414:
998:
such systems are a "molecular tweezers" design that has an open and a closed state, a device that could switch from a paranemic-crossover (PX) conformation to a (JX2) conformation with two non-junction juxtapositions of the DNA backbone, undergoing rotational motion in the process, and a two-dimensional array that could dynamically expand and contract in response to control strands. Structures have also been made that dynamically open or close, potentially acting as a molecular cage to release or reveal a functional cargo upon opening. In another example, a DNA origami nanostructure was coupled to T7 RNA polymerase and could thus be operated as a chemical energy-driven motor that can be coupled to a passive follower, which it then drives.
1480:. In both, the sequence of monomers is designed to favor the desired target structure and to disfavor other structures. Nucleic acid design has the advantage of being much computationally easier than protein design, because the simple base pairing rules are sufficient to predict a structure's energetic favorability, and detailed information about the overall three-dimensional folding of the structure is not required. This allows the use of simple heuristic methods that yield experimentally robust designs. Nucleic acid structures are less versatile than proteins in their function because of proteins' increased ability to fold into complex structures, and the limited chemical diversity of the four
733:
488:, in which molecular components spontaneously organize into stable structures; the particular form of these structures is induced by the physical and chemical properties of the components selected by the designers. In DNA nanotechnology, the component materials are strands of nucleic acids such as DNA; these strands are often synthetic and are almost always used outside the context of a living cell. DNA is well-suited to nanoscale construction because the binding between two nucleic acid strands depends on simple
653:
38:
551:, nucleic acid strands are expected in most cases to bind to each other in the conformation that maximizes the number of correctly paired bases. The sequences of bases in a system of strands thus determine the pattern of binding and the overall structure in an easily controllable way. In DNA nanotechnology, the base sequences of strands are rationally designed by researchers so that the base pairing interactions cause the strands to assemble in the desired conformation. While
7091:
1596:
71:
724:
949:
127:
7066:
882:, a coarse map of the Western Hemisphere, and the Mona Lisa painting. Solid three-dimensional structures can be made by using parallel DNA helices arranged in a honeycomb pattern, and structures with two-dimensional faces can be made to fold into a hollow overall three-dimensional shape, akin to a cardboard box. These can be programmed to open and reveal or release a molecular cargo in response to a stimulus, making them potentially useful as programmable
450:
704:
291:, but Seeman's insight was that immobile nucleic acid junctions could be created by properly designing the strand sequences to remove symmetry in the assembled molecule, and that these immobile junctions could in principle be combined into rigid crystalline lattices. The first theoretical paper proposing this scheme was published in 1982, and the first experimental demonstration of an immobile DNA junction was published the following year.
7103:
1610:
83:
814:, and while they lack the electrical conductance of carbon nanotubes, DNA nanotubes are more easily modified and connected to other structures. One of many schemes for constructing DNA nanotubes uses a lattice of curved DX tiles that curls around itself and closes into a tube. In an alternative method that allows the circumference to be specified in a simple, modular fashion using single-stranded tiles, the rigidity of the tube is an
1497:
908:. This allows the construction of materials and devices with a range of functionalities much greater than is possible with nucleic acids alone. The goal is to use the self-assembly of the nucleic acid structures to template the assembly of the nanoparticles hosted on them, controlling their position and in some cases orientation. Many of these schemes use a covalent attachment scheme, using oligonucleotides with
662:
7078:
596:, which contains two parallel double helical domains with individual strands crossing between the domains at two crossover points. Each crossover point is, topologically, a four-arm junction, but is constrained to one orientation, in contrast to the flexible single four-arm junction, providing a rigidity that makes the DX motif suitable as a structural building block for larger DNA complexes.
1181:(FRET). The constructed box was shown to have a unique reclosing mechanism, which enabled it to repeatedly open and close in response to a unique set of DNA or RNA keys. The authors proposed that this "DNA device can potentially be used for a broad range of applications such as controlling the function of single molecules, controlled drug delivery, and molecular computing."
374:'s DNA octahedron, which consisted mostly of one very long strand. Rothemund's DNA origami contains a long strand which folding is assisted by several short strands. This method allowed forming much larger structures than formerly possible, and which are less technically demanding to design and synthesize. DNA origami was the cover story of
1252:, but there are limits of safety and imprecise targeting, in addition to short shelf life in the blood stream. The DNA nanostructure created by the team consists of six strands of DNA to form a tetrahedron, with one strand of RNA affixed to each of the six edges. The tetrahedron is further equipped with targeting protein, three
1448:
After any of the above approaches are used to design the secondary structure of a target complex, an actual sequence of nucleotides that will form into the desired structure must be devised. Nucleic acid design is the process of assigning a specific nucleic acid base sequence to each of a structure's
1376:
The first step in designing a nucleic acid nanostructure is to decide how a given structure should be represented by a specific arrangement of nucleic acid strands. This design step determines the secondary structure, or the positions of the base pairs that hold the individual strands together in the
1276:
drug efflux pump. The results of the experiment showed the DOX was not being pumped out and apoptosis of the cancer cells was achieved. The tetrahedron without DOX was loaded into cells to test its biocompatibility, and the structure showed no cytotoxicity itself. The DNA tetrahedron was also used as
1004:
are a class of nucleic acid nanomachines that exhibit directional motion along a linear track. A large number of schemes have been demonstrated. One strategy is to control the motion of the walker along the track using control strands that need to be manually added in sequence. It is also possible to
636:
has a robust, defined three-dimensional geometry that makes it possible to simulate, predict and design the structures of more complicated nucleic acid complexes. Many such structures have been created, including two- and three-dimensional structures, and periodic, aperiodic, and discrete structures.
401:
after one reviewer praised its originality while another criticized it for its lack of biological relevance. By the early 2010s the field was considered to have increased its abilities to the point that applications for basic science research were beginning to be realized, and practical applications
1389:
Folding structures. An alternative to the tile-based approach, folding approaches make the nanostructure from one long strand, which can either have a designed sequence that folds due to its interactions with itself, or it can be folded into the desired shape by using shorter, "staple" strands. This
997:
conditions by undergoing a twisting motion. This reliance on buffer conditions caused all devices to change state at the same time. Subsequent systems could change states based upon the presence of control strands, allowing multiple devices to be independently operated in solution. Some examples of
1061:
as inputs and outputs, molecular computers use the concentrations of specific chemical species as signals. In the case of nucleic acid strand displacement circuits, the signal is the presence of nucleic acid strands that are released or consumed by binding and unbinding events to other strands in
1040:
where the newly revealed output sequence of one reaction can initiate another strand displacement reaction elsewhere. This in turn allows for the construction of chemical reaction networks with many components, exhibiting complex computational and information processing abilities. These cascades
861:
polyhedra. Subsequent work yielded polyhedra whose synthesis was much easier. These include a DNA octahedron made from a long single strand designed to fold into the correct conformation, and a tetrahedron that can be produced from four DNA strands in one step, pictured at the top of this article.
576:
end state. On the other hand, dynamic DNA nanotechnology focuses on complexes with useful non-equilibrium behavior such as the ability to reconfigure based on a chemical or physical stimulus. Some complexes, such as nucleic acid nanomechanical devices, combine features of both the structural and
1045:
gain from disassembly reactions. Strand displacement cascades allow isothermal operation of the assembly or computational process, in contrast to traditional nucleic acid assembly's requirement for a thermal annealing step, where the temperature is raised and then slowly lowered to ensure proper
571:
DNA nanotechnology is sometimes divided into two overlapping subfields: structural DNA nanotechnology and dynamic DNA nanotechnology. Structural DNA nanotechnology, sometimes abbreviated as SDN, focuses on synthesizing and characterizing nucleic acid complexes and materials that assemble into a
1381:
Tile-based structures. This approach breaks the target structure into smaller units with strong binding between the strands contained in each unit, and weaker interactions between the units. It is often used to make periodic lattices, but can also be used to implement algorithmic self-assembly,
1145:
DNA nanotechnology is moving toward potential real-world applications. The ability of nucleic acid arrays to arrange other molecules indicates its potential applications in molecular scale electronics. The assembly of a nucleic acid structure could be used to template the assembly of molecular
1077:
structure for the reactants, so that when the input strand binds, the newly revealed sequence is on the same molecule rather than disassembling. This allows new opened hairpins to be added to a growing complex. This approach has been used to make simple structures such as three- and four-arm
966:
Dynamic DNA nanotechnology focuses on forming nucleic acid systems with designed dynamic functionalities related to their overall structures, such as computation and mechanical motion. There is some overlap between structural and dynamic DNA nanotechnology, as structures can be formed through
767:
pattern of the individual molecular tiles. The earliest example of this used double-crossover (DX) complexes as the basic tiles, each containing four sticky ends designed with sequences that caused the DX units to combine into periodic two-dimensional flat sheets that are essentially rigid
1013:
to cleave the strands and cause the walker to move forward, which has the advantage of running autonomously. A later system could walk upon a two-dimensional surface rather than a linear track, and demonstrated the ability to selectively pick up and move molecular cargo. In 2018, a
1035:
Cascades of strand displacement reactions can be used for either computational or structural purposes. An individual strand displacement reaction involves revealing a new sequence in response to the presence of some initiator strand. Many such reactions can be linked into a
985:. These structures are initially formed in the same way as the static structures made in structural DNA nanotechnology, but are designed so that dynamic reconfiguration is possible after the initial assembly. The earliest such device made use of the transition between the
295:
402:
in medicine and other fields were beginning to be considered feasible. The field had grown from very few active laboratories in 2001 to at least 60 in 2010, which increased the talent pool and thus the number of scientific advances in the field during that decade.
362:. The idea of using DNA arrays to template the assembly of other molecules such as nanoparticles and proteins, first suggested by Bruche Robinson and Seeman in 1987, was demonstrated in 2002 by Seeman, Kiehl et al. and subsequently by many other groups.
413:
957:
process across region 2, the blue strand is displaced and freed from the complex. Reactions like these are used to dynamically reconfigure or assemble nucleic acid nanostructures. In addition, the red and blue strands can be used as signals in a
1264:, dropped by more than half. This study shows promise in using DNA nanotechnology as an effective tool to deliver treatment using the emerging RNA Interference technology. The DNA tetrahedron was also used in an effort to overcome the phenomena
869:
method. These structures consist of a long, natural virus strand as a "scaffold", which is made to fold into the desired shape by computationally designed short "staple" strands. This method has the advantages of being easy to design, as the
936:. In addition, there are nucleic acid metallization methods, in which the nucleic acid is replaced by a metal which assumes the general shape of the original nucleic acid structure, and schemes for using nucleic acid nanostructures as
346:—a motif that changes its structure in response to an input—was demonstrated in 1999 by Seeman. An improved system, which was the first nucleic acid device to make use of toehold-mediated strand displacement, was demonstrated by
778:
Two-dimensional arrays can be made to exhibit aperiodic structures whose assembly implements a specific algorithm, exhibiting one form of DNA computing. The DX tiles can have their sticky end sequences chosen so that they act as
369:
method for easily and robustly forming folded DNA structures of arbitrary shape. Rothemund had conceived of this method as being conceptually intermediate between Seeman's DX lattices, which used many short strands, and
952:
Dynamic DNA nanotechnology often makes use of toehold-mediated strand displacement reactions. In this example, the red strand binds to the single stranded toehold region on the green strand (region 1), and then in a
334:
in their 2004 paper on the algorithmic self-assembly of a
Sierpinski gasket structure, and for which they shared the 2006 Feynman Prize in Nanotechnology. Winfree's key insight was that the DX tiles could be used as
1277:
barcode for profiling the subcellular expression and distribution of proteins in cells for diagnostic purposes. The tetrahedral-nanostructured showed enhanced signal due to higher labeling efficiency and stability.
106:
330:, published the creation of two-dimensional lattices of DX tiles. These tile-based structures had the advantage that they provided the ability to implement DNA computing, which was demonstrated by Winfree and
1106:, where molecules that are difficult to crystallize in isolation could be arranged within a three-dimensional nucleic acid lattice, allowing determination of their structure. Another application is the use of
1572:, which is well suited to extended two-dimensional structures, but less useful for discrete three-dimensional structures because of the microscope tip's interaction with the fragile nucleic acid structure;
1026:
as the walker advances along the track, allowing autonomous multistep chemical synthesis directed by the walker. The synthetic DNA walkers' function is similar to that of the proteins dynein and kinesin.
611:
process. The overall effect is that one of the strands in the complex is replaced with another one. In addition, reconfigurable structures and devices can be made using functional nucleic acids such as
632:
Structural DNA nanotechnology, sometimes abbreviated as SDN, focuses on synthesizing and characterizing nucleic acid complexes and materials where the assembly has a static, equilibrium endpoint. The
339:, meaning that their assembly could perform computation. The synthesis of a three-dimensional lattice was finally published by Seeman in 2009, nearly thirty years after he had set out to achieve it.
928:
polyamides on a DX array was used to arrange streptavidin proteins in a specific pattern on a DX array. Carbon nanotubes have been hosted on DNA arrays in a pattern allowing the assembly to act as a
1364:
of the target complex is determined, specifying the arrangement of nucleic acid strands within the structure, and which portions of those strands should be bound to each other. The last step is the
391:
DNA nanotechnology was initially met with some skepticism due to the unusual non-biological use of nucleic acids as materials for building structures and doing computation, and the preponderance of
1623:
821:
Forming three-dimensional lattices of DNA was the earliest goal of DNA nanotechnology, but this proved to be one of the most difficult to realize. Success using a motif based on the concept of
1507:
on a DX complex, are used to ascertain whether the desired structures are forming properly. Each vertical lane contains a series of bands, where each band is characteristic of a particular
1296:
modifications and induce ionic currents across the membrane. This first demonstration of a synthetic DNA ion channel was followed by a variety of pore-inducing designs ranging from a single
580:
The complexes constructed in structural DNA nanotechnology use topologically branched nucleic acid structures containing junctions. (In contrast, most biological DNA exists as an unbranched
584:.) One of the simplest branched structures is a four-arm junction that consists of four individual DNA strands, portions of which are complementary in a specific pattern. Unlike in natural
306:
reportedly inspired
Nadrian Seeman to consider using three-dimensional lattices of DNA to orient hard-to-crystallize molecules. This led to the beginning of the field of DNA nanotechnology.
310:
In 1991, Seeman's laboratory published a report on the synthesis of a cube made of DNA, the first synthetic three-dimensional nucleic acid nanostructure, for which he received the 1995
1090:
DNA nanotechnology provides one of the few ways to form designed, complex structures with precise control over nanoscale features. The field is beginning to see application to solve
1661:
Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ (December 2005). "Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication".
1453:, and is thus the most thermodynamically favorable, while incorrectly assembled structures have higher energies and are thus disfavored. This is done either through simple, faster
5415:
Sundah NR, Ho NR, Lim GS, Natalia A, Ding X, Liu Y, et al. (September 2019). "Barcoded DNA nanostructures for the multiplexed profiling of subcellular protein distribution".
1458:
380:
on March 15, 2006. Rothemund's research demonstrating two-dimensional DNA origami structures was followed by the demonstration of solid three-dimensional DNA origami by
Douglas
5380:
Kim KR, Kim DR, Lee T, Yhee JY, Kim BS, Kwon IC, Ahn DR (March 2013). "Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells".
395:
experiments that extended the abilities of the field but were far from actual applications. Seeman's 1991 paper on the synthesis of the DNA cube was rejected by the journal
1449:
constituent strands so that they will associate into a desired conformation. Most methods have the goal of designing sequences so that the target structure has the lowest
592:, causing the junction point to be fixed at a certain position. Multiple junctions can be combined in the same complex, such as in the widely used double-crossover (DX)
247:
features. Several assembly methods are used to make these structures, including tile-based structures that assemble from smaller structures, folding structures using the
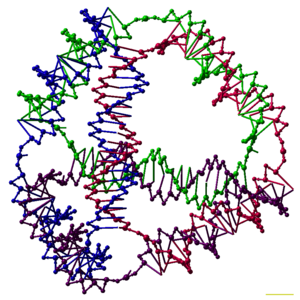
113:
275:
by eliminating the difficult process of obtaining pure crystals. This idea had reportedly come to him in late 1980, after realizing the similarity between the woodcut
5157:
Zadegan RM, Jepsen MD, Thomsen KE, Okholm AH, Schaffert DH, Andersen ES, et al. (November 2012). "Construction of a 4 zeptoliters switchable 3D DNA box origami".
