195:
217:
206:
126:
79:
53:. Other confounding factors such as differences in evolutionary rates at different sites or among different species also generally do not affect the interpretation of a CSI. By determining the presence or absence of CSIs in an out-group species, one can infer whether the ancestral form of the CSI was an insert or deletion and this can be used to develop a rooted phylogenetic relationship among organisms.
436:-based phylogenetic trees. No molecular characteristics unique to the class or its different subgroups are known. A detailed CSI-based study was conducted to better understand the phylogeny of this class. Firstly, a phylogenetic tree based on concatenated sequences of a number of universally-distributed proteins was created. The branching order of the different
326:(formerly Thaumarchaeota). However there are very few molecular markers that can distinguish this group of archaea from the phylum Thermoproteota (formerly Crenarchaeota). A detailed phylogenetic study using the CSI approach was conducted to distinguish these phyla in molecular terms. 6 CSIs were uniquely found in various Nitrososphaerota, namely
26:) in protein sequences provide an important category of molecular markers for understanding phylogenetic relationships. CSIs, brought about by rare genetic changes, provide useful phylogenetic markers that are generally of defined size and they are flanked on both sides by conserved regions to ensure their reliability. While
94:(e.g. genus, family, class, order, phylum) but they are not present in other groups. These CSIs were most likely introduced in an ancestor of the group of species before the members of the taxa diverged. They provide molecular means for distinguishing members of a particular taxon from all other organisms.
60:
constructed from protein sequences. Most CSIs that have been identified have been found to have high predictive value upon addition of new sequences, retaining the specificity for the originally identified clades of species. They can be used to identify both known and even previously unknown species
232:
or other genes/proteins. These trees are not always able to resolve key phylogenetic questions with a high degree of certainty. However in recent years the discovery and analyses of conserved indels (CSIs) in many universally distributed proteins have aided in this quest. The genetic events leading
142:
shows an example of 5aa CSI found in a conserved region that is commonly present in the species belonging to phyla X, Y and Z, but it is absent in other phyla (A, B and C). This signature indicates a specific relationship of taxa X, Y and Z and also A, B and C. Based upon the presence or absence of
367:
are currently distinguished mainly based on their position in the branching of the 16srRNA tree. There are currently very few molecular markers known that can distinguish members of this order from other bacteria. A CSI approach was recently used to elucidate the phylogenetic relationships between
100:
shows an example of 5aa CSI found in all species belonging to the taxon X. This is a distinctive characteristic of this taxon as it is not found in any other species. This signature was likely introduced in a common ancestor of the species from this taxon. Similarly other group-specific signatures
803:
Gupta, Radhey S.; Kanter-Eivin, David A. (9 May 2023). "AppIndels.com server: a web-based tool for the identification of known taxon-specific conserved signature indels in genome sequences. Validation of its usefulness by predicting the taxonomic affiliation of >700 unclassified strains of
249:
were known that could clearly distinguish the species of this phylum from all other bacteria. More than 60 CSIs that were specific for the entire
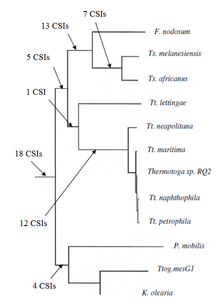
Thermotogota phylum or its different subgroups were discovered. Of these, 18 CSIs are uniquely present in various Thermotogota species and provide
519:
was commonly present in the above orders of the class
Gammaproteobacteria and in some members of the order Oceanospirillales. Another CSI-based study has also identified 4 CSIs that are exclusive to the order Xanthomonadales. Taken together, these two facts show that Xanthomonadales is a
250:
molecular markers for the phylum. Additionally there were many CSIs that were specific for various
Thermotogota subgroups. Another 12 CSIs were specific for a clade consisting of various Thermotogota species except Tt. Lettingae. While 14 CSIs were specific for a clade consisting of the
305:
between these groups. However the number of CSIs that are commonly shared with other taxa is much smaller than those that are specific for
Thermotogota and they do not exhibit any specific pattern. Hence they have no significant effect on the distinction of Thermotogota.
