1207:
tumorigenesis in the urinary tract. As not all NOTCH receptors are equally involved in the urothelial bladder cancer, 90% of samples in one study had some level of NOTCH3 expression, suggesting that NOTCH3 plays an important role in urothelial bladder cancer. A higher level of NOTCH3 expression was observed in high-grade tumors, and a higher level of positivity was associated with a higher mortality risk. NOTCH3 was identified as an independent predictor of poor outcome. Therefore, it is suggested that NOTCH3 could be used as a marker for urothelial bladder cancer-specific mortality risk. It was also shown that NOTCH3 expression could be a prognostic immunohistochemical marker for clinical follow-up of urothelial bladder cancer patients, contributing to a more individualized approach by selecting patients to undergo control cystoscopy after a shorter time interval.
736:. In 1995, Notch1 was shown to be important for coordinating the segmentation of somites in mice. Further studies identified the role of Notch signaling in the segmentation clock. These studies hypothesized that the primary function of Notch signaling does not act on an individual cell, but coordinates cell clocks and keep them synchronized. This hypothesis explained the role of Notch signaling in the development of segmentation and has been supported by experiments in mice and zebrafish. Experiments with Delta1 mutant mice that show abnormal somitogenesis with loss of anterior/posterior polarity suggest that Notch signaling is also necessary for the maintenance of somite borders.
1142:
progressive recruitment of endocrine cell types from a common precursor, acting through two possible mechanisms. One is the "lateral inhibition", which specifies some cells for a primary fate but others for a secondary fate among cells that have the potential to adopt the same fate. Lateral inhibition is required for many types of cell fate determination. Here, it could explain the dispersed distribution of endocrine cells within pancreatic epithelium. A second mechanism is "suppressive maintenance", which explains the role of Notch signaling in pancreas differentiation. Fibroblast growth factor10 is thought to be important in this activity, but the details are unclear.
528:
role in selective recognition of
Serrate/Jagged and EGF domains 6-15 are required for maximal signaling upon ligand stimulation. A crystal structure of the interacting regions of Notch1 and Delta-like 4 (Dll4) provided a molecular-level visualization of Notch-ligand interactions, and revealed that the N-terminal MNNL (or C2) and DSL domains of ligands bind to Notch EGF domains 12 and 11, respectively. The Notch1-Dll4 structure also illuminated a direct role for Notch O-linked fucose and glucose moieties in ligand recognition, and rationalized a structural mechanism for the glycan-mediated tuning of Notch signaling.
781:
1105:(Dll4), which is expressed in the endothelial tip cells. VEGF signaling, which is an important factor for migration and proliferation of endothelial cells, can be downregulated in cells with activated Notch signaling by lowering the levels of Vegf receptor transcript. Zebrafish embryos lacking Notch signaling exhibit ectopic and persistent expression of the zebrafish ortholog of VEGF3, flt4, within all endothelial cells, while Notch activation completely represses its expression.
32:
512:
20:
503:. It was originally thought that these CSL proteins suppressed Notch target transcription. However, further research showed that, when the intracellular domain binds to the complex, it switches from a repressor to an activator of transcription. Other proteins also participate in the intracellular portion of the notch signaling cascade.
888:, deletion of the Notch signaling modulator, Numb, disrupts neuronal maturation in the developing cerebellum, whereas deletion of Numb disrupts axonal arborization in sensory ganglia. Although the mechanism underlying this phenomenon is not clear, together these findings suggest Notch signaling might be crucial in neuronal maturation.
759:. Several key components of the Notch signaling pathway help coordinate key steps in this process. In mice, mutations in Notch1, Dll1 or Dll3, Lfng, or Hes7 result in abnormal somite formation. Similarly, in humans, the following mutations have been seen to lead to development of spondylocostal dysostosis: DLL3, LFNG, or HES7.