1220:
reported the self-assembly of four short strands of synthetic DNA into a cage which can enter cells and survive for at least 48 hours. The fluorescently labeled DNA
896:
Nucleic acid structures can be made to incorporate molecules other than nucleic acids, sometimes called heteroelements, including proteins, metallic nanoparticles,
350:
in 2000. The next advance was to translate this into mechanical motion, and in 2004 and 2005, several DNA walker systems were demonstrated by the groups of Seeman,
4086:
Liu J, Geng Y, Pound E, Gyawali S, Ashton JR, Hickey J, et al. (March 2011). "Metallization of branched DNA origami for nanoelectronic circuit fabrication".
803:, displaying a representation of increasing binary numbers as it grows. These results show that computation can be incorporated into the assembly of DNA arrays.
547:, meaning that they form matching sequences of base pairs, with A only binding to T, and C only to G. Because the formation of correctly matched base pairs is
603:
to allow the nucleic acid complexes to reconfigure in response to the addition of a new nucleic acid strand. In this reaction, the incoming strand binds to a
271:
in the early 1980s. Seeman's original motivation was to create a three-dimensional DNA lattice for orienting other large molecules, which would simplify their
6324:
Lin C, Ke Y, Chhabra R, Sharma J, Liu Y, Yan H (2011). "Synthesis and
Characterization of Self-Assembled DNA Nanostructures". In Zuccheri G, Samorì B (eds.).
2196:
Xiao S, Liu F, Rosen AE, Hainfeld JF, Seeman NC, Musier-Forsyth K, Kiehl RA (August 2002). "Selfassembly of metallic nanoparticle arrays by DNA scaffolding".
4358:
Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ (February 2008). "Reconfigurable, braced, three-dimensional DNA nanostructures".
4453:
Centola, Mathias; Poppleton, Erik; Ray, Sujay; Centola, Martin; Welty, Robb; Valero, Julián; Walter, Nils G.; Šulc, Petr; Famulok, Michael (2023-10-19).
3693:
Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, et al. (May 2009). "Self-assembly of a nanoscale DNA box with a controllable lid".
1308:, this ensemble of synthetic DNA-made counterparts thereby spans multiple orders of magnitude in conductance. The study of the membrane-inserting single
1119:
775:
lattice, and various DX-based arrays making use of a double-cohesion scheme. The top two images at right show examples of tile-based periodic lattices.
311:
206:
2868:
Doye JP, Ouldridge TE, Louis AA, Romano F, Šulc P, Matek C, et al. (December 2013). "Coarse-graining DNA for simulations of DNA nanotechnology".
2981:
Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH (September 2003). "DNA-templated self-assembly of protein arrays and highly conductive nanowires".
385:
1312:
showed that current must also flow on the DNA-lipid interface as no central channel lumen is present in the design that lets ions pass across the
810:
in diameter, essentially two-dimensional lattices which curve back upon themselves. These DNA nanotubes are somewhat similar in size and shape to
1957:
Rothemund PW (2006). "Scaffolded DNA origami: from generalized multicrossovers to polygonal networks". In Chen J, Jonoska N, Rozenberg G (eds.).
2612:
Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L (December 2004). "Building programmable jigsaw puzzles with RNA".
1628:
1356:
so that individual nucleic acid strands will assemble into the desired structures. This process usually begins with specification of a desired
371:
1204:. There has additionally been interest in expressing these artificial structures in engineered living bacterial cells, most likely using the
849:
of a polyhedron with a DNA junction at each vertex. The earliest demonstrations of DNA polyhedra were very work-intensive, requiring multiple
224:
in the early 1980s, and the field began to attract widespread interest in the mid-2000s. This use of nucleic acids is enabled by their strict
359:
1022:
was shown to walk along a DNA-path, guided by the generated RNA strand. Additionally, a linear walker has been demonstrated that performs
255:, but the same principles have been used with other types of nucleic acids as well, leading to the occasional use of the alternative name
4979:
Zadegan RM, Jepsen MD, Hildebrandt LL, Birkedal V, Kjems J (April 2015). "Construction of a fuzzy and
Boolean logic gates based on DNA".
3304:
Barish RD, Rothemund PW, Winfree E (December 2005). "Two computational primitives for algorithmic self-assembly: copying and counting".
465:
domains, on the top and the bottom in this image. There are two crossover points where the strands cross from one domain into the other.
3168:
Mao C, Sun W, Seeman NC (16 June 1999). "Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy".
1329:
99:
4585:Škugor M, Valero J, Murayama K, Centola M, Asanuma H, Famulok M (May 2019). "Orthogonally Photocontrolled Non-Autonomous DNA Walker".
543:(T). Nucleic acids have the property that two molecules will only bind to each other to form a double helix if the two sequences are
6822:
1462:
1208:
RNA for the assembly, although it is unknown whether these complex structures are able to efficiently fold or assemble in the cell's
4629:
Tian Y, He Y, Chen Y, Yin P, Mao C (July 2005). "A DNAzyme that walks processively and autonomously along a one-dimensional track".
1386:. This was the dominant design strategy used from the mid-1990s until the mid-2000s, when the DNA origami methodology was developed.
1150:, providing a method for nanometer-scale control of the placement and overall architecture of the device analogous to a molecular
1169:, researchers were able to construct a small multi-switchable 3D DNA Box Origami. The proposed nanoparticle was characterized by
933:
1465:
thermodynamic model, which is more accurate but slower and more computationally intensive. Geometric models are used to examine
1005:
control individual steps of a DNA walker by irradiation with light of different wavelengths. Another approach is to make use of
693:
image of the assembled array. The individual DX tiles are clearly visible within the assembled structure. The field is 150
1184:
There are potential applications for DNA nanotechnology in nanomedicine, making use of its ability to perform computation in a
1050:
function of the initiator species, where less than one equivalent of the initiator can cause the reaction to go to completion.
6270:
Walter NG (1 February 2003). "Probing RNA Structural
Dynamics and Function by Fluorescence Resonance Energy Transfer (FRET)".
6120:
Ellington A, Pollard JD (1 May 2001). "Purification of
Oligonucleotides Using Denaturing Polyacrylamide Gel Electrophoresis".
1280:
Applications for DNA nanotechnology in nanomedicine also focus on mimicking the structure and function of naturally occurring
251:
method, and dynamically reconfigurable structures using strand displacement methods. The field's name specifically references
6375:
6341:
5243:
5134:
2595:
2569:
1984:
1727:
5782:"A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane"
3755:
Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H (June 2009). "Scaffolded DNA origami of a DNA tetrahedron molecular container".
1232:
after two days. This experiment showed the potential of drug delivery inside the living cells using the DNA ‘cage’. A DNA
6852:
4402:
Douglas SM, Bachelet I, Church GM (February 2012). "A logic-gated nanorobot for targeted transport of molecular payloads".
3583:
Shih WM, Quispe JD, Joyce GF (February 2004). "A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron".
2409:
1554:
1543:
1504:
1162:
1122:; using DNA origami is advantageous because, unlike liquid crystals, they are tolerant of the detergents needed to suspend
600:
322:, were not rigid enough to form extended three-dimensional lattices. Seeman developed the more rigid double-crossover (DX)
1562:
1178:
283:
and an array of DNA six-arm junctions. Several natural branched DNA structures were known at the time, including the DNA
5301:
4262:
Yan H, Zhang X, Shen Z, Seeman NC (January 2002). "A robust DNA mechanical device controlled by hybridization topology".
916:
functional groups as a chemical handle to bind the heteroelements. This covalent binding scheme has been used to arrange
679:
544:
438:
229:
1433:
step where a temperature change is required to trigger the assembly and favor proper formation of the desired structure.
878:, as most other DNA nanotechnology methods do. DNA origami was first demonstrated for two-dimensional shapes, such as a
6291:
6237:
6191:
6141:
6087:
4210:
Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL (August 2000). "A DNA-fuelled molecular machine made of DNA".
6942:
1344:. This development highlights the potential of synthetic DNA nanostructures for personalized drugs and therapeutics.
161:
structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for
3088:
Winfree E, Liu F, Wenzler LA, Seeman NC (August 1998). "Design and self-assembly of two-dimensional DNA crystals".
2053:
Yurke, Bernard; Turberfield, Andrew J.; Mills, Allen P.; Simmel, Friedrich C.; Neumann, Jennifer L. (August 2000).
1573:
1361:
1174:
1023:
2160:
Robinson BH, Seeman NC (August 1987). "The design of a biochip: a self-assembling molecular-scale memory device".
7082:
6719:
Lin C, Liu Y, Rinker S, Yan H (August 2006). "DNA tile based self-assembly: building complex nanoarchitectures".
5975:
Lin C, Liu Y, Rinker S, Yan H (August 2006). "DNA tile based self-assembly: building complex nanoarchitectures".
1466:
1357:
768:
two-dimensional crystals of DNA. Two-dimensional arrays have been made from other motifs as well, including the
442:
417:
These four strands associate into a DNA four-arm junction because this structure maximizes the number of correct
6876:
5071:
Qian L, Winfree E (June 2011). "Scaling up digital circuit computation with DNA strand displacement cascades".
1365:
1340:
that flips lipids in biological membranes orders of magnitudes faster than naturally occurring proteins called
150:, and each vertex is a three-arm junction. The 4 DNA strands that form the 4 tetrahedral faces are color-coded.
4158:
Mao C, Sun W, Shen Z, Seeman NC (January 1999). "A nanomechanical device based on the B-Z transition of DNA".
3140:
Liu F, Sha R, Seeman NC (10 February 1999). "Modifying the surface features of two-dimensional DNA crystals".
2832:
Lu Y, Liu J (December 2006). "Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers".
7023:
6815:
685:. The DX complex at top will combine with other DX complexes into the two-dimensional array shown at bottom.
482:
607:
of a double-stranded complex, and then displaces one of the strands bound in the original complex through a
6899:
6216:
Chory J, Pollard JD (1 May 2001). "Separation of Small DNA Fragments by
Conventional Gel Electrophoresis".
1429:, at a constant temperature. This is in contrast to the thermodynamic approaches, which require a thermal
1272:(DOX) was conjugated with the tetrahedron and was loaded into MCF-7 breast cancer cells that contained the
4314:
Feng L, Park SH, Reif JH, Yan H (September 2003). "A two-state DNA lattice switched by DNA nanoactuator".
7128:
6995:
1532:
1520:
1066:
such as AND, OR, and NOT gates. More recently, a four-bit circuit was demonstrated that can compute the
929:
548:
210:
5265:
Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ (July 2011). "DNA cage delivery to mammalian cells".
1196:
applications. One such system being investigated uses a hollow DNA box containing proteins that induce
981:
DNA complexes have been made that change their conformation upon some stimulus, making them one form of
833:
Researchers have synthesized many three-dimensional DNA complexes that each have the connectivity of a
6749:
Zhang DY, Seelig G (February 2011). "Dynamic DNA nanotechnology using strand-displacement reactions".
4673:
Bath J, Green SJ, Turberfield AJ (July 2005). "A free-running DNA motor powered by a nicking enzyme".
2729:
Zhang DY, Seelig G (February 2011). "Dynamic DNA nanotechnology using strand-displacement reactions".
1073:
Another use of strand displacement cascades is to make dynamically assembled structures. These use a
342:
New abilities continued to be discovered for designed DNA structures throughout the 2000s. The first
7010:
6862:
6857:
6847:
6839:
1539:
1528:
1524:
1309:
1297:
1115:
633:
581:
493:
236:
147:
5600:
1967:
967:
annealing and then reconfigured dynamically, or can be made to form dynamically in the first place.
874:
is predetermined by the scaffold strand sequence, and not requiring high strand purity and accurate
7033:
6977:
6962:
6872:
6808:
5578:
Burns JR, Stulz E, Howorka S (June 2013). "Self-assembled DNA nanopores that span lipid bilayers".
3326:
1577:
1485:
1205:
47:
1515:
The sequences of the DNA strands making up a target structure are designed computationally, using
799:. The third image at right shows this type of array. Another system has the function of a binary
7070:
7018:
6967:
6954:
5016:
4924:
4827:"Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker"
4510:
4039:
3635:
3442:
3402:
1633:
1569:
1325:
1170:
883:
690:
485:
52:
29:
5732:
Göpfrich K, Zettl T, Meijering AE, Hernández-Ainsa S, Kocabey S, Liedl T, Keyser UF (May 2015).
1553:
gel electrophoresis, which gives size and shape information for the nucleic acid complexes. An
318:. It soon became clear that these structures, polygonal shapes with flexible junctions as their
5595:
4040:"Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates"
3321:
3196:
Constantinou PE, Wang T, Kopatsch J, Israel LB, Zhang X, Ding B, et al. (September 2006).
1962:
1336:
between the lipid bilayer leaflets. Utilizing this effect, they designed a synthetic DNA-built
1189:
516:
454:
5124:
4709:
Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, et al. (May 2010).
1102:. The earliest such application envisaged for the field, and one still in development, is in
4038:
Maune HT, Han SP, Barish RD, Bockrath M, Goddard WA, Rothemund PW, Winfree E (January 2010).
3950:"Finite-size, fully addressable DNA tile lattices formed by hierarchical assembly procedures"
2246:
1581:
1508:
1406:
1368:
design, which is the specification of the actual base sequences of each nucleic acid strand.
871:
854:
589:
232:
202:
169:. Researchers in the field have created static structures such as two- and three-dimensional
5828:
Göpfrich K, Li CY, Ricci M, Bhamidimarri SP, Yoo J, Gyenes B, et al. (September 2016).
4882:
Pan J, Li F, Cha TG, Chen H, Choi JH (August 2015). "Recent progress on DNA based walkers".
3497:
Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, et al. (September 2009).
924:
protein molecules into specific patterns on a DX array. A non-covalent hosting scheme using
388:
and Yan demonstrated hollow three-dimensional structures made out of two-dimensional faces.
243:
that will selectively assemble to form complex target structures with precisely controlled
6894:
6758:
6660:
6541:
6449:
6407:
5891:
5793:
5745:
5587:
5533:
5472:
5336:
5325:"Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery"
5080:
5028:
4936:
4838:
4780:
4722:
4638:
4558:
4411:
4367:
4323:
4271:
4219:
4167:
4051:
3961:
3903:
3764:
3702:
3647:
3592:
3510:
3454:
3401:
Rothemund PW, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E (December 2004).
3313:
3259:
3097:
2990:
2887:
2738:
2677:
2621:
2458:
2373:
2320:
2261:
2205:
2125:
2066:
2019:
1909:
1762:
1670:
1558:
1265:
1054:
959:
741:
An example of an aperiodic two-dimensional lattice that assembles into a fractal pattern.
573:
560:
315:
6505:
5632:
Burns JR, Göpfrich K, Wood JW, Thacker VV, Stulz E, Keyser UF, Howorka S (November 2013).
5459:
Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, et al. (November 2012).
5323:
Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, et al. (June 2012).
4455:"A rhythmically pulsing leaf-spring DNA-origami nanoengine that drives a passive follower"
3723:
3499:"From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal"
1846:
1531:, and strands of custom sequences are commercially available. Strands can be purified by
477:
is often defined as the study of materials and devices with features on a scale below 100
8:
7041:
6930:
6594:.—A news article focusing on the history of the field and development of new applications
1535:
1500:
1443:
1430:
1418:
1155:
1037:
497:
240:
131:
6762:
6664:
6545:
6453:
6411:
6174:
Gallagher SR, Desjardins P (1 July 2011). "Quantitation of nucleic acids and proteins".
6066:
Ellington A, Pollard JD (1 May 2001). "Synthesis and
Purification of Oligonucleotides".
