638:
324:
170:
29:
679:
Following synthesis of HIV's doubled stranded DNA genome, integrase binds to the long tandem repeats flanking the genome on both ends. Using its endonucleolytic activity, integrase cleaves a di or trinucleotide from both 3' ends of the genome in a processing known as 3'-processing. The specificity of
687:
through SN2-type nucleophilic attack. The 3' ends are covalently linked to the target DNA. The 5' over hangs of the viral genome are then cleaved using host repair enzymes, those same enzymes are believed to be responsible for the integration of the 5' end into the host genome forming the provirus.
663:
The N-terminus is composed of 50 amino acid residues which contain a conserved histidine, histidine, cytosine, cytosine sequence which chelates zinc ions, furthermore enhancing the enzymatic activity of the catalytic core domain. As metal chelation is vital in integrase efficacy, it is a target for
667:
The catalytic core domain, like the N-terminus, contains highly conserved amino acid residues -Asp64, Asp116, Glu152- as the conserved DDE (Asp-Asp-Glu) motif contributes to the endonuclease and polynucleotide transferase functions of integrase. Mutations in these regions inactivates integrase and
602:
replication. Integration is a "point of no return" for the cell, which becomes a permanent carrier of the viral genome (provirus). Integration is in part responsible for the persistence of retroviral infections. After integration, the viral gene expression and particle production may take place
573:(dimer-of-dimers), with all three domains being important for multimerization and viral DNA binding. In addition, several host cellular proteins have been shown to interact with IN to facilitate the integration process: e.g., the host factor, human chromatin-associated protein
583:(HFV), an agent harmless to humans, has an integrase similar to HIV IN and is therefore a model of HIV IN function; a 2010 crystal structure of the HFV integrase assembled on viral DNA ends has been determined.
2164:
2278:
680:
cleavage is improved through the use of cofactors such as Mn and Mg which interact with the DDE motif of the catalytic core domain, acting as cofactors to integrase function.
1638:
1661:
423:
282:
128:
2174:
1444:
1610:
2148:
2169:
951:
Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, et al. (August 1995). "Solution structure of the DNA binding domain of HIV-1 integrase".
708:, demonstrated that the compound has potent antiviral activity. On October 12, 2007, the Food and Drug Administration (U.S.) approved the integrase inhibitor
2289:
2267:
569:(RSV) have been reported, with the first structures determined in 1994. Biochemical data and structural data suggest that retroviral IN functions as a
2611:
2606:
2253:
2085:
610:
3'-processing, in which two or three nucleotides are removed from one or both 3' ends of the viral DNA to expose an invariant CA dinucleotide.
2663:
2300:
660:, catalytic core domain, and C-terminus, which each have distinct properties and functions contributing to the efficacy of HIV integrase.
2228:
2195:
749:
Beck BJ, Freudenreich O, Worth JL (2010). "Patients with Human
Immunodeficiency Virus Infection and Acquired Immunodeficiency Syndrome".
3191:
3186:
3181:
3176:
3138:
3133:
1756:
3513:
3171:
3166:
2090:
1716:
2642:
2637:
2632:
2356:
2257:
1247:"HIV-1 Reverse Transcriptase/Integrase Dual Inhibitors: A Review of Recent Advances and Structure-activity Relationship Studies"
3894:
603:
immediately or at some point in the future, the timing depends on the activity of the chromosomal locus hosting the provirus.
591:
Integration occurs following production of the double-stranded linear viral DNA by the viral RNA/DNA-dependent DNA polymerase
3657:
1896:
613:
the strand transfer reaction, in which the processed 3' ends of the viral DNA are covalently ligated to host chromosomal DNA.
371:
230:
76:
519:
443:
302:
148:
495:
2469:
2030:
3627:
3561:
2788:
1600:
1394:
766:
641:
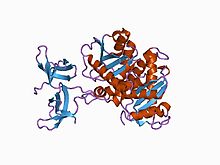
Structural depiction of the HIV catalytic core domain based on the works of Feng, L. and
Kvaratskhelia, M. from the
3255:
2923:
1741:
577:, tightly binds HIV IN and directs the HIV pre-integration complex towards highly expressed genes for integration.
561:
Crystal and NMR structures of the individual domains and 2-domain constructs of integrases from HIV-1, HIV-2,
431:
290:
136:
3774:
2314:
2309:
1869:
1687:
562:
3248:
3243:
2349:
2179:
2067:
2035:
2020:
3876:
3811:
3483:
3459:
3266:
3151:
3057:
2983:
2908:
2803:
2687:
2622:
2542:
2508:
2459:
1702:
626:
503:
598:
The main function of IN is to insert the viral DNA into the host chromosomal DNA, an essential step for
2552:
2479:
2444:
427:
286:
132:
1372:
784:"Non-Enzymatic Functions of Retroviral Integrase: The Next Target for Novel Anti-HIV Drug Development"
33:
solution structure of the n-terminal zn binding domain of hiv-1 integrase (e form), nmr, 38 structures
3434:
3238:
2439:
1578:
384:
243:
89:
3933:
3821:
3747:
3685:
3637:
3589:
3541:
3493:
3372:
3221:
3116:
2877:
2783:
2763:
2746:
2596:
2484:
2419:
1765:
1746:
1677:
1531:
1411:
1366:
2342:
1623:
1193:"The role of HIV integration in viral persistence: no more whistling past the proviral graveyard"
671:
The C-terminus domain binds to host DNA non-specifically and stabilizes the integration complex.
543:
HH-CC zinc-binding domain (a three-helical bundle stabilized by coordination of a Zn(II) cation),
524:
4038:
3996:
3832:
3717:
2586:
2557:
2077:
1850:
1760:
684:
475:
3966:
3857:
3737:
3609:
3469:
1931:
1886:
1588:
1573:
1509:
1489:
1387:
592:
1042:
Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P (July 2009). Luban J (ed.).