117:, within a highly conserved region (82-124 amino acid). This is not present in any other bacteria species and could be used to characterize members of Thermotogota from all other bacteria. Group-specific CSIs were also used to characterize subgroups within Thermotogota.
338:
and a number of uncultured marine
Thermoproteota. 3 CSIs were found that were commonly shared between species belonging to Nitrososphaerota and Thermoproteota. Additionally, a number of CSIs were found that are specific for different orders of Thermoproteota—3 CSIs for
420:, was supported by 9 CSIs. Based on these results, it was proposed that Pasteurellales be divided from its current one family into two different ones. Additionally, the signatures described would provide novel means of identifying undiscovered Pasteurellales species.
233:
to them are postulated to have occurred at important evolutionary branch points and their species distribution patterns provide valuable information regarding the branching order and interrelationships among different bacterial phyla.
108:
Group-specific CSIs have been used in the past to determine the phylogenetic relationship of a number of bacterial phyla and subgroups within it. For example a 3 amino acid insert was uniquely shared by members of the phylum
514:
were found uniquely in various species belonging to the orders
Enterobacteriales, Pasteurellales, Vibrionales, Aeromonadales and Alteromonadales, but were not found in other gammaproteobacteria. Lastly, a 2 aa deletion in
524:
that is ancestral to other
Gammaproteobacteria, which further shows that Xanthomonadales is an independent subdivision, and constitutes one of the deepest-branching lineages within the Gammaproteobacteria clade.
355:. The signatures described provide novel means for distinguishing Thermoproteota and Nitrososphaerota, additionally they could be used as a tool for the classification and identification of related species.
227:
A key issue in bacterial phylogeny is to understand how different bacterial species are related to each other and their branching order from a common ancestor. Currently most phylogenetic trees are based on
85:
Example of a group-specific
Conserved signature indel (CSIs), that is specific for species from taxon X. The dashes in the alignments indicate the presence of an amino acid identical to that on the top
1339:
Naushad, Hafiz Sohail; Gupta, Radhey S. (2011). "Molecular signatures (conserved indels) in protein sequences that are specific for the order
Pasteurellales and distinguish two of its main clades".
101:(not shown) could be shared by either A1 and A2 or B1 and B2, etc., or even by X1 and X2 or by X3 and X4, etc. The groups A, B, C, D and X, in this diagram could correspond to various bacterial or
368:
the species in this order; more than 40 CSIs were discovered that were uniquely shared by all or most of the species. Two major clades are formed within this
Pasteurellales: Clade I, encompassing
146:
Mainline CSIs have been used in the past to determine the phylogenetic relationship of a number of bacterial phyla. The large CSI of about 150-180 amino acids within a conserved region of
186:, Planctomycetota, and Aquificota. This CSI is absent in other ancestral bacterial phyla as well as Archaea. In both cases one can infer that the groups lacking the CSI are ancestral.
1045:
Brown, James R.; Douady, Christophe J.; Italia, Michael J.; Marshall, William E.; Stanhope, Michael J. (2001). "Universal trees based on large combined protein sequence data sets".
143:
such an indel, in out-group species (viz. Archaea), it can be inferred whether the indel is an insert or a deletion, and which of these two groups A, B, C or X, Y, Z is ancestral.
41:
markers of common evolutionary descent. Due to the rarity and highly specific nature of such changes, it is less likely that they could arise independently by either
497:
223:
A concatenated protein tree showing the phylogenetic relationship of the group Pasteurellales. The number of CSIs that support the branching order are indicated .
201:
A concatenated protein tree showing the phylogenetic relationship of the group Thermotogota. The number of CSIs that support the branching order are indicated .
212:
A concatenated protein tree showing the phylogenetic relationship of two phyla of Archaea. The number of CSIs that support the branching order are indicated .
861:
61:
belonging to these groups in different environments. Compared to tree branching orders which can vary among methods, specific CSIs make for more concrete
1129:
Ciccarelli, F. D.; Doerks, T; Von Mering, C; Creevey, CJ; Snel, B; Bork, P (2006). "Toward Automatic Reconstruction of a Highly Resolved Tree of Life".
