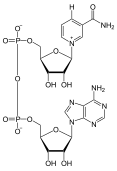
297:, or nicotinamide adenine dinucleotide, is a dinucleotide, containing two nucleotides. One of the nucleotides it contains is an adenine group, while the other is nicotinamide. In order to reduce this molecule, a hydrogen and two electrons must be added to the 6-carbon ring of nicotinamide; one electron is added to the carbon opposite the positively charged nitrogen, causing a rearrangement of bonds within the ring to give nitrogen more electrons; it will lose its positive charge as a result. The other electron is "stolen" from an additional hydrogen, leaving the hydrogen ion in solution.
401:(ALDH) are NAD dependent enzymes that function to remove toxic aldehydes from the body, functioning mostly in the mitochondria of cells. These enzymes are largely responsible for the detoxification of acetylaldehyde, which is an intermediate in the metabolism of ethanol. It has been shown that a mutation in the ALDH2 gene (one of 19 aldehyde dehydrogenase genes) is what leads to the common occurrence in East Asian population of a flushed face after consuming alcohol, due to the build-up of acetaldehyde. This build-up of acetaldehyde also causes headaches and vomiting (
171:
203:
386:
99:
313:
1441:
274:
131:
333:
pathways that convert substrates to more complicated products, using ATP. The reasoning behind having two separate electron carriers for anabolic and catabolic pathways relates to regulation of metabolism. The ratio of NADP to NADPH in the cell is kept rather low, so that NADPH is readily available as a reducing agent; it is more commonly used as a reducing agent than NADP is used as an oxidizing agent.
342:
417:
Deactivation of aldehyde dehydrogenases has been shown to be instrumental in the mechanisms of many cancers. ALDHs function in cell differentiation, proliferation, oxidation, and drug resistance. These enzymes are only one example of the many different types of dehydrogenases in the human body; their
393:
Aldehydes are the natural by-product of many physiological processes, as well as being the consequence of many industrial processes, put out into the environment in the form of smog and motor vehicle exhaust. Build-up of aldehydes in the brain and pericardium can be detrimental to a person's health,
328:
differs from NAD only in the addition of a phosphate group to the adenosine 5-membered carbon ring. The addition of the phosphate does not alter the electron transport abilities of the carrier. The phosphate group creates enough contrast between the two groups that they bind to the active site of
142:
The result of a dehydrogenase catalyzed reaction is not always the acquisition of a positive charge. Sometimes the substrate loses a proton. This may leave free electrons on the substrate that move into a double bond. This happens frequently when an alcohol is the substrate; when the proton on the
241:
O. In this case, the enzyme is taking electrons from the substrate, and using free protons to reduce the oxygen, leaving the substrate with a positive charge. The product is water, instead of hydrogen peroxide as seen above. An example of an oxidase that functions like this is complex IV in the
332:
These two electron carriers are easily distinguished by enzymes and participate in very different reactions. NADP mainly functions with enzymes that catalyze anabolic, or biosynthetic, pathways. Specifically, NADPH will act as a reducing agent in these reactions, resulting in NADP. These are
281:
Dehydrogenase enzymes transfer electrons from the substrate to an electron carrier; what carrier is used depends on the reaction taking place. Common electron acceptors used by this subclass are NAD, FAD, and NADP. Electron carriers are reduced in this process and considered oxidizers of the
367:
The double-bonded nitrogen atoms in FAD make it a good acceptor in taking two hydrogen atoms from a substrate. Because it takes two atoms rather than one, FAD is often involved when a double bond is formed in the newly oxidized substrate. FAD is unique because it is reduced by two electrons
114:
This would be considered an oxidation of the substrate, in which the substrate either loses hydrogen atoms or gains an oxygen atom (from water). The name "dehydrogenase" is based on the idea that it facilitates the removal (de-) of hydrogen (-hydrogen-) and is an enzyme (-ase). Dehydrogenase
138:
A represents the substrate that will be oxidized, while B is the hydride acceptor. Note how when the hydride is transferred from A to B, the A has taken on a positive charge; this is because the enzyme has taken two electrons from the substrate in order to reduce the acceptor to BH.
