:
1230:"Circular RNAs are abundant, conserved, and associated with ALU repeats"
3332:
3206:
1965:
1035:
918:
807:
elements near the 5' splice site and the branchpoint site (see above).
720:
585:
441:
Hepatitis delta virus containing a circular single-stranded RNA genome.
337:
244:
this enables RNA pairing, which in turn facilitates circRNA synthesis.
122:
72:
28:
348:
167:. The intron lariat is subsequently de-branched and quickly degraded.
97:
Because circular RNA do not have 5' or 3' ends, they are resistant to
2337:
976:
939:
922:
863:
815:
716:
685:
561:
1841:
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (September 2014).
1791:"Short intronic repeat sequences facilitate circular RNA production"
641:
3253:"Circular RNA as An Epigenetic Regulator in Chronic Liver Diseases"
2739:
Biochimica et
Biophysica Acta (BBA) - Gene Structure and Expression
734:. SRY, which is highly expressed in murine testes, functions as a
724:
674:
618:
Evolutionary conservation of circularization mechanisms and signals
279:
118:
90:. Some types of circular RNA have also recently shown potential as
87:
2554:
962:
regulation leading to liver fibrosis and autoimmune disease by an
905:
801:
1023:
735:
631:
581:
537:
387:
374:
358:
321:
133:(mRNA) transcript, intervening introns are removed, leaving only
917:
Circular RNA CDR1 has an important role during infection in the
1144:
1061:
1056:
855:
819:
391:
370:
150:
126:
106:
42:
36:
2483:
Models of Life - Dynamics and
Regulation in Biological Systems
336:
In 2013, a higher abundance of circRNAs was discovered. Human
331:
1949:"Circular RNAs: analysis, expression and potential functions"
658:
654:
426:
247:
192:
134:
76:
3072:
2431:
622:
CircRNAs have been identified in various species across the
599:
369:
In 2014, human circRNAs were identified and quantified from
364:
278:
genes, and the recent discovery of the human non-coding RNA
2016:
878:
Circular RNAs play a role in
Alzheimer disease pathogenesis
848:
313:
283:
171:
94:. The biological function of most circular RNA is unclear.
3317:"Circular RNA in cancer development and immune regulation"
2508:"Transcriptome-wide discovery of circular RNAs in Archaea"
2505:
2377:
1276:
873:
disrupting SRSF1's ability to inhibit HIV-1 transcription.
129:. In eukaryotes, as a gene is transcribed from DNA into a
1648:
Daniel C, Silberberg G, Behm M, Öhman M (February 2014).
1647:
979:
target. Another circular RNA hsa_circRNA_002178 leads to
404:
Genome-wide calling of circRNA in Alzheimer disease cases
297:
68:
2847:
2269:
2119:
1593:"Molecular biology. A circuitous route to noncoding RNA"
1467:
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012).
1227:
1029:
748:
653:
circRNAs. For example, both humans and mice encode the
499:
CircRNAs from intermediates in RNA processing reactions
3502:
2771:
2695:
2506:
Danan M, Schwartz S, Edelheit S, Sorek R (April 2012).
1698:
294:), and the human dystrophin gene were also discovered.
234:
3374:"Circular RNA vaccine induces potent T cell responses"
2169:
1843:"Complementary sequence-mediated exon circularization"
1742:"Repetitive elements regulate circular RNA biogenesis"
1466:
1277:
Vromman M, Vandesompele J, Volders PJ (January 2021).
612:
2070:
912:. circSLC8A1-1 overexpression causes sponging of the
560:
In the cell, circRNAs are predominantly found in the
86:
genes. Some circular RNA have been shown to code for
854:
miR-7 plays an important regulatory role in several
839:
and devise therapeutic interventions. For example:
3371:
2980:
2603:
1894:
1532:
925:and therapeutic target. For example, postoperative
555:
445:Circular RNAs can be separated into five classes:
349:
CircRNAs tissue specificity and antagonist activity
156:The 5' splice site sequence is then subjected to a
3443:
2691:
2689:
1336:
1012:
669:
302:
260:
3021:
2924:
2922:
2315:
2223:
2219:
2217:
1533:Guo JU, Agarwal V, Guo H, Bartel DP (July 2014).
1343:(9th ed.). San Francisco: Benjamin Cummings.
520:CircRNAs in eukaryotes produced by back-splicing
361:, which has miRNA binding sites (as seen below).
3559:
2812:
1840:
1586:
1584:
1582:
1580:
265:
82:Many types of circular RNA arise from otherwise
3378:Proceedings of the National Academy of Sciences
3250:
2686:
2639:
2637:
2635:
2012:
2010:
2008:
2006:
2004:
2002:
2000:
1998:
1996:
1994:
1942:
1940:
1404:
802:Circular intronic long non-coding RNAs (ciRNAs)
738:sponge. In the genome, SRY is flanked by long
125:genes are split by non-coding sequences called
23:This is a representative figure showing mature
2919:
2646:"MicroRNA sponges: progress and possibilities"
2265:
2263:
2214:
1946:
1330:
1328:
1326:
1324:
290:genes, the rat androgen binding protein gene (
3439:
3437:
3435:
3433:
3188:
2931:"Circular RNAs: splicing's enigma variations"
2485:. San Francisco: Cambridge University Press.