1241:"The RecA Protein as a Model Molecule for Molecular Systematic Studies of Bacteria: Comparison of Trees of RecAs and 16S rRNAs from the Same Species"
925:"Phylogeny and shared conserved inserts in proteins provide evidence that Verrucomicrobia are the closest known free-living relatives of chlamydiae"
724:
Cutiño-Jiménez, Ania M.; Martins-Pinheiro, Marinalva; Lima, Wanessa C.; Martín-Tornet, Alexander; Morales, Osleidys G.; Menck, Carlos F.M. (2010).
132:
Multi group or Mainline Conserved signature indel (CSI). The dashes in indicate the presence of an amino acid identical to that on the top line.
473:
30:
can be arbitrary inserts or deletions, CSIs are defined as only those protein indels that are present within conserved regions of the protein.
634:"Protein Phylogenies and Signature Sequences: A Reappraisal of Evolutionary Relationships among Archaebacteria, Eubacteria, and Eukaryotes"
835:
265:
Lastly 16 CSIs were reported that were shared by either some or all Thermotogota species or some species from other taxa such as
880:
Gupta, Radhey S.; Bhandari, Vaibhav (2011). "Phylogeny and molecular signatures for the phylum Thermotogae and its subgroups".
136:
Mainline CSIs are those in which a conserved insert or deletion is shared by several major phyla, but absent from other phyla.
496:. Additionally, 4 CSIs were discovered that were unique to most species of the class Gammaproteobacteria. A 2 aa deletion in
1090:"The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification"
1387:"Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria"
1296:
Gupta, Radhey S.; Shami, Ali (2010). "Molecular signatures for the Crenarchaeota and the Thaumarchaeota".
432:
forms one of the largest groups of bacteria. It is currently distinguished from other bacteria solely by
1089:
1431:
62:
1192:"A Phylogenomic Approach to Bacterial Phylogeny: Evidence of a Core of Genes Sharing a Common History"
1151:
334:
302:
174:
homologs (between amino acids 919-1058) is present in various species belonging to Pseudomonadota,
560:"Animals and Fungi are Each Other's Closest Relatives: Congruent Evidence from Multiple Proteins"
534:
768:
Rokas, Antonis; Holland, Peter W.H. (2000). "Rare genomic changes as a tool for phylogenetics".
1146:
1009:"Signature sequences in diverse proteins provide evidence for the late divergence of the Order
516:
502:
390:
328:
147:
1252:
1138:
1008:
571:
42:
726:"Evolutionary placement of Xanthomonadales based on conserved protein signature sequences"
8:
429:
408:
1256:
1142:
575:
1364:
1321:
1273:
1240:
1172:
1070:
989:
905:
90:
Group-specific CSIs are commonly shared by different species belonging to a particular
46:
1216:
1191:
781:
1408:
1356:
1313:
1278:
1221:
1164:
1111:
1062:
1027:
981:
964:
Gupta, Radhey S. (2003). "Evolutionary relationships among photosynthetic bacteria".
946:
897:
855:
785:
747:
703:
663:
658:
649:
633:
599:
594:
559:
511:
489:
465:
445:
352:
246:
229:
114:
57:
1368:
1325:
993:
909:
686:
Gupta, Radhey S.; Griffiths, Emma (2002). "Critical Issues in Bacterial Phylogeny".
