236:
119:
during his doctoral thesis, however he lacked noting the importance of hydrogen ion concentration and mutarotation of glucose. The goal of Henri's thesis was to compare his knowledge of enzyme-catalysed reactions to the recognized laws of physical chemistry. Henri is credited with being the first to write the equation that is now known as the
Michaelis-Menten equation. Using glucose and fructose in the catalytic reactions controlled by maltase and invertase, Leonor Michaelis was the first scientist to distinguish the different types of inhibition by using the pH scale which did not exist in Henri's time.
278:
allosteric site and prevents the enzyme-substrate complex from performing a chemical reaction. This does not affect the Km (affinity) of the enzyme (for the substrate). Non-competitive inhibition differs from uncompetitive inhibition in that it still allows the substrate to bind to the enzyme-inhibitor complex and form an enzyme-substrate-inhibitor complex, this is not true in uncompetitive inhibition, it prevents the substrate from binding to the enzyme inhibitor through conformational change upon allosteric binding.
189:
157:. Although, these are both in the dextrorotatory form, this is where they noted that glucose can change spontaneously, also known as mutarotation. Failing to take this into consideration was one of the main reasons Henri's experiments fell short. Using invertase to catalyze sucrose inversion, they could see how fast the enzyme was reacting by polarimetry; therefore, non-competitive inhibition was found to occur in the reaction where sucrose was inverted with invertase.
1397:
108:, and it is the enzyme the kinetics of which have been supported by Michaelis and Menten to be revolutionary for the kinetics of other enzymes. While expressing the rate of the reaction studied, they derived an equation that described the rate in a way which suggested that it is mostly dependent on the enzyme concentration, as well as on presence of the substrate, but only to a certain extent.
126:, that enzyme they were using had some affinity for both products of this reaction – fructose and glucose. Using Henri's methods, Michaelis and Menten nearly perfected this concept of initial-rate method for steady-state experiments. They were studying inhibition when they found that non-competitive (mixed) inhibition is characterized by its effect on
133:(catalyst rate) while competitive is characterized by its effect on velocity (V). In the Michaelis and Menten experiments they heavily focused on pH effects of invertase using hydrogen ions. Invertase is an enzyme found in extracellular yeast and catalyzed reactions by hydrolysis or inverting a sucrose (mixture of sucrose and fructose) to “
232:, but it is possible for the inhibitor to operate via other means including direct binding to the active site. It differs from competitive inhibition in that the binding of the inhibitor does not prevent binding of substrate, and vice versa, but simply prevents product formation for a limited time.
277:
The primary difference between competitive and non-competitive is that competitive inhibition affects the substrate's ability to bind by binding an inhibitor in place of a substrate, which lowers the affinity of the enzyme for the substrate. In non-competitive inhibition, the inhibitor binds to an
196:
Non-competitive inhibition models a system where the inhibitor and the substrate may both be bound to the enzyme at any given time. When both the substrate and the inhibitor are bound, the enzyme-substrate-inhibitor complex cannot form product and can only be converted back to the enzyme-substrate
118:
laid the groundwork for the discoveries in enzyme kinetics that
Michaelis and Menten are known for. Brown theoretically envisioned the mechanism now accepted for enzyme kinetics, but did not have the quantitative data to make a claim. Victor Henri made significant contributions to enzyme kinetics
224:
inhibiting hexokinase in the brain. Carbons 2 and 4 on glucose-6-phosphate contain hydroxyl groups that attach along with the phosphate at carbon 6 to the enzyme-inhibitor complex. The substrate and enzyme are different in their group combinations that an inhibitor attaches to. The ability of
63:
and a friend Peter Rona built a compact lab, in the hospital, and over the course of five years – Michaelis successfully became published over 100 times. During his research in the hospital, he was the first to view the different types of inhibition; specifically using fructose and glucose as
305:
because the inhibitor binds to both the enzyme and the enzyme-substrate complex equally so that the equilibrium is maintained. However, since some enzyme is always inhibited from converting the substrate to product, the effective enzyme concentration is lowered.