5895:
5878:
Ohmann A, Li CY, Maffeo C, Al Nahas K, Baumann KN, Göpfrich K, et al. (June 2018).
5797:
5749:
5591:
5537:
5476:
5340:
5084:
5032:
4940:
4842:
4784:
4726:
4642:
4562:
4485:
4415:
4371:
4327:
4275:
4223:
4171:
4055:
3965:
3907:
3892:"Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs"
3768:
3706:
3651:
3596:
3514:
3458:
3317:
3263:
3101:
2994:
2891:
2742:
2681:
2625:
2462:
2377:
2324:
2265:
2209:
2129:
2070:
2023:
1913:
1766:
1674:
1260:
found on some tumors. The result showed that the gene expression targeted by the RNAi,
6626:
6599:
6514:
6489:
6470:
6437:
6305:
6251:
6197:
6155:
6101:
5912:
5879:
5854:
5829:
5708:
5683:
5658:
5633:
5554:
5521:
5493:
5460:
5440:
5357:
5324:
5219:
5194:
5104:
5052:
4960:
4859:
4826:
4801:
4768:
4743:
4710:
4610:
4435:
4295:
4243:
4191:
3924:
3891:
3871:
3827:
3736:
3671:
3616:
3531:
3498:
3478:
3280:
3247:
3222:
3197:
3121:
3014:
2911:
2877:
2808:
2783:
2698:
2665:
2645:
2479:
2446:
2341:
2308:
2285:
2221:
2090:
1930:
1897:
1855:
1830:
1694:
1516:
1470:
1426:
1413:
in addition to the final product. This is done using starting materials which adopt a
1410:
1213:
1131:
1095:
1006:
788:
496:. These qualities make the assembly of nucleic acid structures easy to control through
392:
355:
194:
16:
The design and manufacture of artificial nucleic acid structures for technological uses
6419:
6042:
6017:
5959:
5942:
5880:"A synthetic enzyme built from DNA flips 10 lipids per second in biological membranes"
5733:
5520:
Göpfrich K, Li CY, Mames I, Bhamidimarri SP, Ricci M, Yoo J, et al. (July 2016).
4549:
Sherman WB, Seeman NC (July 2004). "A precisely controlled DNA biped walking device".
3799:
3064:
3037:
2958:
2931:
2536:
2509:
1774:
7107:
6774:
6736:
6706:
6676:
6631:
6585:
6557:
6519:
6475:
6423:
6371:
6347:
6337:
6297:
6287:
6243:
6233:
6187:
6147:
6137:
6105:
6093:
6083:
6047:
5992:
5917:
5859:
5809:
5761:
5713:
5684:"Bilayer-spanning DNA nanopores with voltage-switching between open and closed state"
5663:
5613:
5559:
5498:
5444:
5432:
5397:
5362:
5282:
5224:
5174:
5130:
5096:
5044:
4996:
4952:
4899:
4864:
4806:
4748:
4690:
4654:
4602:
4530:
4490:
4472:
4454:
4427:
4383:
4339:
4287:
4235:
4183:
4139:
4103:
4067:
4019:
3979:
3929:
3890:
Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC (July 2006).
3875:
3863:
3831:
3819:
3780:
3728:
3663:
3608:
3536:
3470:
3422:
3382:
3339:
3285:
3227:
3113:
3069:
3018:
3006:
2963:
2903:
2849:
2813:
2754:
2703:
2637:
2591:
2565:
2541:
2484:
2389:
2346:
2277:
2177:
2141:
2082:
2035:
1980:
1935:
1860:
1778:
1723:
1686:
1638:
1615:
1450:
1402:
1324:
reorient to face towards the membrane-inserted part of the DNA. Researchers from the
1281:
1217:
1166:
1019:
917:
815:
800:
796:
769:
753:, DNA arrays that display a representation of the Sierpinski gasket on their surfaces
746:
585:
397:
319:
288:
182:
170:
87:
6255:
6201:
6159:
5108:
5056:
4614:
4299:
3482:
2915:
1698:
1057:
capable of complex computation. Unlike traditional electronic computers, which use
555:
is the dominant material used, structures incorporating other nucleic acids such as
7051:
6766:
6728:
6698:
6668:
6621:
6611:
6577:
6549:
6509:
6501:
6465:
6457:
6415:
6329:
6309:
6279:
6225:
6183:
6179:
6129:
6075:
6037:
6029:
5984:
5954:
5907:
5899:
5849:
5841:
5801:
5753:
5703:
5695:
5653:
5645:
5605:
5549:
5541:
5488:
5480:
5424:
5389:
5352:
5344:
5274:
5214:
5206:
5166:
5088:
5036:
4988:
4964:
4944:
4891:
4854:
4846:
4796:
4788:
4738:
4730:
4682:
4646:
4594:
4566:
4522:
4480:
4462:
4439:
4419:
4375:
4331:
4279:
4247:
4227:
4195:
4175:
4131:
4095:
4059:
4011:
3969:
3919:
3911:
3855:
3811:
3772:
3740:
3718:
3710:
3675:
3655:
3620:
3600:
3564:
3526:
3518:
3462:
3414:
3374:
3331:
3275:
3267:
3217:
3209:
3177:
3149:
3125:
3105:
3059:
3049:
2998:
2953:
2943:
2895:
2841:
2803:
2795:
2746:
2693:
2685:
2649:
2629:
2531:
2521:
2474:
2466:
2381:
2336:
2328:
2289:
2269:
2225:
2213:
2169:
2133:
2094:
2074:
2027:
1972:
1925:
1917:
1850:
1842:
1770:
1678:
1237:
1185:
1123:
1058:
954:
608:
593:
376:
323:
284:
244:
6689:
Feldkamp U, Niemeyer CM (March 2006). "Rational design of DNA nanoarchitectures".
6581:
6553:
3948:
Park SH, Pistol C, Ahn SJ, Reif JH, Lebeck AR, Dwyer C, LaBean TH (January 2006).
3365:
Feldkamp U, Niemeyer CM (March 2006). "Rational design of DNA nanoarchitectures".
2385:
2031:
1580:
are often used in this case. Extended three-dimensional lattices are analyzed by
1041:
are made energetically favorable through the formation of new base pairs, and the
825:, a balance between tension and compression forces, was finally reported in 2009.
492:
rules which are well understood, and form the specific nanoscale structure of the
7000:
6987:
6925:
6283:
6229:
6133:
6079:
5781:
5757:
5682:
Seifert A, Göpfrich K, Burns JR, Fertig N, Keyser UF, Howorka S (February 2015).
5545:
4895:
3636:"Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns"
3555:
Zhang Y, Seeman NC (1 March 1994). "Construction of a DNA-truncated octahedron".
3054:
2948:
2845:
2526:
1353:
1301:
1257:
1139:
1111:
1103:
994:
846:
811:
763:
and combined into larger two-dimensional periodic lattices containing a specific
732:
272:
6333:
3846:
Endo M, Sugiyama H (October 2009). "Chemical approaches to DNA nanotechnology".
2417:
1804:
500:. This property is absent in other materials used in nanotechnology, including
7095:
6889:
6831:
5903:
4467:
3441:
Yin P, Hariadi RF, Sahu S, Choi HM, Park SH, Labean TH, Reif JH (August 2008).
1797:
1601:
1477:
1273:
1244:. Delivery of the interfering RNA for treatment has showed some success using
1147:
710:, a model of a DNA tile used to make another two-dimensional periodic lattice.
505:
474:
331:
268:
221:
166:
162:
75:
37:
6794:
5428:
4792:
4122:
Deng Z, Mao C (August 2004). "Molecular lithography with DNA nanostructures".
3815:
2799:
2217:
2173:
652:
7122:
6909:
4769:"A bio-hybrid DNA rotor-stator nanoengine that moves along predefined tracks"
4476:
3999:
3823:
2086:
1383:
1332:
then demonstrated that such a DNA-induced toroidal pore can facilitate rapid
1313:
1289:
1091:
1010:
875:
783:, allowing them to perform computation. A DX array whose assembly encodes an
613:
347:
190:
186:
6528:—A more comprehensive review including both old and new results in the field
5845:
5484:
5092:
5040:
4423:
3466:
3002:
2633:
1976:
1682:
1134:. Further, DNA origami structures have aided in the biophysical studies of
1130:
have been used as nanoscale assembly lines to move nanoparticles and direct
7046:
6937:
6884:
6778:
6740:
6732:
6710:
6702:
6680:
6672:
6635:
6589:
6561:
6532:
Service RF (June 2011). "DNA nanotechnology. DNA nanotechnology grows up".
6523:
6479:
6427:
6351:
6301:
6247:
6151:
6097:
6051:
5996:
5988:
5921:
5863:
5813:
5805:
5765:
5717:
5667:
5649:
5634:"Lipid-bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor"
5617:
5563:
5502:
5436:
5401:
5366:
5286:
5228:
5178:
5100:
5048:
5000:
4992:
4956:
4903:
4868:
4850:
4810:
4752:
4694:
4686:
4658:
4650:
4606:
4598:
4534:
4494:
4431:
4387:
4343:
4335:
4291:
4239:
4143:
4135:
4107:
4071:
4063:
4023:
4015:
3983:
3974:
3949:
3933:
3867:
3859:
3784:
3732:
3667:
3612:
3540:
3474:
3426:
3386:
3378:
3343:
3289:
3231:
3073:
3010:
2967:
2907:
2853:
2817:
2758:
2707:
2689:
2641:
2545:
2488:
2393:
2364:
Service RF (June 2011). "DNA nanotechnology. DNA nanotechnology grows up".
2350:
2281:
2145:
2137:
2054:
2039:
2010:
Service RF (June 2011). "DNA nanotechnology. DNA nanotechnology grows up".
1939:
1921:
1864:
1782:
1690:
1550:
1523:
software. The nucleic acids themselves are then synthesized using standard
1305:
982:
925:
921:
865:
Nanostructures of arbitrary, non-regular shapes are usually made using the
784:
764:
509:
462:
351:
327:
303:
280:
214:
158:
135:
5348:
4187:
3117:
2181:
1565:(FRET) are sometimes used to characterize the structure of the complexes.
1070:
of the integers 0–15, using a system of gates containing 130 DNA strands.
787:
operation has been demonstrated; this allows the DNA array to implement a
6616:
6033:
5461:"Synthetic lipid membrane channels formed by designed DNA nanostructures"
4379:
3798:
Zaborova, O. V.; Voinova, A. D.; Shmykov, B. D.; Sergeyev, V. G. (2021).
1624:
International
Society for Nanoscale Science, Computation, and Engineering
1391:
1341:
1293:
1288:
introduced a pore-shaped DNA origami structure that can self-insert into
1269:
1233:
1221:
1201:
1107:
1067:
1063:
976:
937:
897:
866:
366:
343:
248:
4948:
4734:
3714:
3659:
3604:
3568:
3522:
2332:
2273:
1394:, which allows forming nanoscale two- and three-dimensional shapes (see
588:, each arm in the artificial immobile four-arm junction has a different
126:
6370:. Sausalito, Calif: University Science Books. pp. 84–86, 396–407.
5393:
2899:
1481:
1333:
1261:
1193:
1151:
1099:
1015:
1001:
850:
842:
834:
822:
760:
682:
604:
524:
520:
267:
The conceptual foundation for DNA nanotechnology was first laid out by
220:
The conceptual foundation for DNA nanotechnology was first laid out by
198:
178:
7090:
6770:
6568:
Service RF (June 2011). "DNA nanotechnology. Next step: DNA robots?".
6461:
5699:
5609:
5278:
5210:
5170:
4570:
4526:
4099:
3915:
3776:
3418:
3335:
3271:
3246:
Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, Seeman NC (April 2005).
3181:
3153:
2750:
2470:
1595:
1469:
of the nanostructures and to ensure that the complexes are not overly
1300:, to small tile-based structures, and large DNA origami transmembrane
948:
70:
6917:
6783:—A review of DNA systems making use of strand displacement mechanisms
5731:
4231:
3213:
2307:
Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM (May 2009).
2078:
1454:
1414:
1209:
1197:
1079:
1074:
1047:
905:
807:
780:
723:
703:
694:
489:
478:
449:
418:
336:
225:
143:
4283:
2586:
Long EC (1996). "Fundamentals of nucleic acids". In Hecht SM (ed.).
1538:
if needed, and precise concentrations determined via any of several
674:, schematic diagram. Each bar represents a double-helical domain of
5734:"DNA-Tile Structures Induce Ionic Currents through Lipid Membranes"
1496:
1158:
because of the coupling of computation to its material properties.
858:
617:
532:
430:
174:
7102:
4179:
3889:
3400:
3109:
2882:
1609:
1557:
can assess whether a structure incorporates all desired strands.
82:
6800:
6715:—A review coming from the viewpoint of secondary structure design
2562:
Self-assembly: the science of things that put themselves together
1720:
Self-assembly: the science of things that put themselves together
1245:
1229:
1042:
792:
772:
621:
540:
536:
528:
501:
434:
426:
422:
6438:"Structural DNA nanotechnology: growing along with Nano Letters"
4978:
2447:"Structural DNA nanotechnology: growing along with Nano Letters"
1898:"Challenges and opportunities for structural DNA nanotechnology"
1417:
structure; these then assemble into the final conformation in a
512:, which lack the capability for specific assembly on their own.
441:
for a more realistic model of the four-arm junction showing its
1961:. Natural Computing Series. New York: Springer. pp. 3–21.
1337:
1253:
1225:
1135:
879:
661:
294:
6398:
Seeman NC (June 2004). "Nanotechnology and the double helix".
6328:. Methods in Molecular Biology. Vol. 749. pp. 1–11.
5522:"Ion Channels Made from a Single Membrane-Spanning DNA Duplex"
5015:
Seelig G, Soloveichik D, Zhang DY, Winfree E (December 2006).
3800:"Solid Lipid Nanoparticles for the Nucleic Acid Encapsulation"
3797:
3195:
2309:"Self-assembly of DNA into nanoscale three-dimensional shapes"
1753:
Seeman NC (June 2004). "Nanotechnology and the double helix".
6432:—An article written for laypeople by the founder of the field
5830:"Large-Conductance Transmembrane Porin Made from DNA Origami"
5014:
4767:
Valero J, Pal N, Dhakal S, Walter NG, Famulok M (June 2018).
2611:
1321:
1317:
1249:
1224:
were found to remain intact in the laboratory cultured human
1062:
displacement complexes. This approach has been used to make
990:
986:
913:
909:
901:
165:
rather than as the carriers of genetic information in living
6600:"Structural DNA nanotechnology: from design to applications"
5302:"Researchers achieve RNA interference, in a lighter package"
1722:. New York: Chapman & Hall/CRC. pp. 201, 242, 259.
1256:
molecules, which lead the DNA nanoparticles to the abundant
1200:, or cell death, that will only open when in proximity to a
806:
DX arrays have been made to form hollow nanotubes 4–20
6365:
5827:
5519:
4584:
4209:
3403:"Design and characterization of programmable DNA nanotubes"
2052:
1377:
desired shape. Several approaches have been demonstrated:
1240:(RNAi) in a mouse model, reported a team of researchers in
838:
6745:—A minireview specifically focusing on tile-based assembly
6016:
Dirks RM, Lin M, Winfree E, Pierce NA (15 February 2004).
5780:
Burns JR, Seifert A, Fertig N, Howorka S (February 2016).
5681:
5156:
4452:
4357:
1154:. DNA nanotechnology has been compared to the concept of
1046:
formation of the desired structure. They can also support
1018:
DNA that uses rolling circle transcription by an attached
624:, which can bind to specific proteins or small molecules.
5779:
5458:
3035:
1549:
The fully formed target structures can be verified using
1241:
675:
556:
552:
458:
252:
139:
5631:
4708:
2867:
2306:
1660:
940:
masks, transferring their pattern into a solid surface.
6651:
Bath J, Turberfield AJ (May 2007). "DNA nanomachines".
3692:
3245:
3087:
3038:"Algorithmic self-assembly of DNA Sierpinski triangles"
2116:
Bath J, Turberfield AJ (May 2007). "DNA nanomachines".