2165:
2163:
2161:
2159:
2157:
1577:
215:in the genome, which often occurs in cancers.
75:closed continuous loop. In circular RNA, the
3246:
3244:
3242:
2928:
2632:
2311:
2309:
1991:
1937:
1353:
568:. It is unclear how circular RNAs exit the
463:Viroids and the hepatitis delta virus (HDV)
381:
3314:
2373:
2371:
2369:
2367:
2365:
2363:
2260:
2176:Frontiers in Cell and Developmental Biology
1890:
1888:
1886:
1788:
1590:
1462:
1460:
1458:
1456:
1454:
1452:
1450:
1321:
1050:
997:are good to use as cancer biomarker agent.
826:
332:Discovery of a higher abundance of circRNAs
3430:
3321:Journal of Cellular and Molecular Medicine
3310:
3308:
3306:
3195:Journal of Cellular and Molecular Medicine
2976:
2974:
2882:
2643:
2154:
248:Impact of RNA editing on circRNA formation
3538:
3528:
3479:
3461:
3413:
3348:
3286:
3268:
3239:
3222:
3165:
3147:
3106:
3049:
3039:
2998:
2954:
2789:
2736:
2713:
2669:
2580:
2531:
2457:
2427:
2425:
2306:
2197:
2187:
2096:
1974:
1964:
1920:
1858:
1814:
1765:
1716:
1675:
1665:
1624:
1560:
1550:
1502:
1492:
1430:
1304:
1294:
1253:
1170:
1113:
1103:
904:binds to the transcription factors E2F1,
730:Another notable circular miRNA sponge is
600:CircRNAs are more stable than linear RNAs
365:CircRNAs and ENCODE Ribozero RNA-seq data
198:
2360:
1883:
1447:
1405:Celotto AM, Graveley BR (October 2001).
1034:Circular RNA plays an important role in
640:
590:
486:their function is yet to be determined.
436:
170:
18:
3446:"Circular RNA Regulation of Myogenesis"
3303:
3129:
2983:"Circular intronic long noncoding RNAs"
2971:
2480:
1528:
1526:
1524:
1522:
760:Potential roles include the following:
696:CDR1as/CiRS-7 is encoded in the genome
179:
3560:
2422:
1836:
1834:
1739:
1223:
1221:
1219:
1217:
298:Genome-wide identification of circRNAs
252:RNAs can undergo base modification by
2066:
2064:
1334:
1215:
1213:
1211:
1209:
1207:
1205:
1203:
1201:
1199:
1197:
1030:Circular RNA in Myogenesis regulation
1000:
749:Other possible functions for circRNAs
530:
432:
2644:Ebert MS, Sharp PA (November 2010).
1519:
235:Alu elements impact circRNA splicing
47:B.4) lariat-driven circularization (
3130:Wang, Ying; Liu, Bin (2020-05-17).
1947:Barrett SP, Salzman J (June 2016).
1831:
1789:Liang D, Wilusz JE (October 2014).
613:Plausible functions of circular RNA
312:, with 2,748 scrambled isoforms in
286:gene, the human and rat cytochrome
13:
2929:Hentze MW, Preiss T (April 2013).
2061:
1591:Wilusz JE, Sharp PA (April 2013).
1194:
203:
71:which, unlike linear RNA, forms a
41:B.3) debranching resistant intron
14:
3579:
3191:"Circular RNA in renal diseases"
3132:"Circular RNA in Diseased Heart"
1186:
953:
933:
898:
556:Location of circRNAs in the cell
39:pairing driven circularization;
31:of circRNAs which might include:
3496:
3365:
3182:
3123:
3066:
3015:
2876:
2841:
2806:
2765:
2730:
2597:
2548:
2499:
2474:
2113:
1782:
1733:
1692:
1641:
1013:Circular RNA in immune response
1005:Circular RNA has a function in
670:CDR1as/CiRS-7 as a miR-7 sponge
515:and to promote proper folding.
303:Scrambled isoforms and circRNAs
261:Characteristics of circular RNA
112:
67:) is a type of single-stranded
1398:
1347:
1270:
1138:
1079:
1026:to express genes of interest.
508:Noncoding circRNAs in archaea
468:rolling circle RNA replication
1:
2751:10.1016/S0167-4781(99)00150-5
2715:10.1161/CIRCRESAHA.115.306319
2618:10.1016/j.tibtech.2008.09.006
1758:10.1080/2159256X.2015.1045682
1073:
712:sites meet this requirement.
706:Circular RNA Sponge for miR-7
266:Early discoveries of circRNAs
3041:10.1371/journal.pgen.1001233
3000:10.1016/j.molcel.2013.08.017
2827:10.1016/0378-1119(95)00639-7
2791:10.1016/0092-8674(93)90279-y
2284:10.1016/0092-8674(82)90414-7
2238:10.1016/0092-8674(81)90142-2
1901:Basic Research in Cardiology
1718:10.1016/j.celrep.2014.12.019
1494:10.1371/journal.pone.0030733
1163:10.1016/j.molcel.2017.02.021
27:and corresponding canonical
7:
2130:10.1007/978-981-13-1426-1_4
1283:Briefings in Bioinformatics
1105:10.1016/j.ncrna.2022.09.011
1068:
572:through a relatively small
10:
3584:
2573:10.1038/s41598-020-79781-2
1913:10.1007/s00395-022-00940-9
1860:10.1016/j.cell.2014.09.001
1423:10.1093/genetics/159.2.599
1054:
595:Human cell nucleus diagram
33:B.1) direct back splicing;
16:Type of RNA found in cells
2862:10.1101/2022.11.04.515166
2189:10.3389/fcell.2021.617281
2089:10.1038/s41593-019-0501-5
1552:10.1186/s13059-014-0409-z
1147:"Translation of CircRNAs"
969:
536:with exons that code for
382:CircRNAs and CIRCexplorer
175:Pre-mRNA to mRNA splicing
1740:Wilusz JE (2015-05-04).