1398:
1348:
1305:
1268:
1260:
1211:
1203:
1176:
1156:
1101:
1074:
1054:
973:
936:
889:
813:
777:
737:
695:
653:
645:
589:
579:
323:
194:
941:
924:
485:
481:
469:
461:
441:
437:
370:
344:
159:
742:
725:
723:
507:
449:
364:
315:
286:
278:
151:
1352:
1309:
977:
893:
1425:
584:
493:
477:
457:
294:
290:
1160:
1106:
1412:
1403:
1386:
1360:
1317:
1225:
1168:
1115:
1066:
1031:
985:
950:
901:
817:
789:
751:
707:
699:
348:
340:
282:
242:
216:
175:
110:
50:
38:
1282:
667:
603:
245:
was characterized based on the CSI approach. Previously no biochemical or
166:
species. This CSI is absent in other ancestral bacterial phyla as well as
453:
376:
183:
179:
155:
125:
1264:
298:
270:
205:
163:
102:
1207:
444:
Gammaproteobacteria (from most recent to the earliest diverging) was:
78:
521:
274:
433:
1058:
150:(between amino acids 529-751), is commonly shared between various
1391:
International Journal of Systematic and Evolutionary Microbiology
1128:
1094:
International Journal of Systematic and Evolutionary Microbiology
806:
International Journal of Systematic and Evolutionary Microbiology
319:
266:
167:
91:
34:
27:
842:. Gupta lab. Archived from the original on 15 September 2011
301:. The shared presence of some of these CSIs could be due to
171:
500:
was uniquely shared by all gammaproteobacteria except for
1044:
56:
CSIs are discovered by looking for shared changes in a
49:(i.e. homoplasy) and therefore are likely to represent
836:"Conserved Inserts and Deletions in Protein Sequences"
189:
241:
Recently the phylogenetic relationship of the group
170:. Similarly a large CSI of about 100 amino acids in
398:, was supported by 13 CSIs. Clade II, encompassing
1189:
802:
1423:
860:: CS1 maint: bot: original URL status unknown (
719:
717:
1380:
1378:
564:Proceedings of the National Academy of Sciences
258:genera and 18 CSIs were specific for the genus
1087:
1006:
922:
685:
553:
551:
549:
1384:
1000:
879:
714:
33:The CSIs that are restricted to a particular
1375:
1338:
767:
681:
679:
677:
627:
625:
623:
621:
619:
617:
615:
613:
37:or group of species, generally provide good
829:
827:
546:
65:that are computationally cheaper to apply.
1332:
1295:
1007:Griffiths, Emma; Gupta, Radhey S. (2004).
638:Microbiology and Molecular Biology Reviews
318:were recently placed into a new phylum of
120:
1402:
1385:Gao, B.; Mohan, R.; Gupta, R. S. (2009).
1289:
1272:
1215:
1190:Daubin, V.; Gouy, M; Perrière, G (2002).
1150:
1105:
957:
940:
875:
873:
871:
741:
674:
657:
610:
593:
583:
20:Conserved signature inserts and deletions
824:
215:
204:
193:
124:
113:(formerly Thermotogae) in the essential
77:
763:
761:
557:
1424:
1232:
1183:
1122:
1081:
1038:
868:
423:
1238:
963:
730:Molecular Phylogenetics and Evolution
631:
923:Griffiths, E.; Gupta, R. S. (2007).
916:
758:
13:
190:Evolutionary studies based on CSIs
14:
1443:
833:
770:Trends in Ecology & Evolution
358:
73:
650:10.1128/MMBR.62.4.1435-1491.1998
400:Actinobacillus pleuropneumoniae
236:
16:Shared protein sequence feature
1245:Journal of Molecular Evolution
796:
688:Theoretical Population Biology
1:
782:10.1016/S0169-5347(00)01967-4
540:
386:Mannheimia succiniciproducens
115:50S ribosomal protein L7/L12
7:
1239:Eisen, Jonathan A. (1995).
942:10.1099/mic.0.2007/009118-0
743:10.1016/j.ympev.2009.09.026
528:
382:Actinobacillus succinogenes
347:, lastly 2 CSIs common for
303:lateral gene transfer (LGT)
10:
1448:
1088:Cavalier-Smith, T (2002).
1020:International Microbiology
309:
1353:10.1007/s10482-011-9628-4
1310:10.1007/s10482-010-9488-3
894:10.1007/s10482-011-9576-z
632:Gupta, Radhey S. (1998).
363:The members of the order
585:10.1073/pnas.90.24.11558
508:RNA polymerase b-subunit
335:Nitrosopumilus maritimus
68:
1341:Antonie van Leeuwenhoek
1298:Antonie van Leeuwenhoek
1161:10.1126/science.1123061
1107:10.1099/00207713-52-1-7
978:10.1023/A:1024999314839
966:Photosynthesis Research
882:Antonie van Leeuwenhoek
558:Baldauf, S. L. (1993).