1425:
below). This approach has the advantage of proceeding
1316:. This indicated that the DNA-induced lipid pore has a
189:. The field is beginning to be used as a tool to solve
181:, and arbitrary shapes, and functional devices such as
5877:
5322:
4923:
Yin P, Choi HM, Calvert CR, Pierce NA (January 2008).
4037:
3633:
3303:
3036:
Rothemund PW, Papadakis N, Winfree E (December 2004).
2980:
1401:
Dynamic assembly. This approach directly controls the
714:, an atomic force micrograph of the assembled lattice.
481:. DNA nanotechnology, specifically, is an example of
6015:
4766:
4401:
3496:
3030:
3028:
2247:"Folding DNA to create nanoscale shapes and patterns"
1284:
with designed DNA nanostructures. In 2012, Langecker
519:
of a nucleic acid molecule consists of a sequence of
6640:—A very recent and comprehensive review in the field
6368:
Nucleic acids: structures, properties, and functions
5192:
4922:
4711:"Molecular robots guided by prescriptive landscapes"
4672:
3998:
Cohen JD, Sadowski JP, Dervan PB (22 October 2007).
2590:. New York: Oxford University Press. pp. 4–10.
2195:
2188:
1896:
Pinheiro AV, Han D, Shih WM, Yan H (November 2011).
1591:
6173:
5870:
5451:
4085:
4000:"Addressing single molecules on DNA nanostructures"
3997:
2564:. New York: Chapman & Hall/CRC. pp. 5, 7.
2239:
2237:
2235:
1890:
1888:
1886:
1884:
1882:
1880:
1878:
1876:
1874:
1161:In a study conducted by a group of scientists from
599:Dynamic DNA nanotechnology uses a mechanism called
209:to determine structures. Potential applications in
207:
nuclear magnetic resonance spectroscopy of proteins
6323:
5514:
5512:
5414:
4261:
3634:Tikhomirov G, Petersen P, Qian L (December 2017).
3440:
3198:"Double cohesion in structural DNA nanotechnology"
3025:
1895:
1568:Nucleic acid structures can be directly imaged by
1053:Strand displacement complexes can be used to make
759:Small nucleic acid complexes can be equipped with
142:tetrahedron. Each edge of the tetrahedron is a 20
6176:Current Protocols Essential Laboratory Techniques
6018:"Paradigms for computational nucleic acid design"
5577:
5264:
4925:"Programming biomolecular self-assembly pathways"
3947:
1422:
527:they contain. In DNA, the four bases present are
228:rules, which cause only portions of strands with
7120:
6688:
6685:—A review of nucleic acid nanomechanical devices
6366:Bloomfield VA, Crothers DM, Tinoco Jr I (2000).
6119:
6065:
4511:"A synthetic DNA walker for molecular transport"
4157:
3364:
2439:
2437:
2435:
2232:
1951:
1949:
1871:
1216:of nucleic acid nanostructures. Scientists at
627:
130:DNA nanotechnology involves forming artificial,
6718:
6650:
5974:
5940:
5509:
4313:
3754:
3582:
2510:"The emergence of complexity: lessons from DNA"
2115:
2110:
2108:
2106:
2104:
1030:
5935:
5933:
5931:
5820:
5195:"From DNA nanotechnology to synthetic biology"
5193:Jungmann R, Renner S, Simmel FC (April 2008).
3838:
3687:
3685:
2784:"An overview of structural DNA nanotechnology"
2159:
1747:
1745:
1743:
1741:
1739:
1629:Comparison of nucleic acid simulation software
469:
6816:
6597:
5724:
5379:
5299:
4917:
4915:
4913:
4628:
4548:
2432:
2055:"A DNA-fuelled molecular machine made of DNA"
1946:
943:
107:
6484:—A review of results in the period 2001–2010
6215:
6010:
6008:
6006:
5943:"Strand design for biomolecular computation"
5241:
4977:Fuzzy and Boolean logic gates based on DNA:
4881:
4394:
3845:
3167:
3139:
2776:
2774:
2772:
2770:
2768:
2723:
2721:
2719:
2717:
2101:
1823:
1821:
1819:
1817:
1815:
1405:of DNA self-assembly, specifying all of the
620:, which can perform chemical reactions, and
157:is the design and manufacture of artificial
6797:—a video introduction to DNA nanotechnology
6748:
6604:International Journal of Molecular Sciences
6272:Current Protocols in Nucleic Acid Chemistry
5941:Brenneman A, Condon A (25 September 2002).
5928:
5674:
5624:
5570:
5070:
3682:
3554:
3359:
3357:
3355:
3353:
2728:
1736:
1655:
1653:
1127:
6823:
6809:
4910:
4508:
2666:"The emerging field of RNA nanotechnology"
2301:
2299:
1330:University of Illinois at Urbana-Champaign
845:, meaning that the DNA duplexes trace the
365:In 2006, Rothemund first demonstrated the
114:
100:
6625:
6615:
6513:
6469:
6041:
6003:
5958:
5911:
5853:
5772:
5707:
5657:
5599:
5553:
5492:
5356:
5244:"DNA cages can unleash meds inside cells"
5218:
5017:"Enzyme-free nucleic acid logic circuits"
4858:
4800:
4742:
4484:
4466:
3973:
3923:
3722:
3530:
3325:
3279:
3221:
3063:
3053:
2957:
2947:
2881:
2807:
2765:
2714:
2697:
2535:
2525:
2478:
2340:
2244:
2004:
2002:
2000:
1998:
1996:
1966:
1956:
1929:
1854:
1812:
1712:
1710:
1708:
1476:Nucleic acid design has similar goals to
1304:. Similar to naturally occurring protein
970:
4515:Journal of the American Chemical Society
3557:Journal of the American Chemical Society
3489:
3407:Journal of the American Chemical Society
3350:
3170:Journal of the American Chemical Society
3142:Journal of the American Chemical Society
2860:
2502:
2500:
2498:
1650:
1495:
1491:
1395:
947:
702:
448:
412:
293:
125:
6567:
6531:
5122:
4121:
2559:
2363:
2296:
2152:
2009:
1959:Nanotechnology: science and computation
1717:
934:carbon nanotube field-effect transistor
408:
235:to bind together to form strong, rigid
7121:
6487:
6435:
6397:
6269:
6218:Current Protocols in Molecular Biology
6122:Current Protocols in Molecular Biology
6068:Current Protocols in Molecular Biology
2929:
2781:
2444:
2407:
1993:
1828:
1805:"DNA cages containing oriented guests"
1752:
1705:
828:
6804:
6506:10.1146/annurev-biochem-060308-102244
4824:
4509:Shin JS, Pierce NA (September 2004).
3443:"Programming DNA tube circumferences"
3248:"Six-helix bundles designed from DNA"
2831:
2495:
1847:10.1146/annurev-biochem-060308-102244
1228:cells despite the attack by cellular
891:
326:, and in 1998, in collaboration with
7077:
5126:Molecular engineering of nanosystems
3202:Organic & Biomolecular Chemistry
2585:
1802:for a statement of the problem, and
1555:electrophoretic mobility shift assay
1371:
1212:. If successful, this could enable
920:on a DX-based array, and to arrange
642:
601:toehold-mediated strand displacement
6598:Zadegan RM, Norton ML (June 2012).
2870:Physical Chemistry Chemical Physics
2663:
2588:Bioorganic chemistry: nucleic acids
2507:
1544:ultraviolet absorbance spectroscopy
1421:reaction, in a specific order (see
1320:shape, rather than cylindrical, as
13:
6830:
6387:
1798:"Current crystallization protocol"
1437:
563:(PNA) have also been constructed.
14:
7140:
6788:
6420:10.1038/scientificamerican0604-64
3804:Reviews and Advances in Chemistry
1775:10.1038/scientificamerican0604-64
1563:Förster resonance energy transfer
1527:methods, usually automated in an
1188:format to make "smart drugs" for
1179:Förster resonance energy transfer
241:rational design of base sequences
7101:
7089:
7076:
7065:
7064:
6358:
4884:Current Opinion in Biotechnology
2834:Current Opinion in Biotechnology
2198:Journal of Nanoparticle Research
1608:
1594:
1574:transmission electron microscopy
1175:transmission electron microscopy
993:forms to respond to a change in
731:
722:
660:
651:
239:structures. This allows for the
81:
69:
36:
6316:
6262:
6208:
6166:
6112:
6058:
5967:
5408:
5373:
5316:
5293:
5258:
5235:
5185:
5150:
5115:
5063:
5007:
4971:
4875:
4817:
4759:
4701:
4665:
4621:
4577:
4541:
4501:
4446:
4350:
4306:
4254:
4202:
4150:
4114:
4078:
4030:
3990:
3940:
3882:
3791:
3747:
3627:
3575:
3547:
3433:
3393:
3296:
3238:
3188:
3160:
3132:
3080:
2974:
2922:
2824:
2780:Structural DNA nanotechnology:
2656:
2604:
2578:
2552:
2400:
2357:
1360:or function. Then, the overall
1085:
678:, with the shapes representing
312:Feynman Prize in Nanotechnology
24:Part of a series of articles on
6184:10.1002/9780470089941.et0202s5
5129:. Springer. pp. 209–212.
5069:Strand displacement cascades:
5013:Strand displacement cascades:
4825:He Y, Liu DR (November 2010).
3724:11858/00-001M-0000-0010-9363-9
2866:Simulation of DNA structures:
2046:
1789:
1459:sequence symmetry minimization
605:single-stranded toehold region
1:
7024:Scanning tunneling microscope
6582:10.1126/science.332.6034.1142
6554:10.1126/science.332.6034.1140
6494:Annual Review of Biochemistry
5960:10.1016/S0304-3975(02)00135-4
5417:Nature Biomedical Engineering
5300:Trafton, Anne (4 June 2012).
2386:10.1126/science.332.6034.1140
2032:10.1126/science.332.6034.1140
1835:Annual Review of Biochemistry
1644:
628:Structural DNA nanotechnology
461:single strands that form two
314:. This was followed by a DNA
217:are also being investigated.
6490:"Nanomaterials based on DNA"
6284:10.1002/0471142700.nc1110s11
6230:10.1002/0471142727.mb0207s47
6134:10.1002/0471142727.mb0212s42
6080:10.1002/0471142727.mb0211s42
5947:Theoretical Computer Science
5758:10.1021/acs.nanolett.5b00189
5546:10.1021/acs.nanolett.6b02039
5242:Lovy, Howard (5 July 2011).
4896:10.1016/j.copbio.2014.11.017
3055:10.1371/journal.pbio.0020424
2949:10.1371/journal.pbio.0020073
2846:10.1016/j.copbio.2006.10.004
2830:Dynamic DNA nanotechnology:
2727:Dynamic DNA nanotechnology:
2527:10.1371/journal.pbio.0020431
1831:"Nanomaterials based on DNA"
1807:. Nadrian Seeman Laboratory.
1423:Strand displacement cascades
1146:electronic elements such as
1031:Strand displacement cascades
670:The assembly of a DX array.
566:
201:, including applications in
7:
6996:Molecular scale electronics
6334:10.1007/978-1-61779-142-0_1
3302:Algorithmic self-assembly:
3034:Algorithmic self-assembly:
2782:Seeman NC (November 2007).
2245:Rothemund PW (March 2006).
1587:
1529:oligonucleotide synthesizer
1382:making them a platform for
1352:DNA nanostructures must be
470:Properties of nucleic acids
453:This double-crossover (DX)
384:in 2009, while the labs of
257:nucleic acid nanotechnology
211:molecular scale electronics
10:
7145:
6795:What is Bionanotechnology?
5904:10.1038/s41467-018-04821-5
4468:10.1038/s41565-023-01516-x
3495:Three-dimensional arrays:
1809:for the proposed solution.
1484:as compared to the twenty
1441:
974:
944:Dynamic DNA nanotechnology
262:
7060:
7032:
7011:Scanning probe microscopy
7009:
6986:
6953:
6908:
6871:
6838:
5429:10.1038/s41551-019-0417-0
4793:10.1038/s41565-018-0109-z
3816:10.1134/S2079978021030055
2800:10.1007/s12033-007-0059-4
1540:nucleic acid quantitation
1525:oligonucleotide synthesis
1486:proteinogenic amino acids
1347:
1116:residual dipolar coupling
634:nucleic acid double helix
494:nucleic acid double helix
7034:Molecular nanotechnology
6978:Solid lipid nanoparticle
6963:Self-assembled monolayer
5246:. fiercedrugdelivery.com
4823:Functional DNA walkers:
4765:Functional DNA walkers:
4707:Functional DNA walkers:
2408:Hopkin K (August 2011).
1578:cryo-electron microscopy
1390:latter method is called
1120:protein NMR spectroscopy
48:Self-assembled monolayer
7019:Atomic force microscope
6968:Supramolecular assembly
6955:Molecular self-assembly
6436:Seeman NC (June 2010).
5846:10.1021/acsnano.6b03759
5485:10.1126/science.1225624
5382:Chemical Communications
5093:10.1126/science.1200520
5041:10.1126/science.1132493
4424:10.1126/science.1214081
3467:10.1126/science.1157312
3003:10.1126/science.1089389
2930:Strong M (March 2004).
2788:Molecular Biotechnology
2664:Guo P (December 2010).
2634:10.1126/science.1104686
2508:Mao C (December 2004).
2445:Seeman NC (June 2010).
2218:10.1023/A:1021145208328
2174:10.1093/protein/1.4.295
1977:10.1007/3-540-30296-4_1
1683:10.1126/science.1120367
1634:Molecular models of DNA
1570:atomic force microscopy
1326:University of Cambridge
1171:atomic force microscopy
1024:DNA-templated synthesis
691:atomic force microscopy
549:energetically favorable
523:distinguished by which
508:is very difficult, and
486:molecular self-assembly
53:Supramolecular assembly
30:Molecular self-assembly
6733:10.1002/cphc.200600260
6703:10.1002/anie.200502358
6673:10.1038/nnano.2007.104
6540:(6034): 1140–1, 1143.
6022:Nucleic Acids Research
5989:10.1002/cphc.200600260
5806:10.1038/nnano.2015.279
5650:10.1002/anie.201305765
4993:10.1002/smll.201402755
4851:10.1038/nnano.2010.190
4687:10.1002/anie.200501262
4651:10.1002/ange.200500703
4599:10.1002/anie.201901272
4336:10.1002/ange.200351818
4136:10.1002/anie.200460257
4064:10.1038/nnano.2009.311
4016:10.1002/anie.200702767
3975:10.1002/ange.200690141
3860:10.1002/cbic.200900286
3379:10.1002/anie.200502358
2932:"Protein nanomachines"
2690:10.1038/nnano.2010.231
2372:(6034): 1140–1, 1143.
2138:10.1038/nnano.2007.104
2018:(6034): 1140–1, 1143.
2008:History/applications:
1922:10.1038/nnano.2011.187
1521:thermodynamic modeling
1512:
1503:methods, such as this
1190:targeted drug delivery
971:Nanomechanical devices
963:
715:
466:
455:supramolecular complex
446:
307:
273:crystallographic study
151:
134:nanostructures out of
7108:Technology portal
6653:Nature Nanotechnology
5884:Nature Communications
5786:Nature Nanotechnology
5349:10.1038/NNANO.2012.73
5329:Nature Nanotechnology
4831:Nature Nanotechnology
4773:Nature Nanotechnology
4459:Nature Nanotechnology
4360:Nature Nanotechnology
4044:Nature Nanotechnology
2670:Nature Nanotechnology
2118:Nature Nanotechnology
1902:Nature Nanotechnology
1800:. Nadrian Seeman Lab.
1582:X-ray crystallography
1509:reaction intermediate
1499:
1492:Materials and methods
1461:, or by using a full
1055:molecular logic gates
951:
855:solid-phase synthesis
706:
452:
416:
297:
203:X-ray crystallography
129:
88:Technology portal
6895:Green nanotechnology
6645:Specific subfields:
6617:10.3390/ijms13067149
6278:: 11.10.1–11.10.23.