3768:
3706:
3393:
2837:
2828:
2750:
2697:
2518:
2409:
1845:
1840:
1720:
1583:
1563:
1472:
535:
All retroviral IN proteins contain three canonical domains, connected by flexible linkers:
510:
410:
269:
115:
8:
3439:
2833:
2823:
2742:
2108:
2100:
1881:
1548:
1484:
1448:
725:
701:
618:
3961:
3838:
1971:
1909:
1860:
1595:
1422:
1337:
1324:
1299:
1271:
1246:
1217:
1192:
1168:
1143:
1119:
1094:
1070:
1043:
1019:
994:
928:
901:
870:
845:
810:
783:
758:
570:
566:
1362:
1144:"Efficient HIV-1 in vitro reverse transcription: optimal capsid stability is required"
4028:
3413:
3161:
3100:
2865:
1986:
1976:
1966:
1891:
1875:
1834:
1820:
1815:
1553:
1516:
1341:
1329:
1276:
1222:
1173:
1124:
1075:
1024:
968:
933:
875:
815:
762:
580:
418:
376:
277:
235:
123:
81:
1262:
352:
198:
57:
4033:
3675:
3281:
3018:
3013:
3008:
3003:
2998:
2993:
2972:
2319:
1981:
1961:
1903:
1799:
1697:
1682:
1628:
1526:
1521:
1380:
1319:
1311:
1266:
1258:
1212:
1204:
1163:
1155:
1114:
1106:
1065:
1055:
1014:
1006:
960:
923:
913:
865:
857:
805:
795:
754:
406:
265:
111:
2334:
995:"Recent advances in the discovery of small-molecule inhibitors of HIV-1 integrase"
211:
3905:
3362:
3352:
3105:
2242:
2232:
2054:
2025:
1651:
1558:
1543:
1060:
487:
364:
223:
69:
1110:
3979:
3862:
3407:
3232:
3127:
2733:
2653:
2210:
2049:
1956:
1948:
1825:
1788:
1315:
1159:
861:
697:
483:
4022:
3928:
3579:
3040:
2677:
2394:
2369:
2365:
1479:
800:
514:
479:
918:
3648:
3600:
3552:
3504:
3425:
3210:
3046:
2723:
2532:
2399:
2373:
1568:
1538:
1467:
1333:
1280:
1226:
1177:
1128:
1079:
1028:
1010:
937:
879:
819:
713:
637:
499:
491:
972:
380:
239:
85:
3988:
3531:
2851:
2814:
2134:
2124:
1725:
1462:
1403:
1044:"Structural basis for functional tetramerization of lentiviral integrase"
709:
964:
3870:
3802:
3399:
3205:
3036:
3032:
1936:
1779:
1692:
1457:
1407:
1402:
657:
554:
550:
540:
467:
902:"Integrase and integration: biochemical activities of HIV-1 integrase"
900:
Delelis O, Carayon K, Saïb A, Deprez E, Mouscadet JF (December 2008).
751:
Massachusetts
General Hospital Handbook of General Hospital Psychiatry
642:
3916:
3911:
3846:
3090:
2967:
2957:
2945:
2718:
2382:
2215:
1665:
1431:
1244:
1208:
517:
bound to the ends of the viral DNA ends has been referred to as the
2498:
2062:
1642:
1357:
730:
622:
359:
218:
64:
617:
Both reactions are catalyzed in the same active site, and involve
2165:
CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase
2013:
1998:
1614:
712:(MK-0518, brand name Isentress). The second integrase inhibitor,
705:
846:"Structural Biology of HIV Integrase Strand Transfer Inhibitors"
3758:
3696:
3444:
3383:
3276:
2774:
2430:
1991:
1426:
463:
438:
323:
297:
169:
143:
28:
3329:
3319:
3314:
3304:
3299:
3294:
3289:
2896:
2891:
2886:
2008:
2003:
1855:
574:
482:
links between) its genetic information into that of the host
1245:
Mahboubi-Rabbani M, Abbasi M, Hajimahdi Z, Zarghi A (2021).
3881:
3712:
3324:
3309:
2738:
2474:
1919:
1914:
1810:
1805:
1504:
1499:
1494:
400:
347:
259:
205:
193:
105:
52:
1297:
3942:
3938:
3921:
3067:
2918:
950:
649:
599:
471:
899:
1298:
Maertens GN, Engelman AN, Cherepanov P (January 2022).
1041:
993:
Choi E, Mallareddy JR, Lu D, Kolluru S (October 2018).
992:
683:
The newly generated 3'OH groups disrupt the host DNA's
486:
it infects. Retroviral INs are not to be confused with
656:
is a 32kDa viral protein consisting of three domains-
2175:
CDP-diacylglycerol—inositol 3-phosphatidyltransferase
748:
2180:
CDP-diacylglycerol—choline O-phosphatidyltransferase
2364:
2170:
CDP-diacylglycerol—serine O-phosphatidyltransferase
843:
523:; IN is a key component in this and the retroviral
844:Jóźwik IK, Passos DO, Lyumkis D (September 2020).
328:Crystal structure of the RSV two-domain integrase.
174:Crystal structure of the RSV two-domain integrase.
4020:
1300:"Structure and function of retroviral integrase"
1092:
3978:
2350:
1388:
1141:
3647:
3599:
3551:
3503:
3987:
2813:
2196:N-acetylglucosamine-1-phosphate transferase
2086:UTP—glucose-1-phosphate uridylyltransferase
1099:Cold Spring Harbor Perspectives in Medicine
716:, was approved in the U.S. in August 2012.
2357:
2343:
1395:
1381:
1251:Iranian Journal of Pharmaceutical Research
1190:
322:
168:
27:
3514:Parainfluenza hemagglutinin-neuraminidase
2290:serine/threonine-specific protein kinases
2268:serine/threonine-specific protein kinases
2091:Galactose-1-phosphate uridylyltransferase
1365:at the U.S. National Library of Medicine
1323:
1270:
1216:
1167:
1118:
1069:
1059:
1018:
927:
917:
869:
809:
799:
691:
664:the development of retroviral therapies.
625:protein-DNA intermediate (in contrast to
586:
1148:Signal Transduction and Targeted Therapy
674:
636:
606:Retroviral INs catalyzes two reactions:
627:Ser/Tyr recombinase-catalyzed reactions
4021:
3757:
3695:
2773:
2429:
1142:Burdick RC, Pathak VK (January 2021).
781:
546:a catalytic core domain (RNaseH fold),
3658:Respiratory syncytial virus G protein
2338:
1376:
1240:
1238:
1236:
1197:The Journal of Clinical Investigation
3745:
3683:
3635:
3587:
3539:
3491:
3055:
2981:
2685:
2540:
2506:
988:
986:
984:
982:
895:
893:
891:
889:
839:
837:
835:
833:
831:
829:
1093:Craigie R, Bushman FD (July 2012).
13:
1290:
1233:
850:Trends in Pharmacological Sciences
759:10.1016/b978-1-4377-1927-7.00026-1
14:
4050:
3562:Mumps hemagglutinin-neuraminidase
1601:Glucose-1,6-bisphosphate synthase
1350:
979:
886:
826:
2924:Hepatitis B virus DNA polymerase
1742:Ribose-phosphate diphosphokinase
2755:(non-enveloped circular ds-DNA)
1356:PDB-101 Molecule of the Month:
1263:10.22037/ijpr.2021.115446.15370
1184:
1191:Maldarelli F (February 2016).