1667:10.1186/gb-2014-15-2-r28
1051:Viroids as circular RNAs
827:Circular RNA and disease
452:Classes of Circular RNAs
415:clinical dementia rating
117:In contrast to genes in
3530:10.1073/pnas.73.11.3852
3398:10.1073/pnas.2302191120
2905:10.1126/science.7536344
2606:Trends in Biotechnology
1795:Genes & Development
1746:Mobile Genetic Elements
1617:10.1126/science.1238522
1376:10.1126/science.1068037
1092:Non-Coding RNA Research
942:, acute kidney injury,
831:As with most topics in
544:complementary inverted
490:also generate circRNAs
213:Tandem exon duplication
3099:10.1126/sciadv.adh9170
2512:Nucleic Acids Research
2438:Nucleic Acids Research
1807:10.1101/gad.251926.114
1246:10.1261/rna.035667.112
690:competitive inhibitors
646:
596:
475:CircRNAs from introns
442:
199:Non-canonical splicing
176:
52:
3270:10.3390/cells10081945
2947:10.1038/emboj.2013.53
2856:: 2022.11.04.515166.
645:Three domains of life
644:
594:
440:
417:(CDR) at death after
174:
22:
3463:10.3390/cells8080885
3149:10.3390/cells9051240
2702:Circulation Research
990:human papillomavirus
944:diabetic nephropathy
766:RNA-binding proteins
419:false discovery rate
318:embryonic stem cells
185:Alternative splicing
180:Alternative splicing
3521:1976PNAS...73.3852S
3390:2023PNAS..12002191A
3384:(20): e2302191120.
3091:2023SciA....9H9170B
2897:1995Sci...268..415C
2662:10.1261/rna.2414110
2524:10.1093/nar/gkr1009
2450:10.1093/nar/gkv1367
2400:10.1038/nature11993
2392:2013Natur.495..384H
2330:1983Natur.301..578Z
2077:Nature Neuroscience
2039:10.1038/nature11928
2031:2013Natur.495..333M
1609:2013Sci...340..440W
1485:2012PLoSO...730733S
1368:2002Sci...296...79Y
927:atrial fibrillation
914:cardiac hypertrophy
888:Alzheimer's disease
860:Parkinson's disease
771:Protein production
715:An experiment with
630:sequenced RNA from
580:breaks down during
411:Alzheimer's disease
3333:10.1111/jcmm.16102
3207:10.1111/jcmm.15295
2561:Scientific Reports
2481:Sneppen K (2014).
1966:10.1242/dev.128074
1296:10.1093/bib/bbz175
1007:autoimmune disease
1001:Autoimmune disease
986:Epstein Barr virus
647:
597:
531:Length of circRNAs
443:
433:Classes of CircRNA
177:
53:
3515:(11): 3852–3856.
3201:(12): 6523–6533.
2891:(5209): 415–417.
2656:(11): 2043–2050.
2492:978-1-107-06190-3
2386:(7441): 384–388.
2324:(5901): 578–583.
2139:978-981-13-1425-4
2083:(11): 1903–1912.
2025:(7441): 333–338.
1959:(11): 1838–1847.
1801:(20): 2233–2247.
1603:(6131): 440–441.
1335:Reece JB (2010).
833:molecular biology
626:. In 2011, Danan
528:
527:
57:molecular biology
3575:
3553:
3552:
3542:
3532:
3500:
3494:
3493:
3483:
3465:
3441:
3428:
3427:
3417:
3369:
3363:
3362:
3352:
3327:(6): 1785–1798.
3312:
3301:
3300:
3290:
3272:
3248:
3237:
3236:
3226:
3186:
3180:
3179:
3169:
3151:
3127:
3121:
3120:
3110:
3085:(36): eadh9170.
3079:Science Advances
3070:
3064:
3063:
3053:
3043:
3034:(12): e1001233.
3019:
3013:
3012:
3002:
2978:
2969:
2968:
2958:
2935:The EMBO Journal
2926:
2917:
2916:
2880:
2874:
2873:
2845:
2839:
2838:
2821:(1–2): 245–248.
2810:
2804:
2803:
2793:
2784:(5): 1019–1030.
2769:
2763:
2762:
2734:
2728:
2727:
2717:
2693:
2684:
2683:
2673:
2641:
2630:
2629:
2601:
2595:
2594:
2584:
2552:
2546:
2545:
2535:
2518:(7): 3131–3142.
2503:
2497:
2496:
2478:
2472:
2471:
2461:
2444:(3): 1370–1383.