352:, or flavin adenine dinucleotide, is a prosthetic group (a non-polypeptide unit bound to a protein that is required for function) that consists of an adenine nucleotide and a flavin mononucleotide. FAD is a unique electron acceptor. Its fully reduced form is FADH
405:
symptoms) if not broken down quickly enough, another reason why those with acetaldehyde DH deficiencies have bad reactions to alcohol. Importantly, a lack of this enzyme has been linked to an increase in the risk of myocardial
587:"Enzyme Nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes by the Reactions they Catalyse"
566:
pointed out that the oxidized form of NAD is negatively charged, and that NAD is an inappropriate symbol for an anion
However, NAD and, similarly, NADP remain in almost universal use and alternatives such as
178:
In the above case, the dehydrogenase has transferred a hydride while releasing a proton, H, but dehydrogenases can also transfer two hydrogens, using FAD as an electron acceptor. This would be depicted as
143:
oxygen leaves, the free electrons on the oxygen will be used to create a double bond, as seen in the oxidation of ethanol to acetaldehyde carried out by alcohol dehydrogenase in the image on the right.
214:
are easily distinguishable if one considers the electron acceptor. An oxidase will remove electrons from a substrate as well, but only uses oxygen as its electron acceptor. One such reaction is: AH
1094:
van den Hoogen, Christel; van der Horst, Geertje; Cheung, Henry; Buijs, Jeroen T.; Lippitt, Jenny M.; Guzmán-Ramírez, Natalia; Hamdy, Freddie C.; Eaton, Colby L.; Thalmann, George N. (2010-06-15).
418:
wide array of functions, and the impact that their deactivation or mutations has upon crucial cell processes underscores the importance of all dehydrogenases in maintaining body homeostasis.
90:
is the electron acceptor. The systematic name of an oxidoreductase is "donor:acceptor oxidoreductase", but, when possible, it is more conveniently named as "donor dehydrogenase".
70:
Oxidoreductases, enzymes that catalyze oxidation-reduction reactions, constitute Class EC 1 of the IUBMB classification of enzyme-catalyzed reactions. Any of these may be called
115:
reactions come most commonly in two forms: the transfer of a hydride and release of a proton (often with water as a second reactant), and the transfer of two hydrogens.
309:, that break down energy molecules to produce ATP. The ratio of NAD to NADH is kept very high in the cell, keeping it readily available to act as an oxidizing agent.
150:
to the substrate and a proton to the environment. The net result on the substrate is the addition of one oxygen atom. This is seen for example in the oxidation of
810:
50:. Like all catalysts, they catalyze reverse as well as forward reactions, and in some cases this has physiological significance: for example,
325:
174:
Reaction catalyzed by succinate dehydrogenase, note the double bond formed between the two central carbons when two hydrogens are removed
756:
Ying, Weihai (2008-02-01). "NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences".
939:
494:
199:
The distinction between the subclasses of oxidoreductases that catalyze oxidation reactions lies in their electron acceptors.
735:
397:
Considering how prevalent aldehydes are, there must be an enzyme to facilitate their oxidation to a less volatile compound.
1096:"High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer"
356:(known as the hydroquinone form), but FAD can also be partially oxidized as FADH by either reducing FAD or oxidizing FADH
1160:
668:
527:
484:
294:
106:
Dehydrogenases oxidize a substrate by transferring hydrogen to an electron acceptor, common electron acceptors being
32:
437:
187:. A double bond is normally formed in between the two atoms that the hydrogens were taken from, as in the case of
1316:
349:
111:
43:
134:
Alcohol dehydrogenase oxidizes ethanol, with the help of the electron carrier NAD, yielding acetaldehyde
1431:
1069:
427:
159:
1417:
1404:
1391:
1378:
1365:
1352:
1339:
1301:
488:
191:. The two hydrogens have been transferred to the carrier or the other product, with their electrons.
459:(used to convert NADH back to NAD in anaerobic glycolysis, and in the back reaction to produce NADH)
1461:
1311:
1265:
1208:
818:
517:
243:
1213:
1030:"Population genetic studies on aldehyde dehydrogenase isozyme deficiency and alcohol sensitivity"
684:
Yoshikawa, Shinya; Shimada, Atsuhiro (2015-01-20). "Reaction
Mechanism of Cytochrome c Oxidase".
533:
442:
283:
188:
504:
462:
398:
629:
1234:
1153:
456:
432:
51:
47:
1306:
539:
8:
1270:
123:
Sometimes a dehydrogenase catalyzed reaction will look like this: AH + B ↔ A + BH when a
947:
389:
The mechanism of an aldehyde dehydrogenase, note the use of NAD as an electron acceptor.