1135:
1086:
1035:
944:
775:
753:. Elsevier. pp. 353–370.
742:
696:In November 2005, data from a
1:
3775:Vesiculovirus matrix proteins
2315:Protein-histidine tele-kinase
2310:Protein-histidine pros-kinase
2189:Glycosyl-1-phosphotransferase
736:
668:prevents genome integration.
395:Available protein structures:
254:Available protein structures:
100:Available protein structures:
22:Integrase Zinc binding domain
2021:RNA-dependent RNA polymerase
1304:Nature Reviews. Microbiology
1061:10.1371/journal.ppat.1000515
530:
317:Integrase DNA binding domain
7:
3812:Structure and genome of HIV
2869:(circular partially ds-DNA)
1928:RNA-directed DNA polymerase
1796:DNA-directed DNA polymerase
1111:10.1101/cshperspect.a006890
719:
504:site-specific recombination
502:integrase, as discussed in
10:
4055:
2445:Herpesvirus glycoprotein B
2281:: protein-dual-specificity
1316:10.1038/s41579-021-00586-9
1160:10.1038/s41392-020-00458-3
862:10.1016/j.tips.2020.06.003
700:of an investigational HIV
3955:
3892:
3819:
3810:
3801:
3736:
3674:
3626:
3578:
3530:
3482:
3457:
3424:
3382:
3370:
3361:
3346:
3264:
3219:
3204:
3149:
3114:
3099:
3084:
3031:
2966:
2953:
2944:
2906:
2875:
2864:
2801:
2761:
2732:
2714:
2676:
2651:
2620:
2594:
2585:
2574:
2531:
2497:
2457:
2440:HHV capsid portal protein
2417:
2408:
2390:
2381:
2299:
2277:
2252:
2227:
2208:
2188:
2157:
2147:
2117:
2099:
2076:
2048:
1947:
1787:
1778:
1755:
1715:
1660:
1637:
1609:
1443:
1419:
788:Frontiers in Microbiology
632:
437:
417:
399:
394:
390:
370:
358:
346:
338:
333:
321:
316:
296:
276:
258:
253:
249:
229:
217:
204:
192:
184:
179:
167:
162:
142:
122:
104:
99:
95:
75:
63:
51:
43:
38:
26:
21:
2258:protein-serine/threonine
2158:Phosphatidyltransferases
1747:Thiamine diphosphokinase
1367:Medical Subject Headings
801:10.3389/fmicb.2011.00210
3435:Influenza hemagglutinin
919:10.1186/1742-4690-5-114
685:phosphodiester linkages
525:pre-integration complex
3997:Gag-onc fusion protein
2078:Nucleotidyltransferase
1761:nucleotidyltransferase
1688:Nucleoside-diphosphate
1011:10.4155/fsoa-2018-0060
692:Antiretroviral therapy
645:
587:Function and mechanism
511:macromolecular complex
3967:murine leukemia virus
3858:Reverse transcriptase
3738:Indiana vesiculovirus
3610:Measles hemagglutinin
1932:Reverse transcriptase
1095:"HIV DNA integration"
675:Integration mechanism
640:
593:reverse transcriptase
496:used in biotechnology
163:Integrase core domain
2698:Early 35 kDa protein
1721:diphosphotransferase
1703:Thiamine-diphosphate
1410:-containing groups (
621:without involving a
553:DNA-binding domain (
456:Retroviral integrase
2303:: protein-histidine
2221:; protein acceptor)
2109:mRNA capping enzyme
2101:Guanylyltransferase
965:10.1021/bi00031a002
726:Integrase inhibitor
702:integrase inhibitor
619:transesterification
3962:Rous sarcoma virus
1579:Phosphoinositide 3
1423:phosphotransferase
646:
567:Rous Sarcoma Virus
4016:
4015:
4012:
4011:
4008:
4007:
4004:
4003:
3951:
3950:
3797:
3796:
3793:
3792:
3789:
3788:
3785:
3784:
3732:
3731:
3728:
3727:
3670:
3669:
3666:
3665:
3622:
3621:
3618:
3617:
3574:
3573:
3570:
3569:
3526:
3525:
3522:
3521:
3478:
3477:
3453:
3452:
3342:
3341:
3338:
3337:
3200:
3199:
3080:
3079:
3076:
3075:
3027:
3026:
2940:
2939:
2936:
2935:
2932:
2931:
2860:
2859:
2847:
2846:
2797:
2796:
2710:
2709:
2706:
2705:
2672:
2671:
2570:
2569:
2566:
2565:
2527:
2526:
2493:
2492:
2453:
2452:
2332:
2331:
2328:
2327:
2204:
2203:
2143:
2142:
2044:
2043:
1957:Template-directed
1711:
1710:
1678:Phosphomevalonate
1358:135 HIV Integrase
999:Future Science OA
959:(31): 9826–9833.
782:Masuda T (2011).