216:
is an amino acid which is synthesized from pyruvate also inhibits the enzyme pyruvate kinase during glycolysis. Alanine is a non-competitive inhibitor, therefore it binds away from the active site to the substrate in order for it to still be the final product.
593:
Strelow J, Dewe W, Iversen PW, Brooks HB, Radding JA, McGee J, Weidner J (2004). "Mechanism of Action Assays for
Enzymes". In Markossian S, Grossman A, Brimacombe K, Arkin M, Auld D, Austin CP, et al. (eds.).
533:
410:
197:
complex or the enzyme-inhibitor complex. Non-competitive inhibition is distinguished from general mixed inhibition in that the inhibitor has an equal affinity for the enzyme and the enzyme-substrate complex.
562:, and 6-hydroxyflavone. Computer docking simulation and constructed mutants substituted indicate that the noncompetitive binding site of 6-hydroxyflavone is the reported allosteric binding site of
89:) on any given graph; this inhibitor binds to a site that has specificity for the certain molecule. Michaelis determined that when the inhibitor is bound, the enzyme would become inactivated.
274:
the Vmax is reduced during the addition of a non-competitive inhibitor, which is shown in the plot by a change in both the slope and y-intercept when a non-competitive inhibitor is added.
47:
The inhibitor may bind to the enzyme whether or not the substrate has already been bound, but if it has a higher affinity for binding the enzyme in one state or the other, it is called a
40:
where the inhibitor reduces the activity of the enzyme and binds equally well to the enzyme whether or not it has already bound the substrate. This is unlike
772:
180:
Many sources continue to conflate these two terms, or state the definition of allosteric inhibition as the definition for non-competitive inhibition.
137:.” The main reason for using invertase was that it could be easily assayed and experiments could be done in quicker manner. Sucrose rotates in
122:
Particularly during their work on describing the rate of this reaction they also tested and extrapolated on the idea of another scientist,
418:
315:
972:
165:
It is important to note that while all non-competitive inhibitors bind the enzyme at allosteric sites (i.e. locations other than its
291:
286:
In the presence of a non-competitive inhibitor, the apparent enzyme affinity is equivalent to the actual affinity. In terms of
169:)—not all inhibitors that bind at allosteric sites are non-competitive inhibitors. In fact, allosteric inhibitors may act as
1306:
1150:
1109:
1258:
1075:
251:
1267:
270:). When a non-competitive inhibitor is added the Vmax is changed, while the Km remains unchanged. According to the
1181:
965:
647:. Proceedings of the Beilstein ESCEC Symposium - Celebrating the 100th Anniversary of Michaelis Menten-Kinetics.
146:
142:
1227:
1134:
776:
1321:
1155:
1354:
1346:
228:
The most common mechanism of non-competitive inhibition involves reversible binding of the inhibitor to an
1250:
1240:
1222:
1163:
1067:
302:
1427:
1387:
1104:
1046:
958:
225:
glucose-6-phosphate to bind at different places at the same time makes it a non-competitive inhibitor.
44:, where binding affinity for the substrate in the enzyme is decreased in the presence of an inhibitor.
1432:
1370:
1193:
287:
267:
1059:
153:
is released in reactions catalyzed by invertase which is very unstable and spontaneously changes to
1326:
1188:
559:
259:
235:
174:
1301:
1279:
1245:
1004:
819:"The non-competitive inhibition of brain hexokinase by glucose-6-phosphate and related compounds"
20:
1262:
1232:
1198:
1171:
1083:
999:
170:
73:
41:
1311:
229:
636:
271:
134:
8:
1422:
1176:
1091:
919:(3). American Society for Pharmacology & Experimental Therapeutics (ASPET): 629–634.
221:
104:
into two products – fructose and glucose. The enzyme involved in this reaction is called
600:. Eli Lilly & Company and the National Center for Advancing Translational Sciences.
1014:
936:
886:
754:
700:
838:
1284:
928:
890:
878:
843:
746:
692:
601:
247:
243:
111:
758:
704:
149:. This made tracking the inversion of sugar relatively simple. They also found that
1417:
1019:
985:
920:
870:
833:
736:
684:
652:
60:
48:
37:
950:
940:
741:
724:
688:
675:
Michaelis L, Menten MM (September 2013). "The kinetics of invertin action. 1913".