4380:10.1038/nnano.2008.3
2662:RNA nanotechnology:
2610:RNA nanotechnology:
1559:Fluorescent labeling
1266:multidrug resistance
1236:was used to deliver
1165:and CDNA centers in
960:molecular logic gate
930:molecular electronic
561:peptide nucleic acid
409:Fundamental concepts
316:truncated octahedron
7042:Molecular assembler
6763:2011NatCh...3..103Z
6665:2007NatNa...2..275B
6546:2011Sci...332.1140S
6454:2010NanoL..10.1971S
6412:2004SciAm.290f..64S
6400:Scientific American
5896:2018NatCo...9.2426O
5798:2016NatNa..11..152B
5750:2015NanoL..15.3134G
5644:(46): 12069–12072.
5592:2013NanoL..13.2351B
5538:2016NanoL..16.4665G
5477:2012Sci...338..932L
5341:2012NatNa...7..389L
5165:(11): 10050–10053.
5123:Rietman EA (2001).
5085:2011Sci...332.1196Q
5079:(6034): 1196–1201.
5033:2006Sci...314.1585S
5027:(5805): 1585–1588.
4949:10.1038/nature06451
4941:2008Natur.451..318Y
4843:2010NatNa...5..778H
4785:2018NatNa..13..496V
4735:10.1038/nature09012
4727:2010Natur.465..206L
4643:2005AngCh.117.4429T
4563:2004NanoL...4.1203S
4521:(35): 10834–10835.
4416:2012Sci...335..831D
4372:2008NatNa...3...93G
4328:2003AngCh.115.4478F
4276:2002Natur.415...62Y
4224:2000Natur.406..605Y
4172:1999Natur.397..144M
4056:2010NatNa...5...61M
3966:2006AngCh.118.6759P
3908:2006NanoL...6.1502Z
3769:2009NanoL...9.2445K
3715:10.1038/nature07971
3707:2009Natur.459...73A
3660:10.1038/nature24655
3652:2017Natur.552...67T
3605:10.1038/nature02307
3597:2004Natur.427..618S
3569:10.1021/ja00084a006
3523:10.1038/nature08274
3515:2009Natur.461...74Z
3459:2008Sci...321..824Y
3413:(50): 16344–16352.
3318:2005NanoL...5.2586B
3264:2005NanoL...5..661M
3102:1998Natur.394..539W
2995:2003Sci...301.1882Y
2989:(5641): 1882–1884.
2892:2013PCCP...1520395D
2876:(47): 20395–20414.
2743:2011NatCh...3..103Z
2682:2010NatNa...5..833G
2626:2004Sci...306.2068C
2620:(5704): 2068–2072.
2560:Pelesko JA (2007).
2463:2010NanoL..10.1971S
2410:"Profile: 3-D seer"
2378:2011Sci...332.1140S
2333:10.1038/nature08016
2325:2009Natur.459..414D
2274:10.1038/nature04586
2266:2006Natur.440..297R
2210:2002JNR.....4..313X
2162:Protein Engineering
2130:2007NatNa...2..275B
2071:2000Natur.406..605Y
2024:2011Sci...332.1140S
1914:2011NatNa...6..763P
1767:2004SciAm.290f..64S
1755:Scientific American
1718:Pelesko JA (2007).
1675:2005Sci...310.1661G
1669:(5754): 1661–1665.
1536:gel electrophoresis
1501:Gel electrophoresis
1444:Nucleic acid design
1396:Discrete structures
1362:secondary structure
1354:rationally designed
1156:programmable matter
1007:restriction enzymes
829:Discrete structures
577:dynamic subfields.
498:nucleic acid design
7129:DNA nanotechnology
7096:Science portal
6973:DNA nanotechnology
6488:Seeman NC (2010).
6326:DNA Nanotechnology
6074:: 2.11.1–2.11.25.
6034:10.1093/nar/gkh291
5826:DNA ion channels:
5778:DNA ion channels:
5730:DNA ion channels:
5680:DNA ion channels:
5630:DNA ion channels:
5576:DNA ion channels:
5518:DNA ion channels:
5457:DNA ion channels:
5394:10.1039/c3cc38693g
4921:Kinetic assembly:
4120:Nanoarchitecture:
4084:Nanoarchitecture:
4036:Nanoarchitecture:
3996:Nanoarchitecture:
3946:Nanoarchitecture:
3888:Nanoarchitecture:
2900:10.1039/C3CP53545B
2420:on 10 October 2011
2194:Nanoarchitecture:
2158:Nanoarchitecture:
1829:Seeman NC (2010).
1517:molecular modeling
1513:
1467:tertiary structure
1411:reaction mechanism
1214:directed evolution
1132:chemical synthesis
1096:structural biology
964:
918:gold nanoparticles
892:Templated assembly
789:cellular automaton
716:
586:Holliday junctions
467:
447:
443:tertiary structure
393:proof of principle
356:Andrew Turberfield
308:
195:structural biology
183:molecular machines
155:DNA nanotechnology
152:
76:Science portal
58:DNA nanotechnology
7116:
7115:
6771:10.1038/nchem.957
6697:(12): 1856–1876.
6691:Angewandte Chemie
6462:10.1021/nl101262u
6377:978-0-935702-49-1
6343:978-1-61779-141-3
5700:10.1021/nn5039433
5638:Angewandte Chemie
5610:10.1021/nl304147f
5471:(6109): 932–936.
5388:(20): 2010–2012.
5279:10.1021/nn2005574
5211:10.2976/1.2896331
5171:10.1021/nn303767b
5136:978-0-387-98988-4
4987:(15): 1811–1817.
4935:(7176): 318–322.
4721:(7295): 206–210.
4681:(28): 4358–4361.
4675:Angewandte Chemie
4637:(28): 4355–4358.
4631:Angewandte Chemie
4593:(21): 6948–6951.
4587:Angewandte Chemie
4571:10.1021/nl049527q
4527:10.1021/ja047543j
4410:(6070): 831–834.
4322:(36): 4342–4346.
4316:Angewandte Chemie
4218:(6796): 605–608.
4166:(6715): 144–146.
4130:(31): 4068–4070.
4124:Angewandte Chemie
4100:10.1021/nn1035075
4010:(42): 7956–7959.
4004:Angewandte Chemie
3954:Angewandte Chemie
3916:10.1021/nl060994c
3854:(15): 2420–2443.
3777:10.1021/nl901165f
3591:(6975): 618–621.
3453:(5890): 824–826.
3419:10.1021/ja044319l
3373:(12): 1856–1876.
3367:Angewandte Chemie
3336:10.1021/nl052038l
3312:(12): 2586–2592.
3272:10.1021/nl050084f
3208:(18): 3414–3419.
3182:10.1021/ja9900398
3176:(23): 5437–5443.
3154:10.1021/ja982824a
3096:(6693): 539–544.
2751:10.1038/nchem.957
2597:978-0-19-508467-2
2571:978-1-58488-687-7
2471:10.1021/nl101262u
2319:(7245): 414–418.
2260:(7082): 297–302.
2065:(6796): 605–608.
1986:978-3-540-30295-7
1729:978-1-58488-687-7
1639:Nanobiotechnology
1616:Technology portal
1372:Structural design
1366:primary structure
1282:membrane proteins
1218:Oxford University
1192:, as well as for
1167:Aarhus University
1124:membrane proteins
1020:T7 RNA polymerase
816:emergent property
797:Sierpinski gasket
791:that generates a
770:Holliday junction
747:Sierpinski gasket
643:Extended lattices
457:consists of five
289:Holliday junction
124:
123:
7136:
7106:
7105:
7094:
7093:
7080:
7079:
7068:
7067:
7052:Mechanosynthesis
6943:characterization
6825:
6818:
6811:
6802:
6801:
6782:
6751:Nature Chemistry
6744:
6727:(8): 1641–1647.
6714:
6684:
6639:
6629:
6619:
6610:(6): 7149–7162.
6593:
6565:
6527:
6517:
6483:
6473:
6448:(6): 1971–1978.
6431:
6382:
6381:
6362:
6356:
6355:
6320:
6314:
6313:
6266:
6260:
6259:
6212:
6206:
6205:
6170:
6164:
6163:
6116:
6110:
6109:
6062:
6056:
6055:
6045:
6028:(4): 1392–1403.
6012:
6001:
6000:
5983:(8): 1641–1647.
5971:
5965:
5964:
5962:
5937:
5926:
5925:
5915:
5876:DNA scramblase:
5874:
5868:
5867:
5857:
5840:(9): 8207–8214.
5824:
5818:
5817:
5776:
5770:
5769:
5744:(5): 3134–3138.
5728:
5722:
5721:
5711:
5694:(2): 1117–1126.
5678:
5672:
5671:
5661:
5628:
5622:
5621:
5603:
5586:(6): 2351–2356.
5574:
5568:
5567:
5557:
5532:(7): 4665–4669.
5516:
5507:
5506:
5496:
5455:
5449:
5448:
5412:
5406:
5405:
5377:
5371:
5370:
5360:
5320:
5314:
5313:
5311:
5309:
5297:
5291:
5290:
5273:(7): 5427–5432.
5262:
5256:
5255:
5253:
5251:
5239:
5233:
5232:
5222:
5189:
5183:
5182:
5154:
5148:
5147:
5145:
5143:
5119:
5113:
5112:
5067:
5061:
5060:
5011:
5005:
5004:
4975:
4969:
4968:
4919:
4908:
4907:
4879:
4873:
4872:
4862:
4821:
4815:
4814:
4804:
4763:
4757:
4756:
4746:
4705:
4699:
4698:
4669:
4663:
4662:
4625:
4619:
4618:
4581:
4575:
4574:
4557:(7): 1203–1207.
4545:
4539:
4538:
4505:
4499:
4498:
4488:
4470:
4450:
4444:
4443:
4398:
4392:
4391:
4354:
4348:
4347:
4310:
4304:
4303:
4258:
4252:
4251:
4232:10.1038/35020524
4206:
4200:
4199:
4154:
4148:
4147:
4118:
4112:
4111:
4094:(3): 2240–2247.
4082:
4076:
4075:
4034:
4028:
4027:
3994:
3988:
3987:
3977:
3944:
3938:
3937:
3927:
3902:(7): 1502–1504.
3886:
3880:
3879:
3842:
3836:
3835:
3810:(3–4): 178–188.
3795:
3789:
3788:
3763:(6): 2445–2447.
3751:
3745:
3744:
3726:
3689:
3680:
3679:
3631:
3625:
3624:
3579:
3573:
3572:
3563:(5): 1661–1669.
3551:
3545:
3544:
3534:
3493:
3487:
3486:
3437:
3431:
3430:
3397:
3391:
3390:
3361:
3348:
3347:
3329:
3300:
3294:
3293:
3283:
3242:
3236:
3235:
3225:
3214:10.1039/b605212f
3192:
3186:
3185:
3164:
3158:
3157:
3136:
3130:
3129:
3084:
3078:
3077:
3067:
3057:
3032:
3023:
3022:
2978:
2972:
2971:
2961:
2951:
2926:
2920:
2919:
2885:
2864:
2858:
2857:
2828:
2822:
2821:
2811:
2778:
2763:
2762:
2731:Nature Chemistry
2725:
2712:
2711:
2701:
2660:
2654:
2653:
2608:
2602:
2601:
2582:
2576:
2575:
2556:
2550:
2549:
2539:
2529:
2504:
2493:
2492:
2482:
2457:(6): 1971–1978.
2441:
2430:
2429:
2427:
2425:
2416:. Archived from
2404:
2398:
2397:
2361:
2355:
2354:
2344:
2303:
2294:
2293:
2251:
2241:
2230:
2229:
2192:
2186:
2185:
2156:
2150:
2149:
2112:
2099:
2098:
2079:10.1038/35020524
2050:
2044:
2043:
2006:
1991:
1990:
1970:
1953:
1944:
1943:
1933:
1892:
1869:
1868:
1858:
1825:
1810:
1808:
1801:
1793:
1787:
1786:
1749:
1734:
1733:
1714:
1703:
1702:
1657:
1618:
1613:
1612:
1604:
1599:
1598:
1463:nearest-neighbor
1457:methods such as
1358:target structure
1322:lipid headgroups
1292:via hydrophobic
1258:folate receptors
1238:RNA Interference
1110:rods to replace
1059:electric current
955:branch migration
857:steps to create
812:carbon nanotubes
735:
726:
664:
655:
609:branch migration
594:structural motif
324:structural motif
285:replication fork
171:crystal lattices
116:
109:
102:
86:
85:
74:
73:
40:
21:
20:
7144:
7143:
7139:
7138:
7137:
7135:
7134:
7133:
7119:
7118:
7117:
7112:
7100:
7088:
7056:
7028:
7005:
7001:Nanolithography
6988:Nanoelectronics
6982:
6949:
6904:
6867:
6858:Popular culture
6834:
6829:
6791:
6786:
6643:
6390:
6388:Further reading
6385:
6378:
6363:
6359:
6344:
6321:
6317:
6294:
6267:
6263:
6240:
6213:
6209:
6194:
6178:. Vol. 5.
6171:
6167:
6144:
6117:
6113:
6090:
6063:
6059:
6013:
6004:
5972:
5968:
5938:
5929:
5875:
5871:
5825:
5821:
5777:
5773:
5729:
5725:
5679:
5675:
5629:
5625:
5601:10.1.1.659.7660
5575:
5571:
5517:
5510:
5456:
5452:
5413:
5409:
5378:
5374:
5321:
5317:
5307:
5305:
5298:
5294:
5263:
5259:
5249:
5247:
5240:
5236:
5190:
5186:
5155:
5151:
5141:
5139:
5137:
5120:
5116:
5068:
5064:
5012:
5008:
4976:
4972:
4920:
4911:
4880:
4876:
4837:(11): 778–782.
4822:
4818:
4764:
4760:
4706:
4702:
4670:
4666:
4626:
4622:
4582:
4578:
4546:
4542:
4506:
4502:
4451:
4447:
4399:
4395:
4355:
4351:
4311:
4307:
4284:10.1038/415062a
4270:(6867): 62–65.
4259:
4255:
4207:
4203:
4155:
4151:
4119:
4115:
4083:
4079:
4035:
4031:
3995:
3991:
3945:
3941:
3887:
3883:
3843:
3839:
3796:
3792:
3752:
3748:
3701:(7243): 73–76.
3690:
3683:
3646:(7683): 67–71.
3632:
3628:
3581:DNA polyhedra:
3580:
3576:
3553:DNA polyhedra:
3552:
3548:
3509:(7260): 74–77.
3494:
3490:
3439:DNA nanotubes:
3438:
3434:
3399:DNA nanotubes:
3398:
3394:
3362:
3351:
3301:
3297:
3243:
3239:
3193:
3189:
3165:
3161:
3137:
3133:
3085:
3081:
3033:
3026:
2979:
2975:
2927:
2923:
2865:
2861:
2829:
2825:
2779:
2766:
2726:
2715:
2676:(12): 833–842.
2661:
2657:
2609:
2605:
2598:
2583:
2579:
2572:
2557:
2553:
2505:
2496:
2442:
2433:
2423:
2421:
2405:
2401:
2362:
2358:
2304:
2297:
2249:
2242:
2233:
2193:
2189:
2157:
2153:
2113:
2102:
2051:
2047:
2007:
1994:
1987:
1968:10.1.1.144.1380
1954:
1947:
1908:(12): 763–772.