535:Molecular phylogenetics
510:and a 1 aa deletion in
121:Multi-group or mainline
1404:10.1099/ijs.0.002741-0
818:10.1099/ijsem.0.005844
700:10.1006/tpbi.2002.1589
517:leucyl-tRNA synthetase
503:Francisella tularensis
414:Mannheimia haemolytica
391:Haemophilus influenzae
224:
213:
202:
133:
87:
512:ribosomal protein L16
506:. A 4 aa deletion in
329:Cenarchaeum symbiosum
219:
208:
197:
128:
81:
498:AICAR transformylase
418:Haemophilus parasuis
404:Actinobacillus minor
1257:1995JMolE..41.1105E
1143:2006Sci...311.1283C
840:Bacterial Phylogeny
804:Bacillus species".
576:1993PNAS...9011558B
570:(24): 11558–11562.
430:Gammaproteobacteria
424:Gammaproteobacteria
409:Haemophilus ducreyi
1265:10.1007/bf00173192
522:monophyletic group
396:Haemophilus somnus
225:
214:
203:
134:
88:
47:parallel evolution
1432:Molecular biology
1208:10.1101/gr.187002
490:Cardiobacteriales
466:Oceanospirillales
446:Enterobacteriales
353:Desulfurococcales
247:molecular markers
58:phylogenetic tree
1439:
1417:
1416:
1406:
1382:
1373:
1372:
1336:
1330:
1329:
1293:
1287:
1286:
1276:
1236:
1230:
1229:
1219:
1187:
1181:
1180:
1154:
1137:(5765): 1283–7.
1126:
1120:
1119:
1109:
1085:
1079:
1078:
1042:
1036:
1035:
1017:
1004:
998:
997:
961:
955:
954:
944:
920:
914:
913:
877:
866:
865:
859:
851:
849:
847:
831:
822:
821:
800:
794:
793:
765:
756:
755:
745:
721:
712:
711:
683:
672:
671:
661:
629:
608:
607:
597:
587:
555:
324:Nitrososphaerota
252:Fervidobacterium
63:circumscriptions
1447:
1446:
1442:
1441:
1440:
1438:
1437:
1436:
1422:
1421:
1420:
1383:
1376:
1337:
1333:
1294:
1290:
1237:
1233:
1196:Genome Research
1188:
1184:
1152:10.1.1.381.9514
1127:
1123:
1086:
1082:
1047:Nature Genetics
1043:
1039:
1015:
1005:
1001:
972:(1–3): 173–83.
962:
958:
921:
917:
878:
869:
853:
852:
845:
843:
834:Gupta, Radhey.
832:
825:
801:
797:
776:(11): 454–459.
766:
759:
722:
715:
684:
675:
630:
611:
556:
547:
543:
531:
486:Xanthomonadales
482:Methylococcales
470:Pseudomonadales
462:Alteromonadales
426:
371:Aggregatibacter
361:
345:Thermoproteales
312:
239:
192:
160:Planctomycetota
123:
76:
71:
17:
12:
11:
5:
1445:
1435:
1434:
1419:
1418:
1374:
1331:
1288:
1251:(6): 1105–23.
1231:
1202:(7): 1080–90.
1182:
1121:
1080:
1037:
999:
956:
935:(8): 2648–54.
915:
867:
823:
795:
757:
713:
673:
644:(4): 1435–91.
609:
544:
542:
539:
538:
537:
530:
527:
450:Pasteurellales
425:
422:
365:Pasteurellales
360:
359:Pasteurellales
357:
316:Thermoproteota
311:
308:
287:Fusobacteriota
279:Pseudomonadota
238:
235:
191:
188:
152:Pseudomonadota
122:
119:
75:
74:Group-specific
72:
70:
67:
15:
9:
6:
4:
3:
2:
1444:
1433:
1430:
1429:
1427:
1414:
1410:
1405:
1400:
1397:(2): 234–47.