581:Human foamy virus
453:
452:
449:
448:
444:structure summary
312:
311:
308:
307:
303:structure summary
158:
157:
154:
153:
149:structure summary
4046:
3985:
3984:
3976:
3975:
3817:
3816:
3808:
3807:
3755:
3754:
3743:
3742:
3693:
3692:
3681:
3680:
3676:Zaire ebolavirus
3645:
3644:
3633:
3632:
3597:
3596:
3585:
3584:
3549:
3548:
3537:
3536:
3501:
3500:
3489:
3488:
3380:
3379:
3368:
3367:
3359:
3358:
3282:3C-like protease
3217:
3216:
3112:
3111:
3097:
3096:
3053:
3052:
2979:
2978:
2973:Duplornaviricota
2964:
2963:
2951:
2950:
2873:
2872:
2811:
2810:
2771:
2770:
2759:
2758:
2730:
2729:
2683:
2682:
2592:
2591:
2583:
2582:
2538:
2537:
2504:
2503:
2427:
2426:
2415:
2414:
2406:
2405:
2388:
2387:
2359:
2352:
2345:
2336:
2335:
2320:Histidine kinase
2243:tyrosine kinases
2233:protein-tyrosine
2225:
2224:
2155:
2154:
1962:RNA polymerase I
1785:
1784:
1776:
1775:
1629:Aspartate kinase
1624:Phosphoglycerate
1441:
1440:
1397:
1390:
1383:
1374:
1373:
1345:
1327:
1285:
1284:
1274:
1242:
1231:
1230:
1220:
1209:10.1172/JCI80564
1188:
1182:
1181:
1171:
1139:
1133:
1132:
1122:
1090:
1084:
1083:
1073:
1063:
1039:
1033:
1032:
1022:
990:
977:
976:
948:
942:
941:
931:
921:
897:
884:
883:
873:
841:
824:
823:
813:
803:
779:
773:
772:
746:
643:protein database
488:phage integrases
392:
391:
326:
314:
313:
251:
250:
172:
160:
159:
97:
96:
31:
19:
18:
16:Class of enzymes
4054:
4053:
4049:
4048:
4047:
4045:
4044:
4043:
4019:
4018:
4017:
4000:
3947:
3906:transactivators
3888:
3781:
3724:
3662:
3614:
3566:
3518:
3474:
3449:
3420:
3363:Influenza virus
3353:Negarnaviricota
3350:
3348:
3334:
3260:
3208:
3196:
3145:
3106:Kitrinoviricota
3103:
3088:
3086:
3072:
3044:
3023:
2970:
2955:
2928:
2902:
2868:
2856:
2843:
2793:
2754:
2736:
2722:
2716:
2702:
2668:
2647:
2616:
2578:
2577:(Duplodnaviria,
2576:
2575:circular ds-DNA
2562:
2523:
2489:
2449:
2398:
2392:
2377:
2363:
2333:
2324:
2295:
2273:
2248:
2219:
2213:
2209:2.7.10-2.7.13:
2200:
2184:
2151:: miscellaneous
2139:
2113:
2095:
2072:
2055:exoribonuclease
2052:
2040:
2026:Polyadenylation
1943:
1769:
1763:
1751:
1733:
1729:
1723:
1707:
1669:
1656:
1633:
1605:
1435:
1429:
1421:
1415:
1401:
1353:
1348:
1293:
1291:Further reading
1288:
1243:
1234:
1189:
1185:
1140:
1136:
1091:
1087:
1054:(7): e1000515.
1040:
1036:
991:
980:
949:
945:
898:
887:
842:
827:
780:
776:
769:
747:
743:
739:
722:
694:
677:
635:
589:
533:
329:
175:
34:
17:
12:
11:
5:
4052:
4042:
4041:
4036:
4031:
4014:
4013:
4010:
4009:
4006:
4005:
4002:
4001:
3999:
3994:
3992:
3982:
3980:Fusion protein
3973:
3972:
3971:
3970:
3969:
3964:
3953:
3952:
3949:
3948:
3946:
3945:
3936:
3931:
3926:
3925:
3924:
3919:
3914:
3901:
3899:
3890:
3889:
3887:
3886:
3885:
3884:
3879:
3867:
3866:
3865:
3863:HIV-1 protease
3860:
3855:
3843:
3842:
3841:
3828:
3826:
3814:
3805:
3799:
3798:
3795:
3794:
3791:
3790:
3787:
3786:
3783:
3782:
3780:
3779:
3778:
3777:
3769:matrix protein
3764:
3762:
3752:
3740:
3734:
3733:
3730:
3729:
3726:
3725:
3723:
3722:
3721:
3720:
3715:
3707:matrix protein
3702:
3700:
3690:
3678:
3672:
3671:
3668:
3667:
3664:
3663:
3661:
3660:
3654:
3652:
3642:
3630:
3624:
3623:
3620:
3619:
3616:
3615:
3613:
3612:
3606:
3604:
3594:
3582:
3576:
3575:
3572:
3571:
3568:
3567:
3565:
3564:
3558:
3556:
3546:
3534:
3528:
3527:
3524:
3523:
3520:
3519:
3517:
3516:
3510:
3508:
3498:
3486:
3480:
3479:
3476:
3475:
3473:
3472:
3466:
3464:
3455:
3454:
3451:
3450:
3448:
3447:
3442:
3437:
3431:
3429:
3422:
3421:
3419:
3418:
3417:
3416:
3408:viral envelope
3404:
3403:
3402:
3394:matrix protein
3389:
3387:
3377:
3365:
3356:
3349:negative-sense
3344:
3343:
3340:
3339:
3336:
3335:
3333:
3332:
3327:
3322:
3317:
3312:
3307:
3302:
3297:
3292:
3287:
3286:
3285:
3273:
3271:
3262:
3261:
3259:
3258:
3253:
3252:
3251:
3246:
3241:
3233:viral envelope
3228:
3226:
3214:
3202:
3201:
3198:
3197:
3195:
3194:
3189:
3184:
3179:
3174:
3169:
3164:
3158:
3156:
3147:
3146:
3144:
3143:
3142:
3141:
3136:
3128:viral envelope
3123:
3121:
3109:
3094:
3087:positive-sense
3082:
3081:
3078:
3077:
3074:
3073:
3071:
3070:
3064:
3062:
3050:
3029:
3028:
3025:
3024:
3022:
3021:
3016:
3011:
3006:
3001:
2996:
2990:
2988:
2976:
2961:
2948:
2942:
2941:
2938:
2937:
2934:
2933:
2930:
2929:
2927:
2926:
2921:
2915:
2913:
2904:
2903:
2901:
2900:
2899:
2889:
2884:
2882:
2870:
2862:
2861:
2858:
2857:
2855:
2854:
2848:
2845:
2844:
2842:
2841:
2831:
2826:
2820:
2818:
2808:
2799:
2798:
2795:
2794:
2792:
2791:
2786:
2780:
2778:
2768:
2756:
2734:Polyomaviridae
2727:
2712:
2711:
2708:
2707:
2704:
2703:
2701:
2700:
2694:
2692:
2680:
2674:
2673:
2670:
2669:
2667:
2666:
2660:
2658:
2649:
2648:
2646:
2645:
2640:
2635:
2629:
2627:
2618:
2617:
2615:
2614:
2609:
2603:
2601:
2589:
2580:
2579:Varidnaviria?)