1893:
1872:
1826:
1813:
1803:
1796:
1794:
1790:
1750:
1737:
1730:
1715:
1706:
1659:DNA polyhedra:
1658:
1651:
1647:
1614:
1607:
1600:
1593:
1590:
1505:formation assay
1494:
1446:
1440:
1438:Sequence design
1374:
1350:
1334:lipid flip-flop
1290:lipid membranes
1148:molecular wires
1140:protein folding
1118:experiments in
1112:liquid crystals
1104:crystallography
1088:
1033:
979:
973:
946:
894:
884:molecular cages
831:
757:
756:
755:
754:
738:
737:
736:
728:
727:
701:
700:
699:
698:
667:
666:
665:
657:
656:
645:
630:
569:
472:
411:
405:
344:DNA nanomachine
287:and the mobile
265:
138:, such as this
120:
80:
68:
17:
12:
11:
5:
7142:
7132:
7131:
7114:
7113:
7111:
7110:
7098:
7086:
7074:
7061:
7058:
7057:
7055:
7054:
7049:
7044:
7038:
7036:
7030:
7029:
7027:
7026:
7021:
7015:
7013:
7007:
7006:
7004:
7003:
6998:
6992:
6990:
6984:
6983:
6981:
6980:
6975:
6970:
6965:
6959:
6957:
6951:
6950:
6948:
6947:
6946:
6945:
6935:
6934:
6933:
6928:
6920:
6914:
6912:
6906:
6905:
6903:
6902:
6897:
6892:
6890:Nanotoxicology
6887:
6881:
6879:
6869:
6868:
6866:
6865:
6860:
6855:
6850:
6844:
6842:
6836:
6835:
6832:Nanotechnology
6828:
6827:
6820:
6813:
6805:
6799:
6798:
6790:
6789:External links
6787:
6785:
6784:
6757:(2): 103–113.
6746:
6716:
6686:
6659:(5): 275–284.
6647:
6642:
6641:
6595:
6576:(6034): 1142.
6529:
6485:
6433:
6394:
6389:
6386:
6384:
6383:
6376:
6357:
6342:
6315:
6293:978-0471142706
6292:
6261:
6239:978-0471142720
6238:
6207:
6193:978-0470089934
6192:
6165:
6143:978-0471142720
6142:
6111:
6089:978-0471142720
6088:
6057:
6002:
5966:
5927:
5869:
5819:
5792:(2): 152–156.
5771:
5723:
5673:
5623:
5569:
5508:
5450:
5423:(9): 684–694.
5407:
5372:
5335:(6): 389–393.
5315:
5292:
5257:
5234:
5191:Applications:
5184:
5149:
5135:
5121:Applications:
5114:
5062:
5006:
4970:
4909:
4874:
4816:
4779:(6): 496–503.
4758:
4700:
4664:
4620:
4576:
4540:
4500:
4445:
4400:Applications:
4393:
4356:DNA machines:
4349:
4312:DNA machines:
4305:
4260:DNA machines:
4253:
4208:DNA machines:
4201:
4156:DNA machines:
4149:
4113:
4077:
4029:
3989:
3960:(5): 735–739.
3939:
3881:
3837:
3790:
3746:
3681:
3626:
3574:
3546:
3488:
3432:
3392:
3349:
3327:10.1.1.155.676
3295:
3258:(4): 661–665.
3244:Other arrays:
3237:
3194:Other arrays:
3187:
3166:Other arrays:
3159:
3148:(5): 917–922.
3131:
3079:
3024:
2973:
2928:Other arrays:
2921:
2859:
2840:(6): 580–588.
2823:
2794:(3): 246–257.
2764:
2737:(2): 103–113.
2713:
2655:
2603:
2596:
2577:
2570:
2551:
2494:
2431:
2399:
2356:
2295:
2231:
2204:(4): 313–317.
2187:
2168:(4): 295–300.
2151:
2124:(5): 275–284.
2114:DNA machines:
2100:
2045:
1992:
1985:
1945:
1870:
1811:
1788:
1735:
1728:
1704:
1648:
1646:
1643:
1642:
1641:
1636:
1631:
1626:
1620:
1619:
1605:
1602:Science portal
1589:
1586:
1542:methods using
1493:
1490:
1478:protein design
1442:Main article:
1439:
1436:
1435:
1434:
1399:
1387:
1373:
1370:
1349:
1346:
1274:P-glycoprotein
1126:in solution.
1087:
1084:
1078:junctions and
1032:
1029:
1011:deoxyribozymes
975:Main article:
972:
969:
945:
942:
893:
890:
830:
827:
740:
739:
730:
729:
721:
720:
719:
718:
717:
669:
668:
659:
658:
650:
649:
648:
647:
646:
644:
641:
629:
626:
614:deoxyribozymes
568:
565:
506:protein design
475:Nanotechnology
471:
468:
463:double-helical
410:
407:
332:Paul Rothemund
302:(pictured) by
269:Nadrian Seeman
264:
261:
233:base sequences
222:Nadrian Seeman
163:nanotechnology
122:
121:
119:
118:
111:
104:
96:
93:
92:
91:
90:
78:
63:
62:
61:
60:
55:
50:
42:
41:
33:
32:
26:
25:
15:
9:
6:
4:
3:
2:
7141:
7130:
7127:
7126:
7124:
7109:
7104:
7099:
7097:
7092:
7087:
7085:
7084:
7075:
7073:
7072:
7063:
7062:
7059:
7053:
7050:
7048:
7045:
7043:
7040:
7039:
7037:
7035:
7031:
7025:
7022:
7020:
7017:
7016:
7014:
7012:
7008:
7002:
6999:
6997:
6994:
6993:
6991:
6989:
6985:
6979:
6976:
6974:
6971:
6969:
6966:
6964:
6961:
6960:
6958:
6956:
6952:
6944:
6941:
6940:
6939:
6938:Nanoparticles
6936:
6932:
6929:
6927:
6924:
6923:
6921:
6919:
6916:
6915:
6913:
6911:
6910:Nanomaterials
6907:
6901:
6898:
6896:
6893:
6891:
6888:
6886:
6883:
6882:
6880:
6878:
6874:
6870:
6864:
6861:
6859:
6856:
6854:
6853:Organizations
6851:
6849:
6846:
6845:
6843:
6841:
6837:
6833:
6826:
6821:
6819:
6814:
6812:
6807:
6806:
6803:
6796:
6793:
6792:
6780:
6776:
6772:
6768:
6764:
6760:
6756:
6752:
6747:
6742:
6738:
6734:
6730:
6726:
6722:
6717:
6712:
6708:
6704:
6700:
6696:
6692:
6687:
6682:
6678:
6674:
6670:
6666:
6662:
6658:
6654:
6649:
6648:
6646:
6637:
6633:
6628:
6623:
6618:
6613:
6609:
6605:
6601:
6596:
6591:
6587:
6583:
6579:
6575:
6571:
6563:
6559:
6555:
6551:
6547:
6543:
6539:
6535:
6530:
6525:
6521:
6516:
6511:
6507:
6503:
6499:
6495:
6491:
6486:
6481:
6477:
6472:
6467:
6463:
6459:
6455:
6451:
6447:
6443:
6439:
6434:
6429:
6425:
6421:
6417:
6413:
6409:
6405:
6401:
6396:
6395:
6393:
6379:
6373:
6369:
6361:
6353:
6349:
6345:
6339:
6335:
6331:
6327:
6319:
6311:
6307:
6303:
6299:
6295:
6289:
6285:
6281:
6277:
6273:
6265:
6257:
6253:
6249:
6245:
6241:
6235:
6231:
6227:
6223:
6219:
6211:
6203:
6199:
6195:
6189:
6185:
6181:
6177:
6169:
6161:
6157:
6153:
6149:
6145:
6139:
6135:
6131:
6127:
6123:
6115:
6107:
6103:
6099:
6095:
6091:
6085:
6081:
6077:
6073:
6069:
6061:
6053:
6049:
6044:
6039:
6035:
6031:
6027:
6023:
6019:
6011:
6009:
6007:
5998:
5994:
5990:
5986:
5982:
5978:
5970:
5961:
5956:
5952:
5948:
5944:
5936:
5934:
5932:
5923:
5919:
5914:
5909:
5905:
5901:
5897:
5893:
5889:
5885:
5881:
5873:
5865:
5861:
5856:
5851:
5847:
5843:
5839:
5835:
5831:
5823:
5815:
5811:
5807:
5803:
5799:
5795:
5791:
5787:
5783:
5775:
5767:
5763:
5759:
5755:
5751:
5747:
5743:
5739:
5735:
5727:
5719:
5715:
5710:
5705:
5701:
5697:
5693:
5689:
5685:
5677:
5669:
5665:
5660:
5655:
5651:
5647:
5643:
5639:
5635:
5627:
5619:
5615:
5611:
5607:
5602:
5597:
5593:
5589:
5585:
5581:
5573:
5565:
5561:
5556:
5551:
5547:
5543:
5539:
5535:
5531:
5527:
5523:
5515:
5513:
5504:
5500:
5495:
5490:
5486:
5482:
5478:
5474:
5470:
5466:
5462:
5454:
5446:
5442:
5438:
5434:
5430:
5426:
5422:
5418:
5411:
5403:
5399:
5395:
5391:
5387:
5383:
5376:
5368:
5364:
5359:
5354:
5350:
5346:
5342:
5338:
5334:
5330:
5326:
5319:
5303:
5296:
5288:
5284:
5280:
5276:
5272:
5268:
5261:
5245:
5238:
5230:
5226:
5221:
5216:
5212:
5208:
5205:(2): 99–109.
5204:
5200:
5196:
5188:
5180:
5176:
5172:
5168:
5164:
5160:
5153:
5138:
5132:
5128:
5127:
5118:
5110:
5106:
5102:
5098:
5094:
5090:
5086:
5082:
5078:
5074:
5066:
5058:
5054:
5050:
5046:
5042:
5038:
5034:
5030:
5026:
5022:
5018:
5010:
5002:
4998:
4994:
4990:
4986:
4982:
4974:
4966:
4962:
4958:
4954:
4950:
4946:
4942:
4938:
4934:
4930:
4926:
4918:
4916:
4914:
4905:
4901:
4897:
4893:
4889:
4885:
4878:
4870:
4866:
4861:
4856:
4852:
4848:
4844:
4840:
4836:
4832:
4828:
4820:
4812:
4808:
4803:
4798:
4794:
4790:
4786:
4782:
4778:
4774:
4770:
4762:
4754:
4750:
4745:
4740:
4736:
4732:
4728:
4724:
4720:
4716:
4712:
4704:
4696:
4692:
4688:
4684:
4680:
4676:
4671:DNA walkers:
4668:
4660:
4656:
4652:
4648:
4644:
4640:
4636:
4632:
4627:DNA walkers:
4624:
4616:
4612:
4608:
4604:
4600:
4596:
4592:
4588:
4583:DNA walkers:
4580:
4572:
4568:
4564:
4560:
4556:
4552:
4547:DNA walkers:
4544:
4536:
4532:
4528:
4524:
4520:
4516:
4512:
4507:DNA walkers:
4504:
4496:
4492:
4487:
4482:
4478:
4474:
4469:
4464:
4460:
4456:
4449:
4441:
4437:
4433:
4429:
4425:
4421:
4417:
4413:
4409:
4405:
4397:
4389:
4385:
4381:
4377:
4373:
4369:
4365:
4361:
4353:
4345:
4341:
4337:
4333:
4329:
4325:
4321:
4317:
4309:
4301:
4297:
4293:
4289:
4285:
4281:
4277:
4273:
4269:
4265:
4257:
4249:
4245:
4241:
4237:
4233:
4229:
4225:
4221:
4217:
4213:
4205:
4197:
4193:
4189:
4185:
4181:
4180:10.1038/16437
4177:
4173:
4169:
4165:
4161:
4153:
4145:
4141:
4137:
4133:
4129:
4125:
4117:
4109:
4105:
4101:
4097:
4093:
4089:
4081:
4073:
4069:
4065:
4061:
4057:
4053:
4049:
4045:
4041:
4033:
4025:
4021:
4017:
4013:
4009:
4005:
4001:
3993:
3985:
3981:
3976:
3971:
3967:
3963:
3959:
3955:
3951:
3943:
3935:
3931:
3926:
3921:
3917:
3913:
3909:
3905:
3901:
3897:
3893:
3885:
3877:
3873:
3869:
3865:
3861:
3857:
3853:
3849:
3841:
3833:
3829:
3825:
3821:
3817:
3813:
3809:
3805:
3801:
3794:
3786:
3782:
3778:
3774:
3770:
3766:
3762:
3758:
3750:
3742:
3738:
3734:
3730:
3725:
3720:
3716:
3712:
3708:
3704:
3700:
3696:
3688:
3686:
3677:
3673:
3669:
3665:
3661:
3657:
3653:
3649:
3645:
3641:
3637:
3630:
3622:
3618:
3614:
3610:
3606:
3602:
3598:
3594:
3590:
3586:
3578:
3570:
3566:
3562:
3558:
3550:
3542:
3538:
3533:
3528:
3524:
3520:
3516:
3512:
3508:
3504:
3500:
3492:
3484:
3480:
3476:
3472:
3468:
3464:
3460:
3456:
3452:
3448:
3444:
3436:
3428:
3424:
3420:
3416:
3412:
3408:
3404:
3396:
3388:
3384:
3380:
3376:
3372:
3368:
3360:
3358:
3356:
3354:
3345:
3341:
3337:
3333:
3328:
3323:
3319:
3315:
3311:
3307:
3299:
3291:
3287:
3282:
3277:
3273:
3269:
3265:
3261:
3257:
3253:
3249:
3241:
3233:
3229:
3224:
3219:
3215:
3211:
3207:
3203:
3199:
3191:
3183:
3179:
3175:
3171:
3163:
3155:
3151:
3147:
3143:
3135:
3127:
3123:
3119:
3115:
3111:
3110:10.1038/28998
3107:
3103:
3099:
3095:
3091:
3083:
3075:
3071:
3066:
3061:
3056:
3051:
3047:
3043:
3039:
3031:
3029:
3020:
3016:
3012:
3008:
3004:
3000:
2996:
2992:
2988:
2984:
2977:
2969:
2965:
2960:
2955:
2950:
2945:
2941:
2937:
2933:
2925:
2917:
2913:
2909:
2905:
2901:
2897:
2893:
2889:
2884:
2879:
2875:
2871:
2863:
2855:
2851:
2847:
2843:
2839:
2835:
2827:
2819:
2815:
2810:
2805:
2801:
2797:
2793:
2789:
2785:
2777:
2775:
2773:
2771:
2769:
2760:
2756:
2752:
2748:
2744:
2740:
2736:
2732:
2724:
2722:
2720:
2718:
2709:
2705:
2700:
2695:
2691:
2687:
2683:
2679:
2675:
2671:
2667:
2659:
2651:
2647:
2643:
2639:
2635:
2631:
2627:
2623:
2619:
2615:
2607:
2599:
2593:
2589:
2581:
2573:
2567:
2563:
2555:
2547:
2543:
2538:
2533:
2528:
2523:
2519:
2515:
2511:
2503:
2501:
2499:
2490:
2486:
2481:
2476:
2472:
2468:
2464:
2460:
2456:
2452:
2448:
2440:
2438:
2436:
2419:
2415:
2414:The Scientist
2411:
2403:
2395:
2391:
2387:
2383:
2379:
2375:
2371:
2367:
2360:
2352:
2348:
2343:
2338:
2334:
2330:
2326:
2322:
2318:
2314:
2310:
2305:DNA origami:
2302:
2300:
2291:
2287:
2283:
2279:
2275:
2271:
2267:
2263:
2259:
2255:
2248:
2243:DNA origami:
2240:
2238:
2236:
2227:
2223:
2219:
2215:
2211:
2207:
2203:
2199:
2191:
2183:
2179:
2175:
2171:
2167:
2163:
2155:
2147:
2143:
2139:
2135:
2131:
2127:
2123:
2119:
2111:
2109:
2107:
2105:
2096:
2092:
2088:
2084:
2080:
2076:
2072:
2068:
2064:
2060:
2056:
2049:
2041:
2037:
2033:
2029:
2025:
2021:
2017:
2013:
2005:
2003:
2001:
1999:
1997:
1988:
1982:
1978:
1974:
1969:
1964:
1960:
1955:DNA origami:
1952:
1950:
1941:
1937:
1932:
1927:
1923:
1919:
1915:
1911:
1907:
1903:
1899:
1891:
1889:
1887:
1885:
1883:
1881:
1879:
1877:
1875:
1866:
1862:
1857:
1852:
1848:
1844:
1840:
1836:
1832:
1824:
1822:
1820:
1818:
1816:
1806:
1799:
1795:History: See
1792:
1784:
1780:
1776:
1772:
1768:
1764:
1760:
1756:
1748:
1746:
1744:
1742:
1740:
1731:
1725:
1721:
1713:
1711:
1709:
1700:
1696:
1692:
1688:
1684:
1680:
1676:
1672:
1668:
1664:
1656:
1654:
1649:
1640:
1637:
1635:
1632:
1630:
1627:
1625:
1622:
1621:
1617:
1611:
1606:
1603:
1597:
1592:
1585:
1583:
1579:
1575:
1571:
1566:
1564:
1560:
1556:
1552:
1547:
1545:
1541:
1537:
1534:
1530:
1526:
1522:
1518:
1510:
1506:
1502:
1498:
1489:
1487:
1483:
1479:
1474:
1472:
1468:
1464:
1460:
1456:
1452:
1445:
1432:
1428:
1424:
1420:
1416:
1412:
1409:steps in the
1408:
1404:
1400:
1397:
1393:
1388:
1385:
1384:DNA computing
1380:
1379:
1378:
1369:
1367:
1363:
1359:
1355:
1345:
1343:
1339:
1335:
1331:
1327:
1323:
1319:
1315:
1314:lipid bilayer
1311:
1307:
1303:
1299:
1295:
1291:
1287:
1283:
1278:
1275:
1271:
1267:
1263:
1259:
1255:
1251:
1247:
1243:
1239:
1235:
1231:
1227:
1223:
1219:
1215:
1211:
1207:
1203:
1199:
1195:
1191:
1187:
1186:biocompatible
1182:
1180:
1176:
1172:
1168:
1164:
1159:
1157:
1153:
1149:
1143:
1141:
1138:function and
1137:
1133:
1129:
1125:
1121:
1117:
1113:
1109:
1105:
1101:
1097:
1093:
1092:basic science
1083:
1081:
1076:
1071:
1069:
1065:
1060:
1056:
1051:
1049:
1044:
1039:
1028:
1025:
1021:
1017:
1012:
1008:
1003:
999:
996:
992:
988:
984:
978:
968:
961:
956:
950:
941:
939:
935:
931:
927:
923:
919:
915:
911:
907:
903:
899:
889:
887:
885:
881:
877:
876:stoichiometry
873:
872:base sequence
868:
863:
860:
856:
852:
848:
844:
840:
836:
826:
824:
819:
817:
813:
809:
804:
802:
798:
795:known as the
794:
790:
786:
782:
776:
774:
771:
766:
762:
752:
748:
744:
734:
725:
713:
709:
705:
696:
692:
688:
684:
681:
680:complementary
677:
673:
663:
654:
640:
638:
635:
625:
623:
619:
615:
610:
606:
602:
597:
595:
591:
590:base sequence
587:
583:
578:
575:
564:
562:
558:
554:
550:
546:
545:complementary
542:
538:
534:
530:
526:
522:
518:
513:
511:
510:nanoparticles
507:
503:
499:
495:
491:
487:
484:
480:
476:
464:
460:
456:
451:
444:
440:
436:
432:
428:
424:
420:
415:
406:
403:
400:
399:
394:
389:
387:
383:
379:
378:
373:
368:
363:
361:
357:
353:
349:
348:Bernard Yurke
345:
340:
338:
333:
329:
325:
321:
317:
313:
305:
301:
296:
292:
290:
286:
282:
278:
274:
270:
260:
258:
254:
250:
246:
242:
238:
234:
231:
230:complementary
227:
223:
218:
216:
212:
208:
204:
200:
196:
192:
191:basic science
188:
187:DNA computers
184:
180:
176:
172:
168:
164:
160:
156:
149:
145:
141:
137:
136:nucleic acids
133:
128:
117:
112:
110:
105:
103:
98:
97:
95:
94:
89:
84:
79:
77:
72:
67:
66:
65:
64:
59:
56:
54:
51:
49:
46:
45:
44:
43:
39:
35:
34:
31:
28:
27:
23:
22:
19:
7081:
7069:
7047:Nanorobotics
6972:
6885:Nanomedicine
6877:applications
6754:
6750:
6724:
6721:ChemPhysChem
6720:
6694:
6690:
6656:
6652:
6644:
6607:
6603:
6573:
6569:
6537:
6533:
6497:
6493:
6445:
6442:Nano Letters
6441:
6406:(6): 64–75.