1116:
expression. In turn, DLL4 expressing cells down-regulate Vegf receptors in neighboring cells through activation of Notch, thereby preventing their migration into the developing sprout. Likewise, during the sprouting process itself, the migratory behavior of connector cells must be limited to retain a
291:
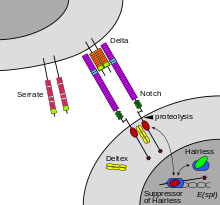
Maturation of the notch receptor involves cleavage at the prospective extracellular side during intracellular trafficking in the Golgi complex. This results in a bipartite protein, composed of a large extracellular domain linked to the smaller transmembrane and intracellular domain. Binding of ligand
536:
It is possible to engineer synthetic Notch receptors by replacing the extracellular receptor and intracellular transcriptional domains with other domains of choice. This allows researchers to select which ligands are detected, and which genes are upregulated in response. Using this technology, cells
527:
Despite the expansive size of the Notch extracellular domain, it has been demonstrated that EGF domains 11 and 12 are the critical determinants for interactions with Delta. Additional studies have implicated regions outside of Notch EGF11-12 in ligand binding. For example, Notch EGF domain 8 plays a
1758:
The mathematical modeling of Notch-Delta signaling thus provides significant insights into lateral inhibition mechanisms and pattern formation in biological systems. It enhances the understanding of cell-cell interaction variations leading to diverse tissue structures, contributing to developmental
244:
The receptor is normally triggered via direct cell-to-cell contact, in which the transmembrane proteins of the cells in direct contact form the ligands that bind the notch receptor. The Notch binding allows groups of cells to organize themselves such that, if one cell expresses a given trait, this
837:
The Notch pathway is essential for maintaining NPCs in the developing brain. Activation of the pathway is sufficient to maintain NPCs in a proliferating state, whereas loss-of-function mutations in the critical components of the pathway cause precocious neuronal differentiation and NPC depletion.
916:
Apart from its role in development, evidence shows that Notch signaling is also involved in neuronal apoptosis, neurite retraction, and neurodegeneration of ischemic stroke in the brain In addition to developmental functions, Notch proteins and ligands are expressed in cells of the adult nervous
466:
sugar then the subsequent addition of a galactose and sialic acid will occur. In the presence of this tetrasaccharide, notch signals strongly when it interacts with the Delta ligand, but has markedly inhibited signaling when interacting with the Jagged ligand. The means by which this addition of
1746:
is modified to include these protrusions, creating a more complex, non-local signaling network. This model is instrumental in exploring pattern formation robustness and biological pattern refinement, considering the stochastic nature of filopodia dynamics and intrinsic noise. The application of
1150:
The role of Notch signaling in the regulation of gut development has been indicated in several reports. Mutations in elements of the Notch signaling pathway affect the earliest intestinal cell fate decisions during zebrafish development. Transcriptional analysis and gain of function experiments
1141:
The formation of the pancreas from endoderm begins in early development. The expression of elements of the Notch signaling pathway have been found in the developing pancreas, suggesting that Notch signaling is important in pancreatic development. Evidence suggests Notch signaling regulates the
295:
Notch and most of its ligands are transmembrane proteins, so the cells expressing the ligands typically must be adjacent to the notch expressing cell for signaling to occur. The notch ligands are also single-pass transmembrane proteins and are members of the DSL (Delta/Serrate/LAG-2) family of
1461:
1206:
Loss of Notch activity is a driving event in urothelial cancer. A study identified inactivating mutations in components of the Notch pathway in over 40% of examined human bladder carcinomas. In mouse models, genetic inactivation of Notch signaling results in Erk1/2 phosphorylation leading to
1175:
during chondrogenesis. Overexpression of Notch signaling inhibits bone morphogenetic protein2-induced osteoblast differentiation. Overall, Notch signaling has a major role in the commitment of mesenchymal cells to the osteoblastic lineage and provides a possible therapeutic approach to bone
1286:
Mathematical modeling in Notch-Delta signaling has become a pivotal tool in understanding pattern formation driven by cell-cell interactions, particularly in the context of lateral-inhibition mechanisms. The
Collier model, a cornerstone in this field, employs a system of coupled
537:
can report or change their behavior in response to contact with user-specified signals, facilitating new avenues of both basic and applied research into cell-cell signaling. Notably, this system allows multiple synthetic pathways to be engineered into a cell in parallel.
345:
Notch 1, for example, has 36 of these repeats. Each EGF-like repeat is composed of approximately 40 amino acids, and its structure is defined largely by six conserved cysteine residues that form three conserved disulfide bonds. Each EGF-like repeat can be modified by
458:
To add another level of complexity, in mammals there are three Fringe GlcNAc-transferases, named lunatic fringe, manic fringe, and radical fringe. These enzymes are responsible for something called a "fringe effect" on notch signaling. If Fringe adds a GlcNAc to the
545:
The Notch signaling pathway is important for cell-cell communication, which involves gene regulation mechanisms that control multiple cell differentiation processes during embryonic and adult life. Notch signaling also has a role in the following processes:
1223:
mutations would provoke Notch signaling pathway activation, fostering the cancer development, but a recent study demonstrated that such an effect cannot be detected. Thus the exact role of Notch signaling in the cancer process awaits further elucidation.