72:. Findings from that experiment allowed for the divergence of non-competitive and
1289:
1096:
555:
205:
818:
1401:
1142:
1129:
1036:
799:
657:
640:
595:
1411:
1272:
188:
192:
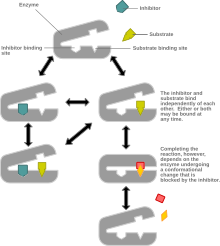
Illustration of a possible mechanism of non-competitive or mixed inhibition.
981:
932:
924:
882:
847:
750:
696:
605:
209:
123:
115:
861:
Waldrop GL (January 2009). "A qualitative approach to enzyme inhibition".
1119:
905:
166:
138:
93:
27:
904:
Si D, Wang Y, Zhou YH, Guo Y, Wang J, Zhou H, et al. (March 2009).
551:
201:
874:
538:
1212:
255:
105:
528:{\displaystyle {apparent\ _{0}}={\frac {_{0}}{1+{\frac {}{K_{I}}}}}}
405:{\displaystyle V_{max}^{app}={\frac {V_{max}}{1+{\frac {}{K_{I}}}}}}
1396:
213:
154:
150:
97:
69:
65:
96:
worked on a reaction that was used to change the composition of
1051:
566:
563:
547:
544:
92:
Like many other scientists of their time, Leonor
Michaelis and
1336:
101:
78:
906:"Mechanism of CYP2C9 inhibition by flavones and flavonols"
220:
Another example of non-competitive inhibition is given by
592:
26:"Non-competitive" redirects here. For other uses, see
1385:
773:"Noncompetitive inhibition and allosteric inhibition"
421:
318:
980:
539:
Example: noncompetitive inhibitors of CYP2C9 enzyme
68:activity. Maltase breaks maltose into two units of
797:
527:
404:
200:For example, in the enzyme-catalyzed reactions of
1409:
641:"One hundred years of Michaelis–Menten kinetics"
674:
800:"The Glycolytic Pathway Is Tightly Controlled"
722:
635:
966:
903:
863:Biochemistry and Molecular Biology Education
731:. A century of Michaelis - Menten kinetics.
854:
204:, accumulation phosphoenol is catalyzed by
973:
959:
837:
816:
810:
740:
656:
588:
586:
584:
582:
76:. Non-competitive inhibition affects the
775:. Biology Online (forum). Archived from
234:
187:
59:During his years working as a physician
860:
798:Berg JM, Tymoczko JL, Stryer L (2002).
301:. This can be seen as a consequence of
1410:
579:
250:without changing the apparent binding
954:
718:
716:
714:
670:
668:
631:
629:
627:
625:
623:
621:
619:
617:
615:
242:This type of inhibition reduces the
826:The Journal of Biological Chemistry
723:Cornish-Bowden A (September 2013).
13:
14:
1444:
817:Crane RK, Sols A (October 1954).
711:
665:
612:
1395:
725:"The origins of enzyme kinetics"
913:Drug Metabolism and Disposition
897:
791:
765:
506:
500:
480:
473:
457:
450:
383:
377:
160:
1:
839:10.1016/S0021-9258(18)65385-2
742:10.1016/j.febslet.2013.06.009
689:10.1016/j.febslet.2013.07.015
572:
543:Noncompetitive inhibitors of
183:
7:
281:
10:
1449:
1151:Dihydropteroate synthetase
1010:Non-competitive inhibition
658:10.1016/j.pisc.2014.12.002
54:
34:Non-competitive inhibition
25:
18:
1371:Steroidogenesis inhibitor
1363:
1335:
1211:
1118:
1035:
1028:
992:
288:Michaelis-Menten kinetics
268:Michaelis-Menten kinetics
1307:Matrix metalloproteinase
1110:Ribonucleotide reductase
1005:Uncompetitive inhibition
560:phenethyl isothiocyanate
303:Le Chatelier's principle
145:whereas invert sugar is
19:Not to be confused with
1076:Dihydrofolate reductase
645:Perspectives in Science
21:Uncompetitive inhibitor
1172:Nucleotidyltransferase
1000:Competitive inhibition
925:10.1124/dmd.108.023416
529:
406:
239:
193:
173:, non-competitive, or
74:competitive inhibition
42:competitive inhibition
1182:Reverse transcriptase
651:(Supplement C): 3–9.