2572:
2571:
2568:
2567:
2564:
2563:
2561:
2560:
2555:
2549:
2547:
2535:
2529:
2528:
2525:
2524:
2522:
2521:
2515:
2513:
2501:
2495:
2494:
2491:
2490:
2488:
2487:
2482:
2477:
2472:
2466:
2464:
2455:
2454:
2451:
2450:
2448:
2447:
2442:
2436:
2434:
2424:
2412:
2410:Herpes simplex
2403:
2385:
2379:
2378:
2366:Viral proteins
2362:
2361:
2354:
2347:
2339:
2330:
2329:
2326:
2325:
2323:
2322:
2317:
2312:
2306:
2304:
2297:
2296:
2294:
2293:
2284:
2282:
2275:
2274:
2272:
2271:
2262:
2260:
2250:
2249:
2247:
2246:
2237:
2235:
2222:
2217:
2211:protein kinase
2206:
2205:
2202:
2201:
2199:
2198:
2192:
2190:
2186:
2185:
2183:
2182:
2177:
2172:
2167:
2161:
2159:
2152:
2145:
2144:
2141:
2140:
2138:
2137:
2132:
2121:
2119:
2115:
2114:
2112:
2111:
2105:
2103:
2097:
2096:
2094:
2093:
2088:
2082:
2080:
2074:
2073:
2071:
2070:
2065:
2059:
2057:
2050:Phosphorolytic
2046:
2045:
2042:
2041:
2039:
2038:
2033:
2028:
2023:
2018:
2017:
2016:
2011:
2006:
1996:
1995:
1994:
1984:
1979:
1974:
1969:
1964:
1959:
1953:
1951:
1949:RNA polymerase
1945:
1944:
1942:
1941:
1940:
1939:
1929:
1925:
1924:
1923:
1922:
1917:
1912:
1901:
1900:
1899:
1894:
1889:
1884:
1873:
1867:
1866:
1865:
1858:
1853:
1848:
1843:
1832:
1831:
1830:
1823:
1818:
1813:
1808:
1797:
1793:
1791:
1789:DNA polymerase
1782:
1773:
1767:
1753:
1752:
1750:
1749:
1744:
1738:
1736:
1731:
1727:
1713:
1712:
1709:
1708:
1706:
1705:
1700:
1695:
1690:
1685:
1680:
1674:
1672:
1667:
1658:
1657:
1655:
1654:
1648:
1646:
1635:
1634:
1632:
1631:
1626:
1620:
1618:
1607:
1606:
1604:
1603:
1598:
1593:
1592:
1591:
1586:
1576:
1574:Diacylglycerol
1571:
1566:
1561:
1556:
1551:
1546:
1541:
1536:
1535:
1534:
1524:
1519:
1514:
1513:
1512:
1507:
1502:
1497:
1492:
1485:Phosphofructo-
1482:
1477:
1476:
1475:
1465:
1460:
1454:
1452:
1438:
1433:
1417:
1416:
1400:
1399:
1392:
1385:
1377:
1371:
1370:
1360:
1352:
1351:External links
1349:
1347:
1346:
1294:
1292:
1289:
1287:
1286:
1257:(2): 333–369.
1232:
1203:(2): 438–447.
1183:
1134:
1105:(7): a006890.
1085:
1048:PLOS Pathogens
1034:
978:
943:
885:
856:(9): 611–626.
825:
774:
767:
740:
738:
735:
734:
733:
728:
721:
718:
693:
690:
676:
673:
634:
631:
615:
614:
611:
588:
585:
559:
558:
547:
544:
532:
529:
466:produced by a
451:
450:
447:
446:
441:
435:
434:
421:
415:
414:
404:
397:
396:
388:
387:
374:
368:
367:
362:
356:
355:
350:
344:
343:
340:
336:
335:
331:
330:
327:
319:
318:
310:
309:
306:
305:
300:
294:
293:
280:
274:
273:
263:
256:
255:
247:
246:
233:
227:
226:
221:
215:
214:
209:
202:
201:
196:
190:
189:
186:
182:
181:
177:
176:
173:
165:
164:
156:
155:
152:
151:
146:
140:
139:
126:
120:
119:
109:
102:
101:
93:
92:
79:
73:
72:
67:
61:
60:
55:
49:
48:
45:
41:
40:
36:
35:
32:
24:
23:
15:
9:
6:
4:
3:
2:
4051:
4040:
4039:Viral enzymes
4037:
4035:
4032:
4030:
4027:
4026:
4024:
3998:
3995:
3993:
3990:
3986:
3983:
3981:
3977:
3974:
3968:
3965:
3963:
3960:
3959:
3958:
3957:
3954:
3944:
3940:
3937:
3935:
3932:
3930:
3927:
3923:
3920:
3918:
3915:
3913:
3910:
3909:
3908:
3907:
3903:
3902:
3900:
3898:
3896:
3891:
3883:
3880:
3878:
3875:
3874:
3873:
3872:
3868:
3864:
3861:
3859:
3856:
3854:
3851:
3850:
3849:
3848:
3844:
3840:
3837:
3836:
3835:
3834:
3830:
3829:
3827:
3825:
3823:
3818:
3815:
3813:
3809:
3806:
3804:
3800:
3776:
3773:
3772:
3771:
3770:
3766:
3765:
3763:
3760:
3756:
3753:
3751:
3749:
3744:
3741:
3739:
3735:
3719:
3716:
3714:
3711:
3710:
3709:
3708:
3704:
3703:
3701:
3698:
3694:
3691:
3689:
3687:
3682:
3679:
3677:
3673:
3659:
3656:
3655:
3653:
3650:
3646:
3643:
3641:
3639:
3634:
3631:
3629:
3625:
3611:
3608:
3607:
3605:
3602:
3598:
3595:
3593:
3591:
3586:
3583:
3581:
3577:
3563:
3560:
3559:
3557:
3554:
3550:
3547:
3545:
3543:
3538:
3535:
3533:
3529:
3515:
3512:
3511:
3509:
3506:
3502:
3499:
3497:
3495:
3490:
3487:
3485:
3484:Parainfluenza
3481:
3471:
3468:
3467:
3465:
3463:
3461:
3456:
3446:
3443:
3441:
3440:Neuraminidase
3438:
3436:
3433:
3432:
3430:
3427:
3423:
3415:
3412:
3411:
3410:
3409:
3405:
3401:
3398:
3397:
3396:
3395:
3391:
3390:
3388:
3385:
3381:
3378:
3376:
3374:
3369:
3366:
3364:
3360:
3357:
3354:
3345:
3331:
3328:
3326:
3323:
3321:
3318:
3316:
3313:
3311:
3308:
3306:
3303:
3301:
3298:
3296:
3293:
3291:
3288:
3283:
3280:
3279:
3278:
3275:
3274:
3272:
3270:
3268:
3263:
3257:
3254:
3250:
3247:
3245:
3242:
3240:
3237:
3236:
3235:
3234:
3230:
3229:
3227:
3225:
3223:
3218:
3215:
3212:
3207:
3203:
3193:
3190:
3188:
3185:
3183:
3180:
3178:
3175:
3173:
3170:
3168:
3165:
3163:
3160:
3159:
3157:
3155:
3153:
3148:
3140:
3137:
3135:
3132:
3131:
3130:
3129:
3125:
3124:
3122:
3120:
3118:
3113:
3110:
3107:
3102:
3098:
3095:
3092:
3083:
3069:
3066:
3065:
3063:
3061:
3059:
3054:
3051:
3048:
3042:
3038:
3034:
3030:
3020:
3017:
3015:
3012:
3010:
3007:
3005:
3002:
3000:
2997:
2995:
2992:
2991:
2989:
2987:
2985:
2980:
2977:
2974:
2969:
2965:
2962:
2959:
2952:
2949:
2947:
2943:
2925:
2922:
2920:
2917:
2916:
2914:
2912:
2910:
2905:
2898:
2895:
2894:
2893:
2890:
2888:
2885:
2883:
2881:
2879:
2874:
2871:
2867:
2863:
2853:
2850:
2849:
2839:
2835:
2832:
2830:
2827:
2825:
2822:
2821:
2819:
2816:
2812:
2809:
2807:
2805:
2800:
2790:
2787:
2785:
2782:
2781:
2779:
2776:
2772:
2769:
2767:
2765:
2760:
2757:
2752:
2748:
2744:
2740:
2735:
2731:
2728:
2725:
2720:
2713:
2699:
2696:
2695:
2693:
2691:
2689:
2684:
2681:
2679:
2678:Baculoviridae
2675:
2665:
2662:
2661:
2659:
2657:
2655:
2650:
2644:
2641:
2639:
2636:
2634:
2631:
2630:
2628:
2626:
2624:
2619:
2613:
2610:
2608:
2605:
2604:
2602:
2600:
2598:
2593:
2590:
2588:
2584:
2581:
2573:
2559:
2556:
2554:
2551:
2550:
2548:
2546:
2544:
2539:
2536:
2534:
2530:
2520:
2517:
2516:
2514:
2512:
2510:
2505:
2502:
2500:
2496:
2486:
2483:
2481:
2478:
2476:
2473:
2471:
2468:
2467:
2465:
2463:
2461:
2456:
2446:
2443:
2441:
2438:
2437:
2435:
2432:
2428:
2425:
2423:
2421:
2416:
2413:
2411:
2407:
2404:
2401:
2396:
2395:Duplodnaviria
2391:linear ds-DNA
2389:
2386:
2384:
2380:
2375:
2371:
2367:
2360:
2355:
2353:
2348:
2346:
2341:
2340:
2337:
2321:
2318:
2316:
2313:
2311:
2308:
2307:
2305:
2302:
2298:
2292:
2291:
2286:
2285:
2283:
2280:
2276:
2270:
2269:
2264:
2263:
2261:
2259:
2255:
2251:
2245:
2244:
2239:
2238:
2236:
2234:
2230:
2226:
2223:
2220:
2212:
2207:
2197:
2194:
2193:
2191:
2187:
2181:
2178:
2176:
2173:
2171:
2168:
2166:
2163:
2162:
2160:
2156:
2153:
2150:
2146:
2136:
2133:
2130:
2126:
2123:
2122:
2120:
2116:
2110:
2107:
2106:
2104:
2102:
2098:
2092:
2089:
2087:
2084:
2083:
2081:
2079:
2075:
2069:
2066:
2064:
2061:
2060:
2058:
2056:
2051:
2047:
2037:
2034:
2032:
2029:
2027:
2024:
2022:
2019:
2015:
2012:
2010:
2007:
2005:
2002:
2001:
2000:
1997:
1993:
1990:
1989:
1988:
1985:
1983:
1980:
1978:
1975:
1973:
1970:
1968:
1965:
1963:
1960:
1958:
1955:
1954:
1952:
1950:
1946:
1938:
1935:
1934:
1933:
1930:
1927:
1926:
1921:
1918:
1916:
1913:
1911:
1908:
1907:
1905:
1902:
1898:
1895:
1893:
1890:
1888:
1885:
1883:
1880:
1879:
1877:
1874:
1871:
1868:
1864:
1863:
1859:
1857:
1854:
1852:
1849:
1847:
1844:
1842:
1839:
1838:
1836:
1833:
1829:
1828:
1824:
1822:
1819:
1817:
1814:
1812:
1809:
1807:
1804:
1803:
1801:
1798:
1795:
1794:
1792:
1790:
1786:
1783:
1781:
1777:
1774:
1771:
1762:
1758:
1754:
1748:
1745:
1743:
1740:
1739:
1737:
1734:
1722:
1718:
1714:
1704:
1701:
1699:
1696:
1694:
1691:
1689:
1686:
1684:
1681:
1679:
1676:
1675:
1673:
1670:
1663:
1659:
1653:
1650:
1649:
1647:
1644:
1640:
1636:
1630:
1627:
1625:
1622:
1621:
1619:
1616:
1612:
1608:
1602:
1599:
1597:
1594:
1590:
1589:Class II PI 3
1587:
1585:
1582:
1581:
1580:
1577:
1575:
1572:
1570:
1567:
1565:
1564:Deoxycytidine
1562:
1560:
1557:
1555:
1552:
1550:
1547:
1545:
1542:
1540:
1537:
1533:
1532:ADP-thymidine
1530:
1529:
1528:
1525:
1523:
1520:
1518:
1515:
1511:
1508:
1506:
1503:
1501:
1498:
1496:
1493:
1491:
1488:
1487:
1486:
1483:
1481:
1478:
1474:
1471:
1470:
1469:
1466:
1464:
1461:
1459:
1456:
1455:
1453:
1450:
1446:
1442:
1439:
1436:
1428:
1424:
1418:
1413:
1409:
1405:
1398:
1393:
1391:
1386:
1384:
1379:
1378:
1375:
1368:
1364:
1361:
1359:
1355:
1354:
1343:
1339:
1335:
1331:
1326:
1321:
1317:
1313:
1309:
1305:
1301:
1296:
1295:
1282:
1278:
1273:
1268:
1264:
1260:
1256:
1252:
1248:
1241:
1239:
1237:
1228:
1224:
1219:
1214:
1210:
1206:
1202:
1198:
1194:
1187:
1179:
1175:
1170:
1165:
1161:
1157:
1153:
1149:
1145:
1138:
1130:
1126:
1121:
1116:
1112:
1108:
1104:
1100:
1096:
1089:
1081:
1077:
1072:
1067:
1062:
1057:
1053:
1049:
1045:
1038:
1030:
1026:
1021:
1016:
1012:
1008:
1005:(9): FSO338.