1203:
1046:
1029:
1005:
972:
873:
840:
789:
206:
Reaction catalyzed by an oxidase, note the reduction of oxygen as the electron acceptor
607:
170:
1125:
1117:
1051:
1010:
992:
921:
913:
878:
860:
781:
773:
731:
701:
664:
563:
466:
146:
Another possibility is that a water molecule will enter the reaction, contributing a
793:
725:
202:
1249:
1244:
1218:
1146:
1107:
1041:
1000:
984:
905:
868:
852:
765:
693:
477:, using NAD. In this reaction, the substrate not only is oxidized but also loses a
385:
306:
62:
in animals, but in yeast it catalyzes the production of ethanol from acetaldehyde.
1112:
1095:
1296:
1280:
1193:
28:
909:
394:
as they can form adducts with important molecules and cause their inactivation.
78:
is also used when the physiological emphasis on reduction of the substrate, and
1445:
1334:
1275:
478:
98:
71:
1455:
1239:
1198:
1121:
996:
917:
864:
777:
450:
410:, while activation has shown the enzyme's ability to reduce damage caused by
147:
1188:
1129:
1014:
882:
785:
705:
608:"Classification and Nomenclature of Enzymes by the Reactions they Catalyse"
151:
59:
36:
1055:
925:
769:
372:
two protons, as opposed to both NAD and NADP, which only take one proton.
1412:
1347:
1183:
988:
155:
586:
162:, a step in the metabolism of ethanol and in the production of vinegar.
130:
74:, especially those in which NAD is the electron acceptor (oxidant), but
498:
474:
407:
254:
1093:
697:
329:
different enzymes, generally catalyzing different types of reactions.
1386:
1360:
856:
510:
446:
249:
Note that oxidases typically transfer the equivalent of dihydrogen (H
75:
31:
that oxidizes a substrate by reducing an electron acceptor, usually
1440:
470:
411:
402:
312:
39:
273:
521:
211:
124:
79:
55:
118:
1399:
1169:
24:
971:
Chen, Che-Hong; Sun, Lihan; Mochly-Rosen, Daria (2010-10-01).
233:
Sometimes an oxidase reaction will look like this: 4A + 4H + O
1373:
559:
107:
257:(another subclass of oxidoreductases) will use a peroxide (H
973:"Mitochondrial aldehyde dehydrogenase and cardiac diseases"
659:
Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. (2016).
1138:
896:
Rivlin, Richard S. (1970-08-27). "Riboflavin
Metabolism".
661:
841:"Sequence-structure analysis of FAD-containing proteins"
194:
341:
1429:
970:
360:. Dehydrogenases typically fully reduce FAD to FADH
265:) as the electron acceptor, rather than an oxygen.
305:NAD is mostly used in catabolic pathways, such as
562:panel on biochemical thermodynamics convened by
286:that are often referred to as "redox cofactors."
1453:
683:
630:"Definitions of Oxidation and Reduction (Redox)"
253:), and the acceptor is a dioxygen. Similarly, a
481:molecule, and is attached to the CoA coenzyme.)
723:
1154:
1027:
658:
165:
119:Transferring a hydride and releasing a proton
838:
724:Alberts, B; Johnson, A; et al. (2002).
316:Nicotinamide Adenine Dinucleotide Phosphate
1161:
1147:
839:Dym, Orly; Eisenberg, David (2001-09-01).
380:
301:Reduction of NAD: NAD + 2H + 2e ↔ NADH + H
102:A reaction catalyzed by a reductase enzyme
1111:
1045:
1004:
872:
495:glyceraldehyde-3-phosphate dehydrogenase
384:
340:
311:
272:
201:
169:
129:
97:
65:
1454:
895:
93:
1142:
966:
964:
627:
268:
805:
803:
755:
751:
749:
747:
719:
717:
715:
654:
652:
650:
600:
581:
579:
577:
195:Identifying a dehydrogenase reaction
364:. The production of FADH is rare.
13:
961:
758:Antioxidants & Redox Signaling
14:
1473:
811:"The physiological role of NADPH"
800:
744:
712:
663:(5th ed.). New York: Wiley.
647:
621:
574:
528:alpha-ketoglutarate dehydrogenase
485:glucose-6-phosphate dehydrogenase
282:substrate. Electron carriers are
277:Nicotinamide Adenine Dinucleotide
1439:
1028:Goedde, HW; Agarwal, DP (1983).
465:(A common enzyme that feeds the
438:Delta12-fatty acid dehydrogenase
421:
1087:
1062:
1021:
932:
898:New England Journal of Medicine
889:
832:
677:
571:have been very little adopted.