6403:
6399:
6391:
6367:
6360:
6325:
6318:
6275:
6271:
6264:
6221:
6217:
6210:
6175:
6168:
6128:: Unit2.12.
6125:
6121:
6114:
6071:
6067:
6060:
6025:
6021:
5980:
5977:ChemPhysChem
5976:
5969:
5950:
5946:
5887:
5883:
5872:
5837:
5833:
5822:
5789:
5785:
5774:
5741:
5738:Nano Letters
5737:
5726:
5691:
5687:
5676:
5641:
5637:
5626:
5583:
5580:Nano Letters
5579:
5572:
5529:
5526:Nano Letters
5525:
5468:
5464:
5453:
5420:
5416:
5410:
5385:
5381:
5375:
5332:
5328:
5318:
5308:22 September
5306:. Retrieved
5295:
5270:
5266:
5260:
5250:22 September
5248:. Retrieved
5237:
5202:
5199:HFSP Journal
5198:
5187:
5162:
5158:
5152:
5140:. Retrieved
5125:
5117:
5076:
5072:
5065:
5024:
5020:
5009:
4984:
4980:
4973:
4932:
4928:
4887:
4883:
4877:
4834:
4830:
4819:
4776:
4772:
4761:
4718:
4714:
4703:
4678:
4674:
4667:
4634:
4630:
4623:
4590:
4586:
4579:
4554:
4551:Nano Letters
4550:
4543:
4518:
4514:
4503:
4458:
4448:
4407:
4403:
4396:
4366:(2): 93–96.
4363:
4359:
4352:
4319:
4315:
4308:
4267:
4263:
4256:
4215:
4211:
4204:
4163:
4159:
4152:
4127:
4123:
4116:
4091:
4087:
4080:
4050:(1): 61–66.
4047:
4043:
4032:
4007:
4003:
3992:
3957:
3953:
3942:
3899:
3896:Nano Letters
3895:
3884:
3851:
3847:
3840:
3807:
3803:
3793:
3760:
3757:Nano Letters
3756:
3749:
3698:
3694:
3643:
3639:
3629:
3588:
3584:
3577:
3560:
3556:
3549:
3506:
3502:
3491:
3450:
3446:
3435:
3410:
3406:
3395:
3370:
3366:
3309:
3306:Nano Letters
3305:
3298:
3255:
3252:Nano Letters
3251:
3240:
3205:
3201:
3190:
3173:
3169:
3162:
3145:
3141:
3134:
3093:
3089:
3082:
3048:(12): e424.
3045:
3042:PLOS Biology
3041:
2986:
2982:
2976:
2939:
2936:PLOS Biology
2935:
2924:
2873:
2869:
2862:
2837:
2833:
2826:
2791:
2787:
2734:
2730:
2673:
2669:
2658:
2617:
2613:
2606:
2587:
2584:Background:
2580:
2561:
2558:Background:
2554:
2520:(12): e431.
2517:
2514:PLOS Biology
2513:
2454:
2451:Nano Letters
2450:
2422:. Retrieved
2418:the original
2413:
2402:
2369:
2365:
2359:
2316:
2312:
2257:
2253:
2201:
2197:
2190:
2165:
2161:
2154:
2121:
2117:
2062:
2058:
2048:
2015:
2011:
1958:
1905:
1901:
1838:
1834:
1791:
1761:(6): 64–75.
1758:
1754:
1719:
1666:
1662:
1567:
1548:
1514:
1475:
1447:
1427:isothermally
1407:intermediate
1375:
1351:
1306:ion channels
1285:
1279:
1183:
1160:
1144:
1094:problems in
1089:
1086:Applications
1072:
1052:
1034:
1000:
983:nanorobotics
980:
965:
922:streptavidin
898:quantum dots
895:
888:
864:
837:, such as a
832:
820:
805:
777:
758:
750:
742:
711:
707:
686:
671:
639:
631:
598:
582:double helix
579:
570:
514:
504:, for which
490:base pairing
473:
404:
396:
390:
386:Jørgen Kjems
381:
375:
372:William Shih
364:
352:Niles Pierce
341:
328:Erik Winfree
309:
304:M. C. Escher
299:
298:The woodcut
281:M. C. Escher
276:
266:
256:
237:double helix
226:base pairing
219:
215:nanomedicine
193:problems in
159:nucleic acid
154:
153:
148:double helix
57:
18:
6224:: Unit2.7.
5890:(1): 2426.
3848:ChemBioChem
3753:DNA boxes:
3691:DNA boxes:
3138:DX arrays:
3086:DX arrays:
1482:nucleotides
1392:DNA origami
1342:scramblases
1294:cholesterol
1270:Doxorubicin
1234:tetrahedron
1206:transcribed
1202:cancer cell
1128:DNA walkers
1108:DNA origami
1068:square root
1064:logic gates
1002:DNA walkers
977:DNA machine
938:lithography
880:smiley face
867:DNA origami
765:tessellated
761:sticky ends
683:sticky ends
574:equilibrium
521:nucleotides
433:matched to
425:matched to
367:DNA origami
360:Chengde Mao
249:DNA origami
6931:Non-carbon
6922:Nanotubes
6918:Fullerenes
6900:Regulation
5973:Overview:
5304:. MIT News
3844:Overview:
2942:(3): E73.
2506:Overview:
1894:Overview:
1827:Overview:
1751:Overview:
1645:References
1533:denaturing
1310:DNA duplex
1298:DNA duplex
1262:luciferase
1222:tetrahedra
1194:diagnostic
1177:(TEM) and
1152:breadboard
1100:biophysics
1080:dendrimers
932:device, a
906:fullerenes
843:octahedron
835:polyhedron
823:tensegrity
781:Wang tiles
525:nucleobase
479:nanometers
439:this image
419:base pairs
337:Wang tiles
199:biophysics
6500:: 65–87.
6392:General:
6364:Methods:
6322:Methods:
6268:Methods:
6214:Methods:
6172:Methods:
6118:Methods:
6106:205152989
6064:Methods:
5953:: 39–58.
5596:CiteSeerX
5445:195825879
4890:: 56–64.
4477:1748-3395
3876:205554125
3832:246946068
3824:2634-8276
3322:CiteSeerX
3019:137635908
2883:1308.3843
2443:History:
2406:History:
2087:1476-4687
1963:CiteSeerX
1841:: 65–87.
1716:History:
1455:heuristic
1431:annealing
1210:cytoplasm
1198:apoptosis
1048:catalytic
1016:catenated
859:catenated
851:ligations
749:fractal.
618:ribozymes
567:Subfields
539:(G), and
517:structure
483:bottom-up
245:nanoscale
179:polyhedra
175:nanotubes
144:base pair
7123:Category
7071:Category
6840:Overview
6779:21258382
6741:16832805
6711:16470892
6681:18654284
6636:22837684
6590:21636755
6562:21636754
6524:20222824
6480:20486672
6428:15195395
6352:21674361
6302:18428904
6256:43406338
6248:18265187
6202:94329398
6160:27187583
6152:18265180
6098:18265179
6052:14990744
6014:Design:
5997:16832805
5939:Design:
5922:29930243
5864:27504755
5834:ACS Nano
5814:26751170
5766:25816075
5718:25338165
5688:ACS Nano
5668:24014236
5618:23611515
5564:27324157
5503:23161995
5437:31285580
5402:23380739
5367:22659608
5287:21696187
5267:ACS Nano
5229:19404476
5179:23030709
5159:ACS Nano
5142:17 April
5109:10053541
5101:21636773
5057:10966324
5049:17158324
5001:25565140
4957:18202654
4904:25498478
4869:20935654
4811:29632399
4753:20463735
4695:15959864
4659:15945114
4615:85446523
4607:30897257
4535:15339155
4495:37857824
4486:10873200
4461:: 1–11.
4432:22344439
4388:18654468
4344:14502706
4300:52801697
4292:11780115
4240:10949296
4144:15300697
4108:21323323
4088:ACS Nano
4072:19898497
4024:17763481
3984:16374784
3934:16834438
3868:19714700
3785:19419184
3733:19424153
3668:29219965
3613:14961116
3541:19727196
3483:12100380
3475:18687961
3427:15600335
3387:16470892
3363:Design:
3344:16351220
3290:15826105
3232:17036134
3074:15583715
3011:14512621
2968:15024422
2916:15324396
2908:24121860
2854:17056247
2818:17952671
2759:21258382
2708:21102465
2642:15604402
2546:15597116
2489:20486672
2424:8 August
2394:21636754
2351:19458720
2282:16541064
2146:18654284
2040:21636754
1940:22056726
1865:20222824
1783:15195395
1699:13678773
1691:16339440
1588:See also
1471:strained
1403:kinetics
1328:and the
1318:toroidal
622:aptamers
572:static,
533:cytosine
502:proteins
320:vertices
132:designed
7083:Commons
6863:Outline
6848:History
6759:Bibcode
6661:Bibcode
6627:3397516
6570:Science
6542:Bibcode
6534:Science
6515:3454582
6471:2901229
6450:Bibcode
6408:Bibcode
6310:9978415
5913:6013447
5892:Bibcode
5855:5043419
5794:Bibcode
5746:Bibcode
5709:4508203
5659:4016739
5588:Bibcode
5555:4948918
5534:Bibcode
5494:3716461
5473:Bibcode
5465:Science
5358:3898745
5337:Bibcode
5220:2645571
5081:Bibcode
5073:Science
5029:Bibcode
5021:Science
4965:4354536
4937:Bibcode
4860:2974042
4839:Bibcode
4802:5994166
4781:Bibcode
4744:2907518
4723:Bibcode
4639:Bibcode
4559:Bibcode
4440:9866509
4412:Bibcode
4404:Science
4368:Bibcode
4324:Bibcode
4272:Bibcode
4248:2064216
4220:Bibcode
4196:4406177
4188:9923675
4168:Bibcode
4052:Bibcode
3962:Bibcode
3925:3465979
3904:Bibcode
3765:Bibcode
3741:4430815
3703:Bibcode
3676:4455780
3648:Bibcode
3621:4419579
3593:Bibcode
3532:2764300
3511:Bibcode
3455:Bibcode
3447:Science
3314:Bibcode
3281:3464188
3260:Bibcode
3223:3491902
3126:4385579
3118:9707114
3098:Bibcode
2991:Bibcode
2983:Science
2888:Bibcode
2809:3479651
2739:Bibcode
2699:3149862
2678:Bibcode
2650:9296608
2622:Bibcode
2614:Science
2480:2901229
2459:Bibcode
2374:Bibcode
2366:Science
2342:2688462
2321:Bibcode
2290:4316391
2262:Bibcode
2226:2257083
2206:Bibcode
2182:3508280
2126:Bibcode
2095:2064216
2067:Bibcode
2020:Bibcode
2012:Science
1931:3334823
1910:Bibcode
1856:3454582
1763:Bibcode
1671:Bibcode
1663:Science
1419:cascade
1415:hairpin
1398:above).
1246:polymer
1230:enzymes
1173:(AFM),
1075:hairpin
1043:entropy
1038:cascade
801:counter
793:fractal
773:rhombus
697:across.