1004:
1000:
996:
989:
987:
985:
983:
974:
970:
966:
962:
958:
954:
947:
939:
935:
930:
925:
920:
915:
911:
907:
906:Retrovirology
903:
896:
894:
892:
890:
881:
877:
872:
867:
863:
859:
855:
851:
847:
840:
838:
836:
834:
832:
830:
821:
817:
812:
807:
802:
797:
793:
789:
785:
778:
770:
768:9781437719277
764:
760:
756:
752:
745:
741:
732:
729:
727:
724:
723:
717:
715:
711:
707:
703:
699:
698:phase 2 study
689:
686:
681:
672:
669:
665:
661:
659:
655:
652:
651:
644:
639:
630:
628:
624:
620:
612:
609:
608:
607:
604:
601:
596:
594:
584:
582:
578:
576:
572:
568:
564:
556:
552:
548:
545:
542:
538:
537:
536:
528:
526:
522:
521:
516:
515:macromolecule
512:
507:
505:
501:
497:
493:
489:
485:
481:
477:
473:
469:
465:
461:
457:
445:
442:
440:
436:
433:
429:
425:
422:
420:
416:
412:
408:
405:
402:
398:
393:
389:
386:
382:
378:
375:
373:
369:
366:
363:
361:
357:
354:
351:
349:
345:
341:
337:
332:
325:
320:
315:
304:
301:
299:
295:
292:
288:
284:
281:
279:
275:
271:
267:
264:
261:
257:
252:
248:
245:
241:
237:
234:
232:
228:
225:
222:
220:
216:
213:
210:
207:
203:
200:
197:
195:
191:
187:
183:
178:
171:
166:
161:
150:
147:
145:
141:
138:
134:
130:
127:
125:
121:
117:
113:
110:
107:
103:
98:
94:
91:
87:
83:
80:
78:
74:
71:
68:
66:
62:
59:
56:
54:
50:
46:
42:
37:
30:
25:
20:
3904:
3893:
3869:
3852:
3845:
3831:
3820:
3767:
3746:
3705:
3684:
3649:glycoprotein
3636:
3601:glycoprotein
3588:
3553:glycoprotein
3540:
3505:glycoprotein
3492:
3458:
3426:glycoprotein
3406:
3392:
3371:
3265:
3256:Nucleocapsid
3231:
3220:
3211:Pisuviricota
3150:
3126:
3115:
3056:
3047:Pisuviricota
2982:
2907:
2876:
2802:
2762:
2724:Monodnaviria
2686:
2652:
2621:
2595:
2587:Epstein–Barr
2541:
2533:Adenoviridae
2507:
2458:
2418:
2400:Varidnaviria
2287:
2265:
2240:
2128:
1861:
1826:
1584:Class I PI 3
1549:Pantothenate
1420:2.7.1-2.7.4:
1404:Transferases
1310:(1): 20–34.
1307:
1303:
1254:
1250:
1200:
1196:
1186:
1151:
1147:
1137:
1102:
1098:
1088:
1051:
1047:
1037:
1002:
998:
956:
953:Biochemistry
952:
946:
909:
905:
853:
849:
791:
787:
777:
750:
744:
714:elvitegravir
695:
682:
678:
670:
666:
662:
653:
648:
647:
616:
605:
597:
590:
579:
560:
534:
518:
508:
492:recombinases
459:
455:
454:
47:Integrase_Zn
3989:oncoprotein
3101:Hepatitis C
2866:Hepatitis B
2852:Agnoprotein
2815:oncoprotein
2789:VP2 and VP3
2135:Transposase
2125:Recombinase
1770:-nucleoside
1596:Sphingosine
710:Raltegravir
334:Identifiers
180:Identifiers
39:Identifiers
4023:Categories
3414:M2 protein
3400:M1 protein
3206:SARS-CoV-2
1937:Telomerase
1780:Polymerase
1554:Mevalonate
1517:Riboflavin
1408:phosphorus
1363:Integrases
912:(1): 114.
737:References
658:N-terminus
551:C-terminal
541:N-terminal
498:, such as
476:integrates
468:retrovirus
407:structures
266:structures
112:structures
3956:Multiple
3853:Integrase
3091:Riboviria
2968:Rotavirus
2958:Riboviria
2719:Riboviria
2129:Integrase
2053:3' to 5'
1698:Guanylate
1693:Uridylate
1683:Adenylate
1527:Thymidine
1522:Shikimate
1342:235787691
1154:(1): 13.
654:integrase
531:Structure
513:of an IN
470:(such as
365:IPR001037
224:IPR001584
70:IPR003308
4029:Virology
3249:Membrane
3244:Envelope
2838:SV40 Tag
2499:Vaccinia
2063:RNase PH
1671:acceptor
1652:Creatine
1645:acceptor
1617:acceptor
1559:Pyruvate
1544:Glycerol
1505:Platelet
1480:Galacto-
1451:acceptor
1334:34244677
1281:34567166
1227:26829624
1178:33436564
1129:22762018
1080:19609359
1029:30416746
938:19091057
880:32624197
820:22016749
731:Integron
720:See also
623:covalent
571:tetramer
555:SH3 fold
520:intasome
480:covalent
462:) is an
424:RCSB PDB
360:InterPro
342:IN_DBD_C
283:RCSB PDB
219:InterPro
129:RCSB PDB
65:InterPro
4034:Enzymes
3580:Measles
3033:Rhinov.
2480:ICP34.5
2014:PrimPol
1999:Primase
1473:Hepatic
1468:Fructo-
1325:8671357
1272:8457747
1218:4731194
1169:7804106
1120:3385939
1071:2705190
1020:6222271
973:7632683
929:2615046
871:7429322
811:3192317
794:: 210.