2429:
2420:
2419:
2375:
2358:
2357:
2338:10.1038/301578a0
2313:
2304:
2303:
2267:
2258:
2257:
2221:
2212:
2211:
2201:
2191:
2167:
2152:
2151:
2117:
2111:
2110:
2100:
2068:
2059:
2058:
2014:
1989:
1988:
1978:
1968:
1944:
1935:
1934:
1924:
1892:
1881:
1880:
1862:
1838:
1829:
1828:
1818:
1786:
1780:
1779:
1769:
1737:
1731:
1730:
1720:
1696:
1690:
1689:
1679:
1669:
1645:
1639:
1638:
1628:
1588:
1575:
1574:
1564:
1554:
1530:
1517:
1516:
1506:
1496:
1464:
1445:
1444:
1434:
1402:
1396:
1395:
1351:
1345:
1344:
1342:
1339:Campbell Biology
1332:
1319:
1318:
1308:
1298:
1274:
1268:
1267:
1257:
1225:
1192:
1191:
1190:
1184:
1174:
1142:
1136:
1135:
1117:
1107:
1083:
814:rather than the
740:inverted repeats
578:nuclear envelope
488:Group II introns
448:
447:
343:sequencing depth
3583:
3582:
3578:
3577:
3576:
3574:
3573:
3572:
3558:
3557:
3556:
3501:
3497:
3442:
3431:
3370:
3366:
3313:
3304:
3249:
3240:
3187:
3183:
3128:
3124:
3071:
3067:
3020:
3016:
2979:
2972:
2927:
2920:
2881:
2877:
2846:
2842:
2811:
2807:
2770:
2766:
2735:
2731:
2708:(10): 884–890.
2694:
2687:
2642:
2633:
2602:
2598:
2553:
2549:
2504:
2500:
2493:
2479:
2475:
2430:
2423:
2376:
2361:
2314:
2307:
2268:
2261:
2222:
2215:
2168:
2155:
2140:
2118:
2114:
2069:
2062:
2015:
1992:
1945:
1938:
1893:
1884:
1839:
1832:
1787:
1783:
1738:
1734:
1697:
1693:
1646:
1642:
1589:
1578:
1531:
1520:
1465:
1448:
1403:
1399:
1362:(5565): 79–92.
1352:
1348:
1333:
1322:
1275:
1271:
1226:
1195:
1185:
1143:
1139:
1084:
1080:
1076:
1071:
1059:
1053:
1032:
1015:
1003:
972:
956:
948:lupus nephritis
936:
901:
896:
880:
845:atherosclerosis
829:
804:
751:
672:
624:domains of life
620:
615:
602:
576:. Because the
558:
533:
480:Group I introns
435:
406:
384:
367:
351:
334:
305:
300:
268:
263:
250:
237:
206:
204:Exon scrambling
201:
182:
115:
92:gene regulators
46:
40:
34:
32:
17:
12:
11:
5:
3581:
3571:
3570:
3555:
3554:
3495:
3429:
3364:
3302:
3238:
3181:
3122:
3065:
3014:
2993:(6): 792–806.
2987:Molecular Cell
2970:
2941:(7): 923–925.
2918:
2875:
2840:
2805:
2764:
2745:(1): 141–158.
2729:
2685:
2631:
2596:
2547:
2498:
2491:
2473:
2421:
2359:
2305:
2278:(1): 147–157.
2259:
2232:(2): 467–476.
2213:
2153:
2138:
2112:
2060:
1990:
1936:
1882:
1853:(1): 134–147.
1830:
1781:
1732:
1711:(2): 170–177.
1691:
1654:Genome Biology
1640:
1576:
1539:Genome Biology
1518:
1446:
1417:(2): 599–608.
1397:
1346:
1320:
1289:(1): 288–297.
1269:
1240:(2): 141–157.
1193:
1157:(1): 9–21.e7.