552:
1:
1113:10.1158/0008-5472.CAN-09-3806
730:. New York: Garland Science.
727:Molecular Biology of the Cell
546:
445:(an enzyme that can convert
7:
1168:
910:10.1056/NEJM197008272830906
375:
345:Flavin Adenine Dinucleotide
54:catalyzes the oxidation of
10:
1478:
428:acetaldehyde dehydrogenase
242:Electron Transport Chain (
166:Transferring two hydrogens
160:acetaldehyde dehydrogenase
27:belonging to the group of
1325:
1317:Michaelis–Menten kinetics
1289:
1258:
1227:
1176:
489:pentose phosphate pathway
1209:Diffusion-limited enzyme
940:"blobs.org - Metabolism"
518:isocitrate dehydrogenase
320:
977:Cardiovascular Research
534:succinate dehydrogenase
520:(uses NAD, also has an
443:glutamate dehydrogenase
399:Aldehyde dehydrogenases
381:Biological implications
289:
189:succinate dehydrogenase
505:sorbitol dehydrogenase
463:pyruvate dehydrogenase
390:
346:
336:
317:
303:
278:
207:
175:
135:
103:
1302:Eadie–Hofstee diagram
1235:Allosteric regulation
770:10.1089/ars.2007.1672
457:lactate dehydrogenase
433:alcohol dehydrogenase
388:
344:
315:
299:
276:
205:
173:
133:
101:
52:alcohol dehydrogenase
1312:Lineweaver–Burk plot
1070:"How Hangovers Work"
540:malate dehydrogenase
66:IUBMB Classification
815:watcut.uwaterloo.ca
628:Clark, Jim (2002).
94:Reactions catalyzed
1271:Enzyme superfamily
1204:Enzyme promiscuity
989:10.1093/cvr/cvq192
491:, producing NADPH)
391:
347:
318:
279:
269:Electron acceptors
210:Dehydrogenase and
208:
176:
136:
104:
1427:
1426:
1106:(12): 5163–5173.
737:978-0-8153-3218-3
698:10.1021/cr500266a
487:(involved in the
1469:
1444:
1443:
1435:
1307:Hanes–Woolf plot
1250:Enzyme activator
1245:Enzyme inhibitor
1219:Enzyme catalysis
1163:
1156:
1149:
1140:
1139:
1134:
1133:
1115:
1091:
1085:
1084:
1082:
1081:
1066:
1060:
1059:
1049:
1025:
1019:
1018:
1008:
968:
959:
958:
956:
955:
946:. Archived from
936:
930:
929:
893:
887:
886:
876:
857:10.1110/ps.12801
851:(9): 1712–1728.
836:
830:
829:
827:
826:
817:. Archived from
807:
798:
797:
753:
742:
741:
721:
710:
709:
692:(4): 1936–1989.
686:Chemical Reviews
681:
675:
674:
656:
645:
644:
642:
640:
625:
619:
618:
616:
614:
604:
598:
597:
595:
593:
583:
572:
556:
453:and vice versa).
127:is transferred.
16:Class of enzymes
1477:
1476:
1472:
1471:
1470:
1468:
1467:
1466:
1462:Oxidoreductases
1452:
1451:
1450:
1438:
1430:
1428:
1423:
1335:Oxidoreductases
1321:
1297:Enzyme kinetics
1285:
1281:List of enzymes
1254:
1223:
1194:Catalytic triad
1172:
1167:
1137:
1100:Cancer Research
1092:
1088:
1079:
1077:
1068:
1067:
1063:
1026:
1022:
969:
962:
953:
951:
938:
937:
933:
894:
890:
845:Protein Science
837:
833:
824:
822:
809:
808:
801:
754:
745:
738:
722:
713:
682:
678:
671:
657:
648:
638:
636:
626:
622:
612:
610:
606:
605:
601:
591:
589:
585:
584:
575:
570:
557:
553:
549:
524:that uses NADP)
424:
383:
378:
363:
359:
355:
339:
323:
292:
271:
264:
260:
252:
240:
236:
229:
225:
221:
217:
197:
186:
182:
168:
121:
96:
89:
68:
29:oxidoreductases
17:
12:
11:
5:
1475:
1465:
1464:
1449:
1448:
1425:
1424:
1422:
1421:
1408:
1395:
1382:
1369:
1356:
1343:
1329:
1327:
1323:
1322:
1320:
1319:
1314:
1309:
1304:
1299:
1293:
1291:
1287:
1286:
1284:
1283:
1278:
1273:
1268:
1262:
1260:
1259:Classification
1256:
1255:
1253:
1252:
1247:
1242:
1237:
1231:
1229:
1225:
1224:
1222:
1221:
1216:
1211:
1206:
1201:
1196:
1191:
1186:
1180:
1178:
1174:
1173:
1166:
1165:
1158:
1151:
1143:
1136:
1135:
1086:
1061:
1034:Am J Hum Genet
1020:
960:
931:
904:(9): 463–472.