921:
1 and 2, which mediate the Notch intramembranous cleavage. To be specific, conditional deletion of presenilins at 3 weeks after birth in excitatory neurons causes learning and memory deficits, neuronal dysfunction, and gradual neurodegeneration. Several
1012:
and by interendocardial signaling pathways to undergo EMT. Notch1 deficiency results in defective induction of EMT. Very few migrating cells are seen and these lack mesenchymal morphology. Notch may regulate this process by activating matrix
1294:
722:. During C. elegans, GLP-1, the C. elegans Notch homolog, interacts with APX-1, the C. elegans Delta homolog. This signaling between particular blastomeres induces differentiation of cell fates and establishes the dorsal-ventral axis.
1021:
expression in the AV canal endocardium while suppressing the VEGF pathway via VEGFR2. In RBPJk/CBF1-targeted mutants, the heart valve development is severely disrupted, presumably because of defective endocardial maturation and
474:
called ADAM10, cleaves the notch protein just outside the membrane. This releases the extracellular portion of notch (NECD), which continues to interact with the ligand. The ligand plus the notch extracellular domain is then
1189:
Aberrant Notch signaling is a driver of T cell acute lymphoblastic leukemia (T-ALL) and is mutated in at least 65% of all T-ALL cases. Notch signaling can be activated by mutations in Notch itself, inactivating mutations in
868:
In adult rodents and in cell culture, Notch3 promotes neuronal differentiation, having a role opposite to Notch1/2. This indicates that individual Notch receptors can have divergent functions, depending on cellular context.
3472:
Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S (November 1991). "Specific EGF repeats of Notch mediate interactions with Delta and
Serrate: implications for Notch as a multifunctional receptor".
3428:
Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S (November 1991). "Specific EGF repeats of Notch mediate interactions with Delta and
Serrate: implications for Notch as a multifunctional receptor".
713:
indicate that Notch signaling has a major role in the induction of mesoderm and cell fate determination. As mentioned previously, C. elegans has two genes that encode for partially functionally redundant Notch homologs,
245:
may be switched off in neighbouring cells by the intercellular notch signal. In this way, groups of cells influence one another to make large structures. Thus, lateral inhibition mechanisms are key to Notch signaling.
917:
system, suggesting a role in CNS plasticity throughout life. Adult mice heterozygous for mutations in either Notch1 or Cbf1 have deficits in spatial learning and memory. Similar results are seen in experiments with
479:
by the ligand-expressing cell. There may be signaling effects in the ligand-expressing cell after endocytosis; this part of notch signaling is a topic of active research. After this first cleavage, an enzyme called
1078:
During development of the aortic arch and the aortic arch arteries, the Notch receptors, ligands, and target genes display a unique expression pattern. When the Notch pathway was blocked, the induction of vascular
2554:
Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J (August 1991). "TAN-1, the human homolog of the
Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms".
1059:, whereby upregulated Wnt expression downregulates Notch signaling, and a subsequent inhibition of ventricular cardiomyocyte proliferation results. This proliferative arrest can be rescued using Wnt inhibitors.
264:, as well as intracellular proteins transmitting the notch signal to the cell's nucleus. The Notch/Lin-12/Glp-1 receptor family was found to be involved in the specification of cell fates during development in
771:
works in conjunction with ARF4 to regulate Notch signaling during this development. However, it remains to be determined whether gamma-secretase has a direct or indirect role in modulating Notch signaling.
7655:
292:
promotes two proteolytic processing events; as a result of proteolysis, the intracellular domain is liberated and can enter the nucleus to engage other DNA-binding proteins and regulate gene expression.
6833:
Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, Weinstein BM (October 2001). "Notch signaling is required for arterial-venous differentiation during embryonic vascular development".
7768:
Götte M, Wolf M, Staebler A, Buchweitz O, Kelsch R, Schüring AN, Kiesel L (July 2008). "Increased expression of the adult stem cell marker
Musashi-1 in endometriosis and endometrial carcinoma".
1299:
335:. There has been at least one report that suggests that some cells can send out processes that allow signaling to occur between cells that are as much as four or five cell diameters apart.
4689:
van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, et al. (December 1996). "Mutations affecting somite formation and patterning in the zebrafish, Danio rerio".
861:
A non-canonical branch of the Notch signaling pathway that involves the phosphorylation of STAT3 on the serine residue at amino acid position 727 and subsequent Hes3 expression increase (
6489:
Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, et al. (February 2007). "Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis".
5169:
Zhong W, Jiang MM, Weinmaster G, Jan LY, Jan YN (May 1997). "Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis".
3141:
Shao L, Luo Y, Moloney DJ, Haltiwanger R (November 2002). "O-glycosylation of EGF repeats: identification and initial characterization of a UDP-glucose: protein O-glucosyltransferase".