597:Assay Guidance Manual
530:
407:
238:
191:
1228:Acetylcholinesterase
1135:Thymidylate synthase
419:
316:
272:Lineweaver-Burk plot
85:value (but not the K
1322:Histone deacetylase
1156:Farnesyltransferase
345:
222:glucose-6-phosphate
1355:Carbonic anhydrase
1347:Dopa decarboxylase
1015:Suicide inhibition
525:
402:
319:
240:
194:
1428:Enzyme inhibitors
1383:
1382:
1379:
1378:
1251:Alpha-glucosidase
1241:Polygalacturonase
1223:Phosphodiesterase
1164:GABA transaminase
1068:Monoamine oxidase
1052:HMG-CoA reductase
986:enzyme inhibition
875:10.1002/bmb.20243
735:(17): 2725–2730.
683:(17): 2712–2720.
523:
520:
449:
400:
397:
248:chemical reaction
112:Adrian John Brown
38:enzyme inhibition
16:Enzyme inhibition
1440:
1433:Pharmacodynamics
1400:
1399:
1391:
1105:Xanthine oxidase
1047:Aldose reductase
1033:
1032:
1020:Mixed inhibition
975:
968:
961:
952:
951:
945:
944:
910:
901:
895:
894:
858:
852:
851:
841:
823:
814:
808:
807:
795:
789:
788:
786:
784:
779:on 25 April 2015
769:
763:
762:
744:
720:
709:
708:
672:
663:
662:
660:
637:Cornish-Bowden A
633:
610:
609:
590:
534:
532:
531:
526:
524:
522:
521:
519:
518:
509:
498:
489:
488:
487:
471:
466:
465:
464:
447:
411:
409:
408:
403:
401:
399:
398:
396:
395:
386:
375:
366:
365:
350:
344:
333:
309:Mathematically,
143:dextroratatory-D
61:Leonor Michaelis
1448:
1447:
1443:
1442:
1441:
1439:
1438:
1437:
1408:
1407:
1406:
1394:
1386:
1384:
1375:
1359:
1331:
1207:
1194:Tyrosine kinase
1114:
1024:
988:
979:
949:
948:
908:
902:
898:
859:
855:
821:
815:
811:
806:(5th ed.).
796:
792:
782:
780:
771:
770:
766:
721:
712:
673:
666:
634:
613:
591:
580:
575:
556:tranylcypromine
541:
514:
510:
499:
497:
490:
483:
479:
472:
470:
460:
456:
422:
420:
417:
416:
391:
387:
376:
374:
367:
355:
351:
349:
334:
323:
317:
314:
313:
300:
295:
284:
265:
230:allosteric site
206:pyruvate kinase
186:
163:
131:
88:
82:
57:
49:mixed inhibitor
31:
24:
17:
12:
11:
5:
1446:
1436:
1435:
1430:
1425:
1420:
1405:
1404:
1381:
1380:
1377:
1376:
1374:
1373:
1367:
1365:
1361:
1360:
1358:
1357:
1350:
1349:
1342:
1340:
1333:
1332:
1330:
1329:
1327:Beta-lactamase
1324:
1317:
1316:
1315:
1314:
1309:
1304:
1294:
1293:
1292:
1287:
1277:
1276:
1275:
1270:
1254:
1253:
1248:
1243:
1236:
1235:
1230:
1225:
1218:
1216:
1209:
1208:
1206:
1205:
1204:
1203:
1202:
1201:
1189:Protein kinase
1186:
1185:
1184:
1179:
1167:
1166:
1159:
1158:
1153:
1146:
1145:
1138:
1137:
1132:
1125:
1123:
1116:
1115:
1113:
1112:
1107:
1100:
1099:
1094:
1087:
1086:
1079:
1078:
1071:
1070:
1063:
1062:
1055:
1054:
1049:
1042:
1040:
1037:Oxidoreductase
1030:
1026:
1025:
1023:
1022:
1017:
1012:
1007:
1002:
996:
994:
990:
989:
978:
977:
970:
963:
955:
947:
946:
896:
853:
832:(2): 597–606.