1396:
1392:
1388:
1381:
1379:
1370:
1366:
1362:
1358:
1354:
1350:
1347:(1): 105–24.
1346:
1342:
1335:
1327:
1323:
1319:
1315:
1311:
1307:
1304:(2): 133–57.
1303:
1299:
1292:
1284:
1280:
1275:
1270:
1266:
1262:
1258:
1254:
1250:
1246:
1242:
1235:
1227:
1223:
1218:
1213:
1209:
1205:
1201:
1197:
1193:
1186:
1178:
1174:
1170:
1166:
1162:
1158:
1153:
1148:
1144:
1140:
1136:
1132:
1125:
1117:
1113:
1108:
1103:
1099:
1095:
1091:
1084:
1076:
1072:
1068:
1064:
1060:
1059:10.1038/90129
1056:
1052:
1048:
1041:
1033:
1029:
1025:
1021:
1014:
1012:
1003:
995:
991:
987:
983:
979:
975:
971:
967:
960:
952:
948:
943:
938:
934:
930:
926:
919:
911:
907:
903:
899:
895:
891:
887:
883:
876:
874:
872:
863:
857:
841:
837:
830:
828:
819:
815:
811:
807:
799:
791:
787:
783:
779:
775:
771:
764:
762:
753:
749:
744:
739:
736:(2): 524–34.
735:
731:
727:
720:
718:
709:
705:
701:
697:
694:(4): 423–34.
693:
689:
682:
680:
678:
669:
665:
660:
655:
651:
647:
643:
639:
635:
628:
626:
624:
622:
620:
618:
616:
614:
605:
601:
596:
591:
586:
581:
577:
573:
569:
565:
561:
554:
552:
550:
545:
536:
533:
532:
526:
523:
518:
513:
509:
505:
504:
499:
495:
494:Thiotrichales
491:
487:
483:
479:
478:Legionellales
475:
471:
467:
463:
459:
458:Aeromonadales
455:
451:
447:
443:
439:
435:
431:
421:
419:
415:
411:
410:
405:
401:
397:
393:
392:
387:
383:
379:
378:
373:
372:
366:
356:
354:
350:
346:
343:, 5 CSIs for
342:
337:
336:
331:
330:
325:
321:
317:
307:
304:
300:
296:
295:Chloroflexota
292:
291:Dictyoglomota
288:
284:
280:
276:
272:
268:
263:
261:
257:
253:
248:
244:
234:
231:
222:
218:
211:
207:
200:
196:
187:
185:
181:
177:
173:
169:
165:
161:
157:
153:
149:
144:
141:
137:
131:
127:
118:
116:
112:
106:
104:
99:
95:
93:
84:
80:
66:
64:
59:
54:
52:
48:
44:
40:
36:
31:
29:
25:
21:
1394:
1390:
1344:
1340:
1334:
1301:
1297:
1291:
1248:
1244:
1234:
1199:
1195:
1185:
1134:
1130:
1124:
1097:
1093:
1083:
1053:(3): 281–5.
1050:
1046:
1040:
1026:(1): 41–52.
1023:
1019:
1010:
1002:
969:
965:
959:
932:
929:Microbiology
928:
918:
885:
881:
844:. Retrieved
839:
809:
805:
798:
773:
769:
733:
729:
691:
687:
641:
637:
567:
563:
501:
474:Chromatiales
427:
417:
413:
407:
403:
399:
395:
389:
385:
381:
375:
369:
362:
349:Sulfolobales
341:Sulfolobales
333:
327:
314:Mesophillic
313:
283:Deinococcota
264:
260:Thermosiphon
259:
255:
251:
243:Thermotogota
240:
237:Thermotogota
226:
220:
209:
198:
176:Bacteroidota
145:
139:
138:
135:
129:
111:Thermotogota
107:
97:
96:
89:
82:
55:
51:synapomorphy
39:phylogenetic
32:
23:
19:
18:
1100:(1): 7–76.
1011:Aquificales
888:(1): 1–34.