1151:Molecular Cell
1137:
1077:
1075:
1072:
1070:
1067:
1055:Main article:
1052:
1049:
1031:
1028:
1014:
1011:
1002:
999:
971:
968:
955:
952:
935:
932:
910:cardiomyocytes
900:
897:
895:
892:
879:
876:
875:
874:
870:
867:
852:
828:
825:
803:
800:
799:
798:
795:
792:
791:
790:
779:
769:
750:
747:
679:messenger RNAs
671:
668:
619:
616:
614:
611:
601:
598:
557:
554:
532:
529:
526:
525:
521:
517:
516:
509:
505:
504:
500:
496:
495:
476:
472:
471:
464:
460:
459:
454:
434:
431:
405:
402:
383:
380:
366:
363:
350:
347:
333:
330:
304:
301:
299:
296:
267:
264:
262:
259:
249:
246:
236:
233:
229:
228:
225:
219:Trans-splicing
216:
205:
202:
200:
197:
181:
178:
169:
168:
161:
154:
149:recognizes an
114:
111:
84:protein-coding
77:3' and 5' ends
15:
9:
6:
4:
3:
2:
3580:
3569:
3566:
3565:
3563:
3550:
3546:
3541:
3536:
3531:
3526:
3522:
3518:
3514:
3510:
3506:
3499:
3491:
3487:
3482:
3477:
3473:
3469:
3464:
3459:
3455:
3451:
3447:
3440:
3438:
3436:
3434:
3425:
3421:
3416:
3411:
3407:
3403:
3399:
3395:
3391:
3387:
3383:
3379:
3375:
3368:
3360:
3356:
3351:
3346:
3342:
3338:
3334:
3330:
3326:
3322:
3318:
3311:
3309:
3307:
3298:
3294:
3289:
3284:
3280:
3276:
3271:
3266:
3262:
3258:
3254:
3247:
3245:
3243:
3234:
3230:
3225:
3220:
3216:
3212:
3208:
3204:
3200:
3196:
3192:
3185:
3177:
3173:
3168:
3163:
3159:
3155:
3150:
3145:
3141:
3137:
3133:
3126:
3118:
3114:
3109:
3104:
3100:
3096:
3092:
3088:
3084:
3080:
3076:
3069:
3061:
3057:
3052:
3047:
3042:
3037:
3033:
3029:
3028:PLOS Genetics
3025:
3018:
3010:
3006:
3001:
2996:
2992:
2988:
2984:
2977:
2975:
2966:
2962:
2957:
2952:
2948:
2944:
2940:
2936:
2932:
2925:
2923:
2914:
2910:
2906:
2902:
2898:
2894:
2890:
2886:
2879:
2871:
2867:
2863:
2859:
2855:
2851:
2844:
2836:
2832:
2828:
2824:
2820:
2816:
2809:
2801:
2797:
2792:
2787:
2783:
2779:
2775:
2768:
2760:
2756:
2752:
2748:
2744:
2740:
2733:
2725:
2721:
2716:
2711:
2707:
2703:
2699:
2692:
2690:
2681:
2677:
2672:
2667:
2663:
2659:
2655:
2651:
2647:
2640:
2638:
2636:
2627:
2623:
2619:
2615:
2611:
2607:
2600:
2592:
2588:
2583:
2578:
2574:
2570:
2566:
2562:
2558:
2551:
2543:
2539:
2534:
2529:
2525:
2521:
2517:
2513:
2509:
2502:
2494:
2488:
2484:
2477:
2469:
2465:
2460:
2455:
2451:
2447:
2443:
2439:
2435:
2428:
2426:
2417:
2413:
2409:
2405:
2401:
2397:
2393:
2389:
2385:
2381:
2374:
2372:
2370:
2368:
2366:
2364:
2355:
2351:
2347:
2343:
2339:
2335:
2331:
2327:
2323:
2319:
2312:
2310:
2301:
2297:
2293:
2289:
2285:
2281:
2277:
2273:
2266:
2264:
2255:
2251:
2247:
2243:
2239:
2235:
2231:
2227:
2220:
2218:
2209:
2205:
2200:
2195:
2190:
2185:
2181:
2177:
2173:
2166:
2164:
2162:
2160:
2158:
2149:
2145:
2141:
2135:
2131:
2127:
2123:
2122:Circular RNAs
2116:
2108:
2104:
2099:
2094:
2090:
2086:
2082:
2078:
2074:
2067:
2065:
2056:
2052:
2048:
2044:
2040:
2036:
2032:
2028:
2024:
2020:
2013:
2011:
2009:
2007:
2005:
2003:
2001:
1999:
1997:
1995:
1986:
1982:
1977:
1972:
1967:
1962:
1958:
1954:
1950:
1943:
1941:
1932:
1928:
1923:
1918:
1914:
1910:
1906:
1902:
1898:
1891:
1889:
1887:
1878:
1874:
1870:
1866:
1861:
1856:
1852:
1848:
1844:
1837:
1835:
1826:
1822:
1817:
1812:
1808:
1804:
1800:
1796:
1792:
1785:
1777:
1773:
1768:
1763:
1759:
1755:
1751:
1747:
1743:
1736:
1728:
1724:
1719:
1714:
1710:
1706:
1702:
1695:
1687:
1683:
1678:
1673:
1668:
1663:
1659:
1655:
1651:
1644:
1636:
1632:
1627:
1622:
1618:
1614:
1610:
1606:
1602:
1598:
1594:
1587:
1585:
1583:
1581:
1572:
1568:
1563:
1558:
1553:
1548:
1544:
1540:
1536:
1529:
1527:
1525:
1523:
1514:
1510:
1505:
1500:
1495:
1490:
1486:
1482:
1479:(2): e30733.