541:thymine
537:guanine
529:adenine
421:, with
398:Science
263:History
6926:Carbon
6873:Impact
6777:
6739:
6709:
6679:
6634:
6624:
6588:
6560:
6522:
6512:
6478:
6468:
6426:
6374:
6350:
6340:
6308:
6300:
6290:
6254:
6246:
6236:
6200:
6190:
6158:
6150:
6140:
6104:
6096:
6086:
6050:
6043:390280
6040:
5995:
5920:
5910:
5862:
5852:
5812:
5764:
5716:
5706:
5666:
5656:
5616:
5598:
5562:
5552:
5501:
5491:
5443:
5435:
5400:
5365:
5355:
5285:
5227:
5217:
5177:
5133:
5107:
5099:
5055:
5047:
4999:
4963:
4955:
4929:Nature
4902:
4867:
4857:
4809:
4799:
4751:
4741:
4715:Nature
4693:
4657:
4613:
4605:
4533:
4493:
4483:
4475:
4438:
4430:
4386:
4342:
4298:
4290:
4264:Nature
4246:
4238:
4212:Nature
4194:
4186:
4160:Nature
4142:
4106:
4070:
4022:
3982:
3932:
3922:
3874:
3866:
3830:
3822:
3783:
3739:
3731:
3695:Nature
3674:
3666:
3640:Nature
3619:
3611:
3585:Nature
3539:
3529:
3503:Nature
3481:
3473:
3425:
3385:
3342:
3324:
3288:
3278:
3230:
3220:
3124:
3116:
3090:Nature
3072:
3065:534809
3062:
3017:
3009:
2966:
2959:368168
2956:
2914:
2906:
2852:
2816:
2806:
2757:
2706:
2696:
2648:
2640:
2594:
2568:
2544:
2537:535573
2534:
2487:
2477:
2392:
2349:
2339:
2313:Nature
2288:
2280:
2254:Nature
2224:
2180:
2144:
2093:
2085:
2059:Nature
2038:
1983:
1965:
1938:
1928:
1863:
1853:
1781:
1726:
1697:
1689:
1551:native
1451:energy
1348:Design
1338:enzyme
1302:porins
1286:et al.
1254:folate
1226:kidney
1136:enzyme
995:buffer
926:Dervan
904:, and
902:amines
745:, the
437:. See
382:et al.
377:Nature
358:, and
6306:S2CID
6252:S2CID
6198:S2CID
6156:S2CID
6102:S2CID
5441:S2CID
5105:S2CID
5053:S2CID
4981:Small
4961:S2CID
4611:S2CID
4436:S2CID
4296:S2CID
4244:S2CID
4192:S2CID
3872:S2CID
3828:S2CID
3737:S2CID
3672:S2CID
3617:S2CID
3479:S2CID
3122:S2CID
3015:S2CID
2912:S2CID
2878:arXiv
2646:S2CID
2286:S2CID
2250:(PDF)
2222:S2CID
2091:S2CID
1695:S2CID
1250:lipid
1163:iNANO
991:Z-DNA
987:B-DNA
914:thiol
910:amide
847:edges
751:Right
712:Right
689:, an
687:Right
535:(C),
531:(A),
300:Depth
277:Depth
167:cells
6875:and
6775:PMID
6737:PMID
6707:PMID
6677:PMID
6632:PMID
6586:PMID
6566:and
6558:PMID
6520:PMID
6476:PMID
6424:PMID
6372:ISBN
6348:PMID
6338:ISBN
6298:PMID
6288:ISBN
6244:PMID
6234:ISBN
6188:ISBN
6148:PMID
6138:ISBN
6094:PMID
6084:ISBN
6048:PMID
5993:PMID
5918:PMID
5860:PMID
5810:PMID
5762:PMID
5714:PMID
5664:PMID
5614:PMID
5560:PMID
5499:PMID
5433:PMID
5398:PMID
5363:PMID
5310:2013
5283:PMID
5252:2013
5225:PMID
5175:PMID
5144:2011
5131:ISBN
5097:PMID
5045:PMID
4997:PMID
4953:PMID
4900:PMID
4865:PMID
4807:PMID
4749:PMID
4691:PMID
4655:PMID
4603:PMID
4531:PMID
4491:PMID
4473:ISSN
4428:PMID
4384:PMID
4340:PMID
4288:PMID
4236:PMID
4184:PMID
4140:PMID
4104:PMID
4068:PMID
4020:PMID
3980:PMID
3930:PMID
3864:PMID
3820:ISSN
3781:PMID
3729:PMID
3664:PMID
3609:PMID
3537:PMID
3471:PMID
3423:PMID
3383:PMID
3340:PMID
3286:PMID
3228:PMID
3114:PMID
3070:PMID
3007:PMID
2964:PMID
2904:PMID
2850:PMID
2814:PMID
2755:PMID
2704:PMID
2638:PMID
2592:ISBN
2566:ISBN
2542:PMID
2485:PMID
2426:2011
2390:PMID
2347:PMID
2278:PMID
2178:PMID
2142:PMID
2083:ISSN
2036:PMID
1981:ISBN
1936:PMID
1861:PMID
1779:PMID
1724:ISBN
1687:PMID
1576:and
1561:and
1519:and
1098:and
989:and
853:and
839:cube
743:Left
708:Left
672:Left
616:and
559:and
515:The
429:and
213:and
205:and
197:and
185:and
146:DNA
6767:doi
6729:doi
6699:doi
6669:doi
6622:PMC
6612:doi
6578:doi
6574:332
6550:doi
6538:332
6510:PMC
6502:doi
6466:PMC
6458:doi
6416:doi
6404:290
6330:doi
6280:doi
6226:doi
6180:doi
6130:doi
6076:doi
6038:PMC
6030:doi
5985:doi
5955:doi
5951:287
5908:PMC
5900:doi
5850:PMC
5842:doi
5802:doi
5754:doi
5704:PMC
5696:doi
5654:PMC
5646:doi
5606:doi
5550:PMC
5542:doi
5489:PMC
5481:doi
5469:338
5425:doi
5390:doi
5353:PMC
5345:doi
5275:doi
5215:PMC
5207:doi
5167:doi
5089:doi
5077:332
5037:doi
5025:314
4989:doi
4945:doi
4933:451
4892:doi
4855:PMC
4847:doi
4797:PMC
4789:doi
4739:PMC
4731:doi
4719:465
4683:doi
4647:doi
4595:doi
4567:doi
4523:doi
4519:126
4481:PMC
4463:doi
4420:doi
4408:335
4376:doi
4332:doi
4280:doi
4268:415
4228:doi
4216:406
4176:doi
4164:397
4132:doi
4096:doi
4060:doi
4012:doi
3970:doi
3920:PMC
3912:doi
3856:doi
3812:doi
3773:doi
3719:hdl
3711:doi
3699:459
3656:doi
3644:552
3601:doi
3589:427
3565:doi
3561:116
3527:PMC
3519:doi
3507:461
3463:doi
3451:321
3415:doi
3411:126
3375:doi
3332:doi
3276:PMC
3268:doi
3218:PMC
3210:doi
3178:doi
3174:121
3150:doi
3146:121
3106:doi
3094:394
3060:PMC
3050:doi
2999:doi
2987:301
2954:PMC
2944:doi
2896:doi
2842:doi
2804:PMC
2796:doi
2747:doi
2694:PMC
2686:doi
2630:doi
2618:306
2532:PMC
2522:doi
2475:PMC
2467:doi
2382:doi
2370:332
2337:PMC
2329:doi
2317:459
2270:doi
2258:440
2214:doi
2170:doi
2134:doi
2075:doi
2063:406
2028:doi
2016:332
1973:doi
1926:PMC
1918:doi
1851:PMC
1843:doi
1771:doi
1759:290
1679:doi
1667:310
1248:or
1242:MIT
1114:in
1009:or
912:or
841:or
785:XOR
676:DNA
557:RNA
553:DNA
459:DNA
279:by
253:DNA
140:DNA
7125::
6773:.
6765:.
6753:.
6735:.
6723:.
6705:.
6695:45
6693:.
6675:.
6667:.
6655:.
6630:.
6620:.
6608:13
6606:.
6602:.
6584:.
6572:.
6556:.
6548:.
6536:.
6518:.
6508:.
6498:79
6496:.
6492:.
6474:.
6464:.
6456:.
6446:10
6444:.
6440:.
6422:.
6414:.
6402:.
6346:.
6336:.
6304:.
6296:.
6286:.
6276:11
6274:.
6250:.
6242:.
6232:.
6222:47
6220:.
6196:.
6186:.
6154:.
6146:.
6136:.
6126:42
6124:.
6100:.
6092:.
6082:.
6072:42
6070:.
6046:.
6036:.
6026:32
6024:.
6020:.
6005:^
5991:.
5979:.
5949:.
5945:.
5930:^
5916:.
5906:.
5898:.
5886:.
5882:.
5858:.
5848:.
5838:10
5836:.
5832:.
5808:.
5800:.
5790:11
5788:.
5784:.
5760:.
5752:.
5742:15
5740:.
5736:.
5712:.
5702:.
5690:.
5686:.
5662:.
5652:.
5642:52
5640:.
5636:.
5612:.
5604:.
5594:.
5584:13
5582:.
5558:.
5548:.
5540:.
5530:16
5528:.
5524:.
5511:^
5497:.
5487:.
5479:.
5467:.
5463:.
5439:.
5431:.
5419:.
5396:.
5386:49
5384:.
5361:.
5351:.
5343:.
5331:.
5327:.
5281:.
5269:.
5223:.
5213:.
5201:.
5197:.
5173:.
5161:.
5103:.
5095:.
5087:.
5075:.
5051:.
5043:.
5035:.
5023:.
5019:.
4995:.
4985:11
4983:.
4959:.
4951:.
4943:.
4931:.
4927:.
4912:^
4898:.
4888:34
4886:.
4863:.
4853:.
4845:.
4833:.
4829:.
4805:.
4795:.
4787:.
4777:13
4775:.
4771:.
4747:.
4737:.
4729:.
4717:.
4713:.
4689:.
4679:44
4677:.
4653:.
4645:.
4635:44
4633:.
4609:.
4601:.
4591:58
4589:.
4565:.
4553:.
4529:.
4517:.
4513:.
4489:.
4479:.
4471:.
4457:.
4434:.
4426:.
4418:.
4406:.
4382:.
4374:.
4362:.
4338:.
4330:.
4320:42
4318:.
4294:.
4286:.
4278:.
4266:.
4242:.
4234:.
4226:.
4214:.
4190:.
4182:.
4174:.
4162:.
4138:.
4128:43
4126:.
4102:.
4090:.
4066:.
4058:.
4046:.
4042:.
4018:.
4008:46
4006:.
4002:.
3978:.
3968:.
3958:45
3956:.
3952:.
3928:.
3918:.
3910:.
3898:.
3894:.
3870:.
3862:.
3852:10
3850:.
3826:.
3818:.
3808:11
3806:.
3802:.
3779:.
3771:.
3759:.
3735:.
3727:.
3717:.
3709:.
3697:.
3684:^
3670:.
3662:.
3654:.
3642:.
3638:.
3615:.
3607:.
3599:.
3587:.
3559:.
3535:.
3525:.
3517:.
3505:.
3501:.
3477:.
3469:.
3461:.
3449:.
3445:.
3421:.
3409:.
3405:.
3381:.
3371:45
3369:.
3352:^
3338:.
3330:.
3320:.
3308:.
3284:.
3274:.
3266:.
3254:.
3250:.
3226:.
3216:.
3204:.
3200:.
3172:.
3144:.
3120:.
3112:.
3104:.
3092:.
3068:.
3058:.
3044:.
3040:.
3027:^
3013:.
3005:.
2997:.
2985:.
2962:.
2952:.
2938:.
2934:.
2910:.
2902:.
2894:.
2886:.
2874:15
2872:.
2848:.
2838:17
2836:.
2812:.
2802:.
2792:37
2790:.
2786:.
2767:^
2753:.
2745:.
2733:.
2716:^
2702:.
2692:.
2684:.
2672:.
2668:.
2644:.
2636:.
2628:.
2616:.
2540:.
2530:.
2516:.
2512:.
2497:^
2483:.
2473:.
2465:.
2455:10
2453:.
2449:.
2434:^
2412:.
2388:.
2380:.
2368:.
2345:.
2335:.
2327:.
2315:.
2311:.
2298:^
2284:.
2276:.
2268:.
2256:.
2252:.
2234:^
2220:.
2212:.
2200:.
2176:.
2164:.
2140:.
2132:.
2120:.
2103:^
2089:.
2081:.
2073:.
2061:.
2057:.
2034:.
2026:.
2014:.
1995:^
1979:.
1971:.
1948:^
1934:.
1924:.
1916:.
1904:.
1900:.
1873:^
1859:.
1849:.
1839:79
1837:.
1833:.
1814:^
1777:.
1769:.
1757:.
1738:^
1707:^
1693:.
1685:.
1677:.
1665:.
1652:^
1584:.
1546:.
1488:.
1473:.
1268:.
1142:.
1082:.
900:,
886:.
818:.
808:nm
695:nm
354:,
259:.
177:,
173:,
6824:e
6817:t
6810:v
6781:.
6769::
6761::
6755:3
6743:.
6731::
6725:7
6713:.
6701::
6683:.
6671::
6663::
6657:2
6638:.
6614::
6592:.
6580::
6564:.
6552::
6544::
6526:.
6504::
6482:.
6460::
6452::
6430:.
6418::
6410::
6380:.
6354:.
6332::
6312:.
6282::
6258:.
6228::
6204:.
6182::
6162:.
6132::
6108:.
6078::
6054:.
6032::
5999:.
5987::
5981:7
5963:.
5957::
5924:.
5902::
5894::
5888:9
5866:.
5844::
5816:.
5804::
5796::
5768:.
5756::
5748::
5720:.
5698::
5692:9
5670:.
5648::
5620:.
5608::
5590::
5566:.
5544::
5536::
5505:.
5483::
5475::
5447:.
5427::
5421:3
5404:.
5392::
5369:.
5347::
5339::
5333:7
5312:.
5289:.
5277::
5271:5
5254:.
5231:.
5209::
5203:2
5181:.
5169::
5163:6
5146:.
5111:.
5091::
5083::
5059:.
5039::
5031::
5003:.
4991::
4967:.
4947::
4939::
4906:.
4894::
4871:.
4849::
4841::
4835:5
4813:.
4791::
4783::
4755:.
4733::
4725::
4697:.
4685::
4661:.
4649::
4641::
4617:.
4597::
4573:.
4569::
4561::
4555:4
4537:.
4525::
4497:.
4465::
4442:.
4422::
4414::
4390:.
4378::
4370::
4364:3
4346:.
4334::
4326::
4302:.
4282::
4274::
4250:.
4230::
4222::
4198:.
4178::
4170::
4146:.
4134::
4110:.
4098::
4092:5
4074:.
4062::
4054::
4048:5
4026:.
4014::
3986:.
3972::
3964::
3936:.
3914::
3906::
3900:6
3878:.
3858::
3834:.
3814::
3787:.
3775::
3767::
3761:9
3743:.
3721::
3713::
3705::
3678:.
3658::
3650::
3623:.
3603::
3595::
3571:.
3567::
3543:.
3521::
3513::
3485:.
3465::
3457::
3429:.
3417::
3389:.
3377::
3346:.
3334::
3316::
3310:5
3292:.
3270::
3262::
3256:5
3234:.
3212::
3206:4
3184:.
3180::
3156:.
3152::
3128:.
3108::
3100::
3076:.
3052::
3046:2
3021:.
3001::
2993::
2970:.
2946::
2940:2
2918:.
2898::
2890::
2880::
2856:.
2844::
2820:.
2798::
2761:.
2749::
2741::
2735:3
2710:.
2688::
2680::
2674:5
2652:.
2632::
2624::
2600:.
2574:.
2548:.
2524::
2518:2
2491:.
2469::
2461::
2428:.
2396:.
2384::
2376::
2353:.
2331::
2323::
2292:.
2272::
2264::
2228:.
2216::
2208::
2202:4
2184:.
2172::
2166:1
2148:.
2136::
2128::
2122:2
2097:.
2077::
2069::
2042:.
2030::
2022::
1989:.
1975::
1942:.
1920::
1912::
1906:6
1867:.
1845::
1785:.
1773::
1765::
1732:.
1701:.
1681::
1673::
1511:.
962:.
445:.
435:G
431:C
427:T
423:A
115:e
108:t
101:v
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.