454:Vibrionales
377:Pasteurella
322:called the
256:Thermosipho
184:Chlamydiota
180:Chlorobiota
156:Chlamydiota
541:References
428:The class
299:eukaryotes
271:Aquificota
164:Aquificota
103:Eukaryotic
43:convergent
1147:CiteSeerX
275:Bacillota
221:Figure 5:
210:Figure 4:
199:Figure 3:
130:Figure 2:
83:Figure 1:
1426:Category
1413:19196760
1369:15114511
1361:21830122
1326:12874800
1318:20711675
1226:12097345
1169:16513982
1116:11837318
1067:11431701
1032:15179606
994:38460308
986:16228576
951:17660429
910:24995263
902:21503713
856:cite web
790:11050348
752:19786109
708:12167362
529:See also
434:16s rRNA
230:16S rRNA
148:Gyrase B
140:Figure 2
98:Figure 1
1283:8587109
1274:3188426
1253:Bibcode
1177:1615592
1139:Bibcode
1131:Science
1075:8516570
846:2 April
668:9841678
604:8265589
572:Bibcode
440:of the
320:Archaea
310:Archaea
267:Archaea
168:Archaea
105:phyla.
1411:
1367:
1359:
1324:
1316:
1281:
1271:
1224:
1217:186629
1214:
1175:
1167:
1149:
1114:
1073:
1065:
1030:
992:
984:
949:
908:
900:
788:
750:
706:
666:
656:
602:
592:
438:orders
297:, and
28:indels
1365:S2CID
1322:S2CID
1173:S2CID
1071:S2CID
1016:(PDF)
990:S2CID
906:S2CID
812:(5).
659:98952
595:48023
442:class
92:taxon
86:line.
69:Types
35:clade
1409:PMID
1357:PMID
1314:PMID
1279:PMID
1222:PMID
1165:PMID
1112:PMID
1063:PMID
1028:PMID
982:PMID
947:PMID
898:PMID
862:link
848:2012
786:PMID
748:PMID
704:PMID
664:PMID
600:PMID
472:>
464:>
460:>
452:>
448:>
416:and
394:and
351:and
254:and
172:RpoB
162:and
24:CSIs
1399:doi
1349:doi
1345:101
1306:doi
1269:PMC
1261:doi
1212:PMC
1204:doi
1157:doi
1135:311
1102:doi
1055:doi
974:doi
937:doi
933:153
890:doi
886:100
814:doi
778:doi
738:doi
696:doi
654:PMC
646:doi
590:PMC
580:doi
45:or
1428::
1407:.
1395:59
1393:.
1389:.
1377:^
1363:.
1355:.
1343:.
1320:.
1312:.
1302:99
1300:.
1277:.
1267:.
1259:.
1249:41
1247:.
1243:.
1220:.
1210:.
1200:12
1198:.
1194:.
1171:.
1163:.
1155:.
1145:.
1133:.
1110:.
1098:52
1096:.
1092:.
1069:.
1061:.
1051:28
1049:.
1022:.
1018:.
988:.
980:.
970:76
968:.
945:.
931:.
927:.
904:.
896:.
884:.
870:^
858:}}
854:{{
838:.
826:^
810:73
808:.
784:.
774:15
772:.
760:^
746:.
734:54
732:.
728:.
716:^
702:.
692:61
690:.
676:^
662:.
652:.
642:62
640:.
636:.
612:^
598:.
588:.
578:.
568:90
566:.
562:.
548:^
492:,
488:,
484:,
480:,
476:,
468:,
456:,
412:,
406:,
402:,
388:,
384:,
380:,
374:,
332:,
293:,
289:,
285:,
281:,
277:,
273:,
269:,
262:.
182:,
178:,
158:,
154:,
1415:.
1401::
1371:.
1351::
1328:.
1308::
1285:.
1263::
1255::
1228:.
1206::
1179:.
1159::
1141::
1118:.
1104::
1077:.
1057::
1034:.
1024:7
1013:"
996:.
976::
953:.
939::
912:.
892::
864:)
850:.
820:.
816::
792:.
780::
754:.
740::
710:.
698::
670:.
648::
606:.
582::
574::
22:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.