1478:
1474:
1470:
1463:
1461:
1459:
1457:
1455:
1453:
1451:
1442:
1438:
1433:
1428:
1424:
1420:
1416:
1412:
1408:
1401:
1393:
1389:
1385:
1381:
1377:
1373:
1369:
1365:
1361:
1357:
1350:
1341:
1340:
1331:
1329:
1327:
1325:
1316:
1312:
1307:
1302:
1297:
1292:
1288:
1284:
1280:
1273:
1265:
1261:
1256:
1251:
1247:
1243:
1239:
1235:
1231:
1224:
1222:
1220:
1218:
1216:
1214:
1212:
1210:
1208:
1206:
1204:
1202:
1200:
1198:
1189:
1182:
1178:
1173:
1168:
1164:
1160:
1156:
1152:
1148:
1141:
1133:
1129:
1125:
1121:
1116:
1111:
1106:
1101:
1097:
1093:
1089:
1082:
1078:
1066:
1063:
1058:
1048:
1046:
1041:
1037:
1027:
1025:
1019:
1010:
1008:
998:
996:
991:
987:
982:
981:breast cancer
978:
967:
965:
961:
954:Liver disease
951:
949:
945:
941:
934:Renal disease
931:
928:
924:
920:
915:
911:
907:
899:Heart disease
891:
889:
885:
871:
868:
865:
861:
857:
853:
850:
846:
842:
841:
840:
838:
834:
824:
821:
817:
813:
808:
796:
793:
788:
784:
780:
777:
773:
772:
770:
767:
763:
762:
761:
758:
756:
746:
743:
741:
737:
733:
728:
726:
722:
718:
713:
709:
707:
703:
700:to the human
699:
694:
691:
687:
682:
680:
676:
667:
664:
660:
656:
652:
643:
639:
635:
633:
629:
625:
610:
607:
593:
589:
587:
583:
579:
575:
571:
567:
563:
553:
550:
547:
541:
539:
522:
519:
518:
514:
510:
507:
506:
501:
498:
497:
493:
489:
485:
481:
477:
474:
473:
469:
465:
462:
461:
458:
455:
453:
450:
449:
446:
439:
430:
428:
424:
420:
416:
412:
401:
398:
393:
389:
379:
376:
372:
362:
360:
356:
346:
344:
339:
329:
327:
323:
319:
315:
311:
295:
293:
289:
285:
281:
277:
273:
258:
255:
245:
242:
241:Alu sequences
232:
226:
224:
220:
217:
214:
211:
210:
209:
196:
194:
191:
186:
173:
166:
165:intron lariat
162:
159:
155:
152:
148:
144:
143:
142:
140:
136:
132:
131:messenger RNA
128:
124:
120:
110:
108:
104:
100:
95:
93:
89:
85:
80:
78:
74:
70:
66:
62:
58:
50:
49:exon skipping
44:
38:
30:
26:
25:mRNA splicing
21:
3512:
3508:
3498:
3453:
3449:
3381:
3377:
3367:
3324:
3320:
3260:
3256:
3198:
3194:
3184:
3139:
3135:
3125:
3082:
3078:
3068:
3031:
3027:
3017:
2990:
2986:
2938:
2934:
2888:
2884:
2878:
2853:
2843:
2818:
2814:
2808:
2781:
2777:
2767:
2742:
2738:
2732:
2705:
2701:
2653:
2649:
2612:(1): 27–36.
2609:
2605:
2599:
2564:
2560:
2550:
2515:
2511:
2501:
2482:
2476:
2441:
2437:
2383:
2379:
2321:
2317:
2275:
2271:
2229:
2225:
2179:
2175:
2121:
2115:
2080:
2076:
2022:
2018:
1956:
1952:
1904:
1900:
1850:
1846:
1798:
1794:
1784:
1752:(3): 39–45.
1749:
1745:
1735:
1708:
1705:Cell Reports
1704:
1694:
1657:
1653:
1643:
1600:
1596:
1542:
1538:
1476:
1472:
1414:
1410:
1400:
1359:
1355:
1349:
1338:
1286:
1282:
1272:
1237:
1233:
1154:
1150:
1140:
1098:(1): 60–74.
1095:
1091:
1081:
1060:
1045:Wnt pathways
1033:
1020:
1016:
1004:
973:
957:
937:
902:
883:
881:
837:pathogenesis
830:
809:
805:
786:
782:
775:
759:
752:
744:
729:
714:
710:
705:
695:
683:
673:
648:
636:
627:
621:
605:
603:
574:nuclear pore
559:
551:
542:
534:
513:exonucleases
491:
483:
456:
451:
444:
407:
385:
368:
354:
352:
335:
326:poly-A tails
306:
269:
251:
238:
230:
222:
207:
189:
183:
164:
158:nucleophilic
116:
113:RNA splicing
96:
81:
64:
61:circular RNA
60:
54:
3263:(8): 1945.
3142:(5): 1240.
1953:Development
966:mechanism.
960:homeostasis
764:Binding to
755:hippocampus
702:CDR1 (gene)
661:genes, two
651:orthologous
457:Description
413:status and
254:RNA editing
239:Repetitive
147:spliceosome
139:spliceosome
103:degradation
99:exonuclease
3456:(8): 885.
2567:(1): 427.
2182:: 617281.
1660:(2): R28.
1545:(7): 409.
1074:References
1036:myogenesis
964:epigenetic
919:myocardium
721:morpholino
663:paralogous
586:cell cycle
355:C. elegans
338:fibroblast
310:leukocytes
223:C. elegans
190:Drosophila
123:eukaryotic
101:-mediated
73:covalently
29:biogenesis
3472:2073-4409
3406:0027-8424
3341:1582-1838
3279:2073-4409
3215:1582-1838
3158:2073-4409
2870:253371318
2416:205233122
1907:(1): 32.