1133:
differentiates into several gastrointestinal epithelial lineages, including endocrine cells. Many studies have indicated that Notch signaling has a major role in endocrine development.
1035:
and in mouse embryonic stem cells indicate that cardiomyogenic commitment and differentiation require Notch signaling inhibition. Active Notch signaling is required in the ventricular
7292:
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. (October 2004). "Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia".
1083:
cell marker expression failed to occur, suggesting that Notch is involved in the differentiation of cardiac neural crest cells into vascular cells during outflow tract development.
1744:
1618:
696:
Notch signaling is required in the regulation of polarity. For example, mutation experiments have shown that loss of Notch signaling causes abnormal anterior-posterior polarity in
1008:, which is required for AV canal maturation. After the AV canal boundary formation, a subset of endocardial cells lining the AV canal are activated by signals emanating from the
8037:
Eisen JS (2010-07-27). "Faculty
Opinions recommendation of Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition".
7208:
Watanabe N, Tezuka Y, Matsuno K, Miyatani S, Morimura N, Yasuda M, et al. (2003). "Suppression of differentiation and proliferation of early chondrogenic cells by Notch".
7412:
Rampias T, Vgenopoulou P, Avgeris M, Polyzos A, Stravodimos K, Valavanis C, et al. (October 2014). "A new tumor suppressor role for the Notch pathway in bladder cancer".
1194:(a negative regulator of Notch1), or rarely by t(7;9)(q34;q34.3) translocation. In the context of T-ALL, Notch activity cooperates with additional oncogenic lesions such as
1711:
755:. This process is highly regulated as somites must have the correct size and spacing in order to avoid malformations within the axial skeleton that may potentially lead to
767:
Notch signaling is known to occur inside ciliated, differentiating cells found in the first epidermal layers during early skin development. Furthermore, it has found that
1243:
1673:
1062:
The downstream effector of Notch signaling, HEY2, was also demonstrated to be important in regulating ventricular development by its expression in the interventricular
1070:. Cardiomyocyte and smooth muscle cell-specific deletion of HEY2 results in impaired cardiac contractility, malformed right ventricle, and ventricular septal defects.
934:
patients resulted in statistically significant worsening of cognition relative to controls, which is thought to be due to its incidental effect on Notch signalling.
1517:
1490:
814:
embryos. In the past decade, advances in mutation and knockout techniques allowed research on the Notch signaling pathway in mammalian models, especially rodents.
7659:
1101:
Activation of Notch takes place primarily in "connector" cells and cells that line patent stable blood vessels through direct interaction with the Notch ligand,
3927:"Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis"
1638:
1581:
1561:
1541:
988:
Studies have revealed that both loss- and gain-of-function of the Notch pathway results in defects in AV canal development. In addition, the Notch target genes
405:-fucose, all notch proteins fail to function properly. As yet, the manner by which the glycosylation of notch affects function is not completely understood.
241:
by Gary Struhl and in cell culture by
Raphael Kopan. Although this model was initially disputed, the evidence in favor of the model was irrefutable by 2001.
2472:"Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei"
1747:
mathematical modeling in Notch-Delta signaling has been particularly illuminating in understanding the patterning of sensory organ precursors (SOPs) in the
112:
Notch signaling is dysregulated in many cancers, and faulty notch signaling is implicated in many diseases, including T-cell acute lymphoblastic leukemia (
6024:
Rones MS, McLaughlin KA, Raffin M, Mercola M (September 2000). "Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis".
908:
cells at the expense of neurons, whereas reduced Notch signaling induces production of ganglion cells, causing a reduction in the number of Muller glia.
2902:
307:
6998:
Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, et al. (January 2000). "Control of endodermal endocrine development by Hes-1".
1456:{\displaystyle {\begin{aligned}{\frac {d}{dt}}n_{i}&=f(\langle d_{i}\rangle )-n_{i}\\{\frac {d}{dt}}d_{i}&=\nu (g(n_{i})-d_{i})\end{aligned}}}
8679:
8231:
6869:
Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, et al. (August 1999). "Notch signalling controls pancreatic cell differentiation".
1239:
113:
8002:
Vasilopoulos G, Painter KJ (March 2016). "Pattern formation in discrete cell tissues under long range filopodia-based direct cell to cell contact".
3219:"The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression"
1108:
Notch signaling may be used to control the sprouting pattern of blood vessels during angiogenesis. When cells within a patent vessel are exposed to
3260:"The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments"
2294:
Austin J, Kimble J (November 1987). "glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans".