888:
831:
799:
764:(2): 179–206.
743:
736:
711:
676:
669:
646:
620:
599:
573:
568:
564:Robert Alberty
550:
548:
545:
544:
543:
537:
531:
525:
508:
507:
502:
492:
482:
479:carbon dioxide
469:by converting
460:
454:
440:
435:
430:
423:
420:
382:
379:
377:
374:
361:
357:
353:
338:
335:
322:
319:
291:
288:
270:
267:
262:
258:
250:
238:
234:
227:
223:
219:
215:
196:
193:
184:
180:
167:
164:
120:
117:
95:
92:
87:
72:dehydrogenases
67:
64:
15:
9:
6:
4:
3:
2:
1474:
1463:
1460:
1459:
1457:
1447:
1442:
1437:
1436:
1433:
1419:
1415:
1414:
1409:
1406:
1402:
1401:
1396:
1393:
1389:
1388:
1383:
1380:
1376:
1375:
1370:
1367:
1363:
1362:
1357:
1354:
1350:
1349:
1344:
1341:
1337:
1336:
1331:
1330:
1328:
1324:
1318:
1315:
1313:
1310:
1308:
1305:
1303:
1300:
1298:
1295:
1294:
1292:
1288:
1282:
1279:
1277:
1276:Enzyme family
1274:
1272:
1269:
1267:
1264:
1263:
1261:
1257:
1251:
1248:
1246:
1243:
1241:
1240:Cooperativity
1238:
1236:
1233:
1232:
1230:
1226:
1220:
1217:
1215:
1212:
1210:
1207:
1205:
1202:
1200:
1199:Oxyanion hole
1197:
1195:
1192:
1190:
1187:
1185:
1182:
1181:
1179:
1175:
1171:
1164:
1159:
1157:
1152:
1150:
1145:
1144:
1141:
1131:
1127:
1123:
1119:
1114:
1109:
1105:
1101:
1097:
1090:
1075:
1074:HowStuffWorks
1071:
1065:
1057:
1053:
1048:
1043:
1040:(4): 769–72.
1039:
1035:
1031:
1024:
1016:
1012:
1007:
1002:
998:
994:
990:
986:
982:
978:
974:
967:
965:
950:on 2016-02-01
949:
945:
944:www.blobs.org
941:
935:
927:
923:
919:
915:
911:
907:
903:
899:
892:
884:
880:
875:
870:
866:
862:
858:
854:
850:
846:
842:
835:
821:on 2016-03-06
820:
816:
812:
806:
804:
795:
791:
787:
783:
779:
775:
771:
767:
763:
759:
752:
750:
748:
739:
733:
729:
728:
720:
718:
716:
707:
703:
699:
695:
691:
687:
680:
672:
670:9781118918401
666:
662:
655:
653:
651:
635:
631:
624:
609:
603:
588:
582:
580:
578:
565:
561:
555:
551:
541:
538:
535:
532:
529:
526:
523:
519:
516:
515:
514:
512:
506:
503:
500:
497:(involved in
496:
493:
490:
486:
483:
480:
476:
472:
468:
464:
461:
458:
455:
452:
451:Ketoglutarate
448:
444:
441:
439:
436:
434:
431:
429:
426:
425:
422:More examples
419:
415:
413:
409:
404:
400:
395:
387:
373:
371:
365:
351:
343:
334:
330:
327:
314:
310:
308:
302:
298:
296:
287:
285:
275:
266:
256:
247:
245:
231:
213:
204:
200:
192:
190:
172:
163:
161:
157:
153:
149:
148:hydroxide ion
144:
140:
132:
128:
126:
116:
113:
109:
100:
91:
85:
81:
77:
73:
63:
61:
57:
53:
49:
45:
41:
38:
34:
30:
26:
22:
21:dehydrogenase
1413:Translocases
1410:
1397:
1384:
1371:
1358:
1348:Transferases
1345:
1332:
1189:Binding site
1103:
1099:
1089:
1078:. Retrieved
1076:. 2004-10-12
1073:
1064:
1037:
1033:
1023:
983:(1): 51–57.