1392:208529258
1132:252890235
977:caspase-1
940:carcinoma
923:biomarker
864:transgene
816:cytoplasm
717:zebrafish
698:antisense
686:Argonaute
675:microRNAs
562:cytoplasm
373:Ribozero
3562:Category
3490:31412632
3424:37155869
3415:10193964
3359:33277969
3297:34440714
3233:32333642
3176:32429565
3117:37672576
3108:10482341
3060:21151960
3009:24035497
2965:23463100
2759:10807004
2724:26377962
2680:20855538
2626:19012978
2591:33432020
2542:22140119
2468:26657629
2408:23446346
2300:14787080
2254:41905828
2208:33614648
2148:30259356
2107:31591557
2047:23446348
1985:27246710
1931:35737129
1877:18390400
1869:25242744
1825:25281217
1776:26442181
1727:25558066
1686:24485196
1635:23620042
1571:25070500
1513:22319583
1473:PLOS ONE
1441:11606537
1411:Genetics
1384:11935017
1315:31998941
1264:23249747
1181:28344080
1124:36380816
1069:See also
1040:myoblast
1024:plasmids
995:exosomes
776:in vitro
725:plasmids
484:in vivo,
119:bacteria
88:proteins
3549:1069269
3517:Bibcode
3481:6721685
3386:Bibcode
3350:8918416
3288:8392363
3224:7299708
3167:7290921
3087:Bibcode
3051:2996334
2956:3616293
2913:7536344
2893:Bibcode
2885:Science
2854:bioRxiv
2835:8566785
2800:7684656
2671:2957044
2582:7801505
2533:3326292
2459:4756822
2388:Bibcode
2354:4348876
2346:6186917
2326:Bibcode
2292:6297745
2246:6162571
2199:7894079
2098:6858549
2055:4416605
2027:Bibcode
1976:4920157
1922:9226085
1816:4201285
1767:4588227
1677:4053975
1626:4063205
1605:Bibcode
1597:Science
1562:4165365
1504:3270023
1481:Bibcode
1432:1461822
1364:Bibcode
1356:Science
1306:7820840
1255:3543092
1172:5387669
1115:9637558
1062:Viroids
858:and in
856:cancers
812:nucleus
787:in vivo
736:miR-138
632:Archaea
582:mitosis
570:nucleus
538:protein
492:in vivo
388:RNA-seq
375:RNA-seq
322:RNase R
316:and H9
127:introns
65:circRNA
3547:
3540:431239
3537:
3488:
3478:
3470:
3422:
3412:
3404:
3357:
3347:
3339:
3295:
3285:
3277:
3231:
3221:
3213:
3174:
3164:
3156:
3115:
3105:
3058:
3048:
3007:
2963:
2953:
2911:
2868:
2833:
2798:
2757:
2722:
2678:
2668:
2624:
2589:
2579:
2540:
2530:
2489:
2466:
2456:
2414:
2406:
2380:Nature
2352:
2344:
2318:Nature
2298:
2290:
2252:
2244:
2206:
2196:
2146:
2136:
2105:
2095:
2053:
2045:
2019:Nature
1983:
1973:
1929:
1919:
1875:
1867:
1823:
1813:
1774:
1764:
1725:
1684:
1674:
1633:
1623:
1569:
1559:
1511:
1501:
1439:
1429:
1390:
1382:
1313:
1303:
1262:
1252:
1179:
1169:
1130:
1122:
1112:
1057:Viroid
970:Cancer
946:, and
820:Pol II
783:et al.
628:et al.
606:et al.
392:RefSeq
371:ENCODE
359:CDR1as
151:intron
107:cancer
43:lariat
37:Intron
3450:Cells
3257:Cells
3136:Cells
2866:S2CID
2412:S2CID
2350:S2CID
2296:S2CID
2250:S2CID
2051:S2CID
1873:S2CID
1388:S2CID
1128:S2CID
906:HIF1α
884:et al
882:Dube
781:Jeck
659:HIPK3
655:HIPK2
566:locus
427:PSEN1
284:ETS-1
280:ANRIL
193:DSCAM
135:exons
35:B.2)
3545:PMID
3486:PMID
3468:ISSN
3420:PMID
3402:ISSN
3355:PMID
3337:ISSN
3293:PMID
3275:ISSN
3229:PMID
3211:ISSN
3172:PMID
3154:ISSN
3113:PMID
3056:PMID
3005:PMID
2961:PMID
2909:PMID
2831:PMID
2815:Gene
2796:PMID
2778:Cell
2755:PMID
2743:1489
2720:PMID
2676:PMID
2622:PMID
2587:PMID
2538:PMID
2487:ISBN
2464:PMID
2404:PMID
2342:PMID
2288:PMID
2272:Cell
2242:PMID
2226:Cell
2204:PMID
2144:PMID
2134:ISBN
2103:PMID
2043:PMID
1981:PMID
1927:PMID
1865:PMID
1847:Cell
1821:PMID
1772:PMID
1723:PMID
1682:PMID
1631:PMID
1567:PMID
1509:PMID
1437:PMID
1380:PMID
1311:PMID
1260:PMID
1177:PMID
1120:PMID
988:and
849:INK4
657:and
425:and
314:HeLa
292:Shbg
288:P450
274:and
145:The
63:(or
3568:RNA
3535:PMC
3525:doi
3476:PMC
3458:doi
3410:PMC
3394:doi
3382:120
3345:PMC
3329:doi
3283:PMC
3265:doi
3219:PMC
3203:doi
3162:PMC
3144:doi
3103:PMC
3095:doi
3046:PMC
3036:doi
2995:doi
2951:PMC
2943:doi
2901:doi
2889:268
2858:doi
2823:doi
2819:167
2786:doi
2747:doi
2710:doi
2706:117
2666:PMC
2658:doi
2650:RNA
2614:doi
2577:PMC
2569:doi
2528:PMC
2520:doi
2454:PMC
2446:doi
2396:doi
2384:495
2334:doi
2322:301
2280:doi
2234:doi
2194:PMC
2184:doi
2126:doi
2093:PMC
2085:doi
2035:doi
2023:495
1971:PMC
1961:doi
1957:143
1917:PMC
1909:doi
1905:117
1855:doi
1851:159
1811:PMC
1803:doi
1762:PMC
1754:doi
1713:doi
1672:PMC
1662:doi
1621:PMC
1613:doi
1601:340
1557:PMC
1547:doi
1499:PMC
1489:doi
1427:PMC
1419:doi
1415:159
1372:doi
1360:296
1301:PMC
1291:doi
1250:PMC
1242:doi
1234:RNA
1167:PMC
1159:doi
1110:PMC
1100:doi
732:SRY
546:Alu
423:APP
397:Alu
276:Sry
272:DCC
69:RNA
55:In
3564::
3543:.