809:
790:
764:
710:
664:
639:(2015-03-01).
611:
577:
576:
574:
571:
540:
537:
536:
535:
517:
513:
508:
505:
502:
496:
493:
486:
482:
478:
475:
469:
463:
459:
455:
452:
446:
443:
440:
437:
434:
431:
428:
425:
413:
412:
394:
390:
385:
382:
379:
373:
370:
364:
361:
358:
354:
348:
343:
340:
337:
332:
329:
326:
322:
298:
293:
283:
280:
263:
185:
182:
162:
159:
147:levorotatory-L
129:
86:
80:
64:inhibitors of
56:
53:
15:
9:
6:
4:
3:
2:
1445:
1434:
1431:
1429:
1426:
1424:
1421:
1419:
1416:
1415:
1413:
1403:
1398:
1393:
1392:
1389:
1372:
1369:
1368:
1366:
1364:Miscellaneous
1362:
1356:
1352:
1351:
1348:
1344:
1343:
1341:
1338:
1334:
1328:
1325:
1323:
1319:
1318:
1313:
1310:
1308:
1305:
1303:
1302:Enkephalinase
1300:
1299:
1298:
1295:
1291:
1288:
1286:
1283:
1282:
1281:
1280:Endopeptidase
1278:
1274:
1271:
1269:
1266:
1265:
1264:
1260:
1256:
1255:
1252:
1249:
1247:
1246:Neuraminidase
1244:
1242:
1238:
1237:
1234:
1231:
1229:
1226:
1224:
1220:
1219:
1217:
1214:
1210:
1200:
1197:
1196:
1195:
1192:
1191:
1190:
1187:
1183:
1180:
1178:
1175:
1174:
1173:
1169:
1168:
1165:
1161:
1160:
1157:
1154:
1152:
1148:
1147:
1144:
1140:
1139:
1136:
1133:
1131:
1127:
1126:
1124:
1121:
1117:
1111:
1108:
1106:
1102:
1101:
1098:
1095:
1093:
1089:
1088:
1085:
1081:
1080:
1077:
1073:
1072:
1069:
1065:
1064:
1061:
1057:
1056:
1053:
1050:
1048:
1044:
1043:
1041:
1038:
1034:
1031:
1027:
1021:
1018:
1016:
1013:
1011:
1008:
1006:
1003:
1001:
998:
997:
995:
991:
987:
983:
976:
971:
969:
964:
962:
957:
956:
953:
942:
938:
934:
930:
926:
922:
918:
914:
907:
900:
892:
888:
884:
880:
876:
872:
868:
864:
857:
849:
845:
840:
835:
831:
827:
820:
813:
805:
801:
794:
778:
774:
768:
760:
756:
752:
748:
743:
738:
734:
730:
726:
719:
717:
715:
706:
702:
698:
694:
690:
686:
682:
678:
671:
669:
659:
654:
650:
646:
642:
638:
632:
630:
628:
626:
624:
622:
620:
618:
616:
607:
603:
599:
598:
589:
587:
585:
583:
578:
570:
568:
565:
561:
557:
553:
549:
546:
515:
511:
503:
494:
491:
484:
476:
467:
461:
453:
444:
441:
438:
435:
432:
429:
426:
423:
415:
414:
392:
388:
380:
371:
368:
362:
359:
356:
352:
346:
341:
338:
335:
330:
327:
324:
320:
312:
311:
310:
307:
304:
296:
289:
279:
275:
273:
269:
261:
257:
253:
249:
245:
237:
233:
231:
226:
223:
218:
215:
211:
207:
203:
198:
190:
181:
178:
176:
175:uncompetitive
172:
168:
158:
156:
152:
148:
144:
140:
136:
132:
125:
120:
117:
113:
109:
107:
103:
99:
95:
90:
84:
83:
75:
71:
67:
62:
52:
50:
45:
43:
39:
36:is a type of
35:
29:
22:
1296:
1263:Exopeptidase
1233:Ribonuclease
1199:Janus kinase
1084:Lipoxygenase
1060:5α-Reductase
1009:
982:Pharmacology
916:
912:
899:
866:
862:
856:
829:
825:
812:
804:Biochemistry
803:
793:
781:. Retrieved
777:the original
767:
732:
729:FEBS Letters
728:
680:
677:FEBS Letters
676:
648:
644:
596:
542:
308:
285:
276:
266:– see
244:maximum rate
241:
227:
219:
199:
195:
179:
177:inhibitors.