706:MK-0518
500:λ phage
478:(forms
474:) that
353:PF00552
199:PF00665
58:PF02022
3759:capsid
3697:capsid
3445:HA-tag
3384:capsid
3347:ss-RNA
3277:ORF1ab
3085:ss-RNA
3045:etc. (
2954:ds-RNA
2775:capsid
2643:EBNA-3
2638:EBNA-2
2633:EBNA-1
2431:capsid
2301:2.7.13
2279:2.7.12
2254:2.7.11
2229:2.7.10
2068:PNPase
2036:PNPase
1992:POLRMT
1987:ssRNAP
1500:Muscle
1463:Gluco-
1427:kinase
1369:(MeSH)
1340:
1332:
1322:
1279:
1269:
1225:
1215:
1176:
1166:
1127:
1117:
1078:
1068:
1027:
1017:
971:
936:
926:
878:
868:
818:
808:
765:
633:In HIV
565:, and
464:enzyme
439:PDBsum
413:
403:
385:SUPFAM
339:Symbol
298:PDBsum
272:
262:
244:SUPFAM
212:CL0219
185:Symbol
144:PDBsum
118:
108:
90:SUPFAM
44:Symbol
3895:VRAPs
3877:gp120
3532:Mumps
3330:ORF9b
3320:ORF7b
3315:ORF7a
3305:ORF3d
3300:ORF3c
3295:ORF3b
3290:ORF3a
3284:(NS5)
3239:Spike
3041:Hep A
3037:Polio
2897:HBeAg
2892:HBcAg
2887:HBsAg
2751:HaPyV
2747:MCPyV
2715:other
2654:ncRNA
2612:LMP-2
2607:LMP-1
2485:ICP47
2470:vmw65
2370:early
2149:2.7.8
2118:Other
1757:2.7.7
1717:2.7.6
1662:2.7.4
1639:2.7.3
1611:2.7.2
1495:Liver
1458:Hexo-
1445:2.7.1
1338:S2CID
575:LEDGF
381:SCOPe
372:SCOP2
240:SCOPe
231:SCOP2
86:SCOPe
77:SCOP2
3882:gp41
3822:VSPs
3748:VSPs
3718:VP24
3713:VP40
3686:VSPs
3638:VSPs
3590:VSPs
3542:VSPs
3494:VSPs
3460:VNPs
3373:VSPs
3325:ORF8
3310:ORF6
3267:VNPs
3222:VSPs
3192:NS5B
3187:NS5A
3182:NS4B
3177:NS4A
3152:VNPs
3117:VSPs
3058:VNPs
3019:NSP6
3014:NSP5
3009:NSP4
3004:NSP3
2999:NSP2
2994:NSP1
2984:VNPs
2909:VNPs
2878:VSPs
2834:LTag
2829:MTag
2824:STag
2804:VNPs
2764:VSPs
2743:MPyV
2739:SV40
2688:VNPs
2664:EBER
2623:VNPs
2597:VSPs
2543:VNPs
2519:B13R
2509:VNPs
2475:ICP8
2460:VNPs
2420:VSPs
2374:late
2372:and
2288:see
2266:see
2241:see
1615:COOH
1414:2.7)
1330:PMID
1277:PMID
1223:PMID
1174:PMID
1125:PMID
1076:PMID
1025:PMID
969:PMID
934:PMID
876:PMID
816:PMID
763:ISBN
509:The
484:cell
432:PDBj
428:PDBe
411:ECOD
401:Pfam
377:1ihw
348:Pfam
291:PDBj
287:PDBe
270:ECOD
260:Pfam
236:2itg
208:clan
206:Pfam
194:Pfam
137:PDBj
133:PDBe
116:ECOD
106:Pfam
82:1wjb
53:Pfam
3943:Vpx
3941:or
3939:Vpu
3934:Vif
3929:Nef
3922:Vpr
3917:Rev
3912:Tat
3871:env
3847:pol
3839:p24
3833:gag
3628:RSV
3470:NS1
3172:NS3
3167:NS2
3068:VPg
2946:RNA
2919:HBx
2784:VP1
2558:E1B
2553:E1A
2383:DNA
2031:PAP
1972:III
1906:/Y
1897:TDT
1878:/X
1870:III
1862:Pfu
1837:/B
1827:Taq
1802:/A
1569:PFP
1539:NAD
1320:PMC
1312:doi
1267:PMC
1259:doi
1213:PMC
1205:doi
1201:126
1164:PMC
1156:doi
1115:PMC
1107:doi
1066:PMC
1056:doi
1015:PMC
1007:doi
961:doi
924:PMC
914:doi
866:PMC
858:doi
806:PMC
796:doi
755:doi
650:HIV
600:HIV
563:SIV
539:an
472:HIV
419:PDB
278:PDB
188:rve
124:PDB
4025::
3803:RT
3162:P7
3139:E2
3134:E1
3039:,
3035:,
2749:,
2745:,
2741:,
2256::
2231::
2216:PO
1977:IV
1967:II
1876:IV
1872:/C
1835:II
1821:T7
1766:PO
1759::
1719::
1666:PO
1664::
1641::
1613::
1449:OH
1447::
1432:PO
1412:EC
1406::
1336:.
1328:.
1318:.
1308:20
1306:.
1302:.
1275:.
1265:.
1255:20
1253:.
1249:.
1235:^
1221:.
1211:.
1199:.
1195:.
1172:.
1162:.
1150:.
1146:.
1123:.
1113:.
1101:.
1097:.
1074:.
1064:.
1050:.
1046:.
1023:.
1013:.
1001:.
997:.
981:^
967:.
957:34
955:.
932:.
922:.
908:.
904:.
888:^
874:.
864:.
854:41
852:.
848:.
828:^
814:.
804:.
790:.
786:.
761:.
704:,
629:.
595:.
557:).
549:a
527:.
506:.
494:)
460:IN
430:;
426:;
409:/
383:/
379:/
289:;
285:;
268:/
242:/
238:/
135:;
131:;
114:/
88:/
84:/
3991::
3897::
3824::
3761::
3750::
3699::
3688::
3651::
3640::
3603::
3592::
3555::
3544::
3507::
3496::
3462::
3428::
3386::
3375::
3355:)
3351:(
3269::
3224::
3213:)
3209:(
3154::
3119::
3108:)
3104:(
3093:)
3089:(
3060::
3049:)
3043:,
2986::
2975:)
2971:(
2960:)
2956:(
2911::
2880::
2840:)
2836:(
2817::
2806::
2777::
2766::
2753:)
2737:(
2726:)
2721:,
2717:(
2690::
2656::
2625::
2599::
2545::
2511::
2462::
2433::
2422::
2402:)
2397:,
2393:(
2376:)
2368:(
2358:e
2351:t
2344:v
2218:4
2214:(
2131:)
2127:(
2009:2
2004:1
1982:V
1920:κ
1915:ι
1910:η
1904:V
1892:μ
1887:λ
1882:β
1856:ζ
1851:ε
1846:δ
1841:α
1816:ν
1811:θ
1806:γ
1800:I
1772:)
1768:4
1764:(
1735:)
1732:7
1730:O
1728:2
1726:P
1724:(
1668:4
1643:N
1510:2
1490:1
1437:)
1434:4
1430:(
1425:/
1396:e
1389:t
1382:v
1344:.
1314::
1283:.
1261::
1229:.
1207::
1180:.
1158::
1152:6
1131:.
1109::
1103:2
1082:.
1058::
1052:5
1031:.
1009::
1003:4
975:.
963::
940:.
916::
910:5
882:.
860::
822:.
798::
792:2
771:.
757::
490:(
458:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.