1258:, a disease characterised by increased expression of notch pathway constituents. Several notch inhibitors, including the gamma-secretase inhibitor
1151:
revealed that Notch signaling targets Hes1 in the intestine and regulates a binary cell fate decision between adsorptive and secretory cell fates.
5028:
Scheer N, Groth A, Hans S, Campos-Ortega JA (April 2001). "An instructive function for Notch in promoting gliogenesis in the zebrafish retina".
8287:
6383:"Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function"
124:. Inhibition of notch signaling inhibits the proliferation of T-cell acute lymphoblastic leukemia in both cultured cells and a mouse model.
2647:
Schroeter EH, Kisslinger JA, Kopan R (May 1998). "Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain".
2158:"Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats"
311:
5066:
Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A (January 2000). "Nuclear Notch1 signaling and the regulation of dendritic development".
4726:"Analysis of Notch function in presomitic mesoderm suggests a gamma-secretase-independent role for presenilins in somite differentiation"
1259:
1246:) as cancer treatments which are in different phases of clinical trials. As of 2013 at least 7 notch inhibitors were in clinical trials.
94:
interaction with a smaller piece of the notch protein composed of a short extracellular region, a single transmembrane-pass, and a small
8081:"The importance of structured noise in the generation of self-organizing tissue patterns through contact-mediated cell-cell signalling"
6963:
Field HA, Dong PD, Beis D, Stainier DY (September 2003). "Formation of the digestive system in zebrafish. II. Pancreas morphogenesis".
7506:"Unraveling the impact of AXIN1 mutations on HCC development: Insights from CRISPR/Cas9 repaired AXIN1-mutant liver cancer cell lines"
2199:"Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors"
842:
protein are able to antagonize Notch effects, resulting in the halting of cell cycle and the differentiation of NPCs. Conversely, the
4142:"Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling"
4025:
1247:
398:
387:
379:
318:. In mammals there are multiple Delta-like and Jagged ligands, as well as possibly a variety of other ligands, such as F3/contactin.
8779:
4638:
Hrabĕ de
Angelis M, McIntyre J, Gossler A (April 1997). "Maintenance of somite borders in mice requires the Delta homologue DII1".
4450:"Noncyclic Notch activity in the presomitic mesoderm demonstrates uncoupling of somite compartmentalization and boundary formation"
4391:"REX-1 expression and p38 MAPK activation status can determine proliferation/differentiation fates in human mesenchymal stem cells"
208:
proteins binding to the extracellular domain induce proteolytic cleavage and release of the intracellular domain, which enters the
2854:"The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization"
8246:
1796:
Artavanis-Tsakonas S, Rand MD, Lake RJ (April 1999). "Notch signaling: cell fate control and signal integration in development".
7504:
Zhang R, Li S, Schippers K, Eimers B, Niu J, Hornung BV, van den Hout MC, van Ijcken WF, Peppelenbosch MP, Smits R (June 2024).
9302:
8181:"Coordinated control of Notch/Delta signalling and cell cycle progression drives lateral inhibition-mediated tissue patterning"
515:
Crystal structure of the Notch1-DLL4 complex depicted as the interaction is predicted to occur between two cells (PDB ID: 4XLW)
6640:
Siekmann AF, Lawson ND (February 2007). "Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries".
5625:"Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration"
116:), cerebral autosomal-dominant arteriopathy with sub-cortical infarcts and leukoencephalopathy (CADASIL), multiple sclerosis,
8672:
8145:
7572:
5730:
4584:
2511:
Struhl G, Fitzgerald K, Greenwald I (July 1993). "Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo".
2140:
688:
The Notch signaling pathway plays an important role in cell-cell communication, and further regulates embryonic development.
3005:"The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD"
2337:
Priess JR, Schnabel H, Schnabel R (November 1987). "The glp-1 locus and cellular interactions in early C. elegans embryos".
2246:
Greenwald IS, Sternberg PW, Horvitz HR (September 1983). "The lin-12 locus specifies cell fates in
Caenorhabditis elegans".
996:
are involved in restricting the expression of two critical developmental regulator proteins, BMP2 and Tbx2, to the AV canal.
1250:
has given promising results in an early clinical trial for breast cancer. Preclinical studies showed beneficial effects of
8132:. Advances in Experimental Medicine and Biology. Vol. 1066. Cham: Springer International Publishing. pp. 79–98.
7355:"NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth"
5580:
Presente A, Andres A, Nye JS (October 2001). "Requirement of Notch in adulthood for neurological function and longevity".
5206:"Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis"
821:(NPC) maintenance and self-renewal. In recent years, other functions of the Notch pathway have also been found, including
492:
of the notch-expressing cell. This releases the intracellular domain of the notch protein (NICD), which then moves to the