980:
976:
952:. Retrieved
948:the original
943:
934:
901:
897:
891:
848:
844:
834:
823:. Retrieved
819:the original
814:
761:
757:
726:
689:
685:
679:
660:
639:February 14,
637:. Retrieved
633:
623:
611:. Retrieved
602:
590:. Retrieved
554:
509:
416:
396:
392:
369:
366:
348:
331:
324:
304:
300:
293:
280:
248:
232:
209:
198:
183:+ B ↔ A + BH
177:
152:acetaldehyde
145:
141:
137:
122:
105:
83:
69:
60:acetaldehyde
20:
18:
1184:Active site
501:, uses NAD)
156:acetic acid
1387:Isomerases
1361:Hydrolases
1228:Regulation
1080:2016-03-06
954:2016-03-01
825:2016-03-06
547:References
542:(uses NAD)
536:(uses FAD)
530:(uses NAD)
513:examples:
499:glycolysis
475:acetyl CoA
408:infarction
307:glycolysis
255:peroxidase
1266:EC number
1122:1538-7445
997:0008-6363
918:0028-4793
865:1469-896X
778:1523-0864
634:Chemguide
511:TCA cycle
467:TCA Cycle
447:glutamate
412:ischaemia
284:coenzymes
237:↔ 4A + 2H
76:reductase
1456:Category
1290:Kinetics
1214:Cofactor
1177:Activity
1130:20516116
1015:20558439
883:11514662
794:42000527
786:18020963
706:25603498
613:30 March
592:29 March
569:oxidized
471:pyruvate
403:hangover
376:Examples
82:is used
42:such as
40:coenzyme
33:NAD/NADP
1446:Biology
1400:Ligases
1170:Enzymes
1056:6881146
1047:1685745
1006:2936126
926:4915004
874:2253189
522:isozyme
222:↔ A + H
212:oxidase
125:hydride
80:oxidase
56:ethanol
1432:Portal
1374:Lyases
1128:
1120:
1054:
1044:
1013:
1003:
995:
924:
916:
881:
871:
863:
792:
784:
776:
734:
704:
667:
86:when O
37:flavin
25:enzyme
23:is an
1326:Types
790:S2CID
560:IUPAC
449:to α-
35:or a
1418:list
1411:EC7
1405:list
1398:EC6
1392:list
1385:EC5
1379:list
1372:EC4
1366:list
1359:EC3
1353:list
1346:EC2
1340:list
1333:EC1
1126:PMID
1118:ISSN
1052:PMID
1011:PMID
993:ISSN
922:PMID
914:ISSN
879:PMID
861:ISSN
782:PMID
774:ISSN
732:ISBN
702:PMID
665:ISBN
641:2016
615:2021
594:2021
326:NADP
321:NADP
112:FAD.
84:only
1108:doi
1042:PMC
1001:PMC
985:doi
906:doi
902:283
869:PMC
853:doi
766:doi
694:doi
690:115
567:NAD
558:An
473:to
370:and
350:FAD
337:FAD
295:NAD
290:NAD
246:).
244:ETC
218:+ O
158:by
154:to
110:or
108:NAD
58:to
48:FMN
46:or
44:FAD
1458::
1124:.
1116:.
1104:70
1102:.
1098:.
1072:.
1050:.
1038:35
1036:.
1032:.
1009:.
999:.
991:.
981:88
979:.
975:.
963:^
942:.
920:.
912:.
900:.
877:.
867:.
859:.
849:10
847:.
843:.
813:.
802:^
788:.
780:.
772:.
762:10
760:.
746:^
714:^
700:.
688:.
649:^
632:.
576:^
414:.
230:.
179:AH
19:A
1434::
1420:)
1416:(
1407:)
1403:(
1394:)
1390:(
1381:)
1377:(
1368:)
1364:(
1355:)
1351:(
1342:)
1338:(
1162:e
1155:t
1148:v
1132:.
1110::
1083:.
1058:.
1017:.
987::
957:.
928:.
908::
885:.
855::
828:.
796:.
768::
740:.
708:.
696::
673:.
643:.
617:.
596:.
362:2
358:2
354:2
263:2
261:O
259:2
251:2
239:2
235:2
228:2
226:O
224:2
220:2
216:2
185:2
181:2
88:2
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.