3533:.
3523:.
3513:73
3511:.
3507:.
3484:.
3474:.
3466:.
3452:.
3448:.
3432:^
3418:.
3408:.
3400:.
3392:.
3380:.
3376:.
3353:.
3343:.
3335:.
3325:26
3323:.
3319:.
3305:^
3291:.
3281:.
3273:.
3261:10
3259:.
3255:.
3241:^
3227:.
3217:.
3209:.
3199:24
3197:.
3193:.
3170:.
3160:.
3152:.
3138:.
3134:.
3111:.
3101:.
3093:.
3081:.
3077:.
3054:.
3044:.
3030:.
3026:.
3003:.
2991:51
2989:.
2985:.
2973:^
2959:.
2949:.
2939:32
2937:.
2933:.
2921:^
2907:.
2899:.
2887:.
2864:.
2852:.
2829:.
2817:.
2794:.
2782:73
2780:.
2776:.
2753:.
2741:.
2718:.
2704:.
2700:.
2688:^
2674:.
2664:.
2654:16
2652:.
2648:.
2634:^
2620:.
2610:27
2608:.
2585:.
2575:.
2565:11
2563:.
2559:.
2536:.
2526:.
2516:40
2514:.
2510:.
2462:.
2452:.
2442:44
2440:.
2436:.
2424:^
2410:.
2402:.
2394:.
2382:.
2362:^
2348:.
2340:.
2332:.
2320:.
2308:^
2294:.
2286:.
2276:31
2274:.
2262:^
2248:.
2240:.
2230:23
2228:.
2216:^
2202:.
2192:.
2178:.
2174:.
2156:^
2142:.
2132:.
2101:.
2091:.
2081:22
2079:.
2075:.
2063:^
2049:.
2041:.
2033:.
2021:.
1993:^
1979:.
1969:.
1955:.
1951:.
1939:^
1925:.
1915:.
1903:.
1899:.
1885:^
1871:.
1863:.
1849:.
1845:.
1833:^
1819:.
1809:.
1799:28
1797:.
1793:.
1770:.
1760:.
1748:.
1744:.
1721:.
1709:10
1707:.
1703:.
1680:.
1670:.
1658:15
1656:.
1652:.
1629:.
1619:.
1611:.
1599:.
1595:.
1579:^
1565:.
1555:.
1543:15
1541:.
1537:.
1521:^
1507:.
1497:.
1487:.
1475:.
1471:.
1449:^
1435:.
1425:.
1413:.
1409:.
1386:.
1378:.
1370:.
1358:.
1323:^
1309:.
1299:.
1287:22
1285:.
1281:.
1258:.
1248:.
1238:19
1236:.
1232:.
1196:^
1175:.
1165:.
1155:66
1153:.
1149:.
1126:.
1118:.
1108:.
1094:.
1090:.
470:.
345:.
121:,
109:.
59:,
51:).
45:;
3551:.
3527::
3519::
3492:.
3460::
3454:8
3426:.
3396::
3388::
3361:.
3331::
3299:.
3267::
3235:.
3205::
3178:.
3146::
3140:9
3119:.
3097::
3089::
3083:9
3062:.
3038::
3032:6
3011:.
2997::
2967:.
2945::
2915:.
2903::
2895::
2872:.
2860::
2837:.
2825::
2802:.
2788::
2761:.
2749::
2726:.
2712::
2682:.
2660::
2628:.
2616::
2593:.
2571::
2544:.
2522::
2495:.
2470:.
2448::
2418:.
2398::
2390::
2356:.
2336::
2328::
2302:.
2282::
2256:.
2236::
2210:.
2186::
2180:9
2150:.
2128::
2109:.
2087::
2057:.
2037::
2029::
1987:.
1963::
1933:.
1911::
1879:.
1857::
1827:.
1805::
1778:.
1756::
1750:5
1729:.
1715::
1688:.
1664::
1637:.
1615::
1607::
1573:.
1549::
1515:.
1491::
1483::
1477:7
1443:.
1421::
1394:.
1374::
1366::
1317:.
1293::
1266:.
1244::
1183:.
1161::
1134:.
1102::
1096:8
789:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.