164:
135:invert sugar
127:
124:Victor Henri
121:
116:Victor Henri
110:
100:and make it
91:
77:
58:
46:
33:
32:
1312:Oxytocinase
1120:Transferase
869:(1): 11–5.
171:competitive
167:active site
161:Terminology
155:β-D-glucose
151:α-D-glucose
139:polarimeter
94:Maud Menten
28:Competition
1423:Metabolism
1412:Categories
573:References
552:nifedipine
202:glycolysis
1213:Hydrolase
1177:Integrase
1092:Aromatase
1029:Substrate
891:205518237
260:substrate
184:Mechanism
106:invertase
1259:Protease
933:19074529
883:21567682
848:13211596
759:12573784
751:23791665
705:43226286
697:23867202
606:22553872
550:include
282:Equation
258:for the
256:catalyst
252:affinity
210:pyruvate
1418:Enzymes
1402:Biology
1285:Trypsin
783:2 April
254:of the
214:Alanine
98:sucrose
70:glucose
66:maltase
55:History
1388:Portal
1339:(EC 4)
1215:(EC 3)
1122:(EC 2)
1082:1.13
1039:(EC 1)
941:285706
939:
931:
889:
881:
846:
757:
749:
703:
695:
604:
567:enzyme
564:CYP2C9
548:enzyme
545:CYP2C9
448:
1337:Lyase
1297:Mixed
1290:Renin
1268:DPP-4
1103:1.17
1097:COX-2
1090:1.14
993:Class
937:S2CID
909:(PDF)
887:S2CID
822:(PDF)
755:S2CID
701:S2CID
246:of a
208:into
1353:4.2
1345:4.1
1320:3.5
1257:3.4
1239:3.2
1221:3.1
1170:2.7
1162:2.6
1149:2.5
1143:PARP
1141:2.4
1130:COMT
1128:2.1
1074:1.5
1066:1.4
1058:1.3
1045:1.1
929:PMID
879:PMID
844:PMID
785:2012
747:PMID
693:PMID
602:PMID
114:and
102:lyse
1273:ACE
921:doi
871:doi
834:doi
830:210
737:doi
733:587
685:doi
681:587
653:doi
297:= K
141:as
130:cat
81:cat
1414::
1261::
984::
935:.
927:.
917:37
915:.
911:.
885:.
877:.
867:37
865:.
842:.
828:.
824:.
802:.
753:.
745:.
727:.
713:^
699:.
691:.
679:.
667:^
643:.
614:^
581:^
569:.
558:,
554:,
290:,
262:(K
212:.
51:.
1390::
974:e
967:t
960:v
943:.
923::
893:.
873::
850:.
836::
787:.
761:.
739::
707:.
687::
661:.
655::
649:4
608:.
516:I
512:K
507:]
504:I
501:[
495:+
492:1
485:0
481:]
477:E
474:[
468:=
462:0
458:]
454:E
451:[
445:t
442:n
439:e
436:r
433:a
430:p
427:p
424:a
393:I
389:K
384:]
381:I
378:[
372:+
369:1
363:x
360:a
357:m
353:V
347:=
342:p
339:p
336:a
331:x
328:a
325:m
321:V
299:m
294:m
292:K
264:m
128:k
87:m
79:k
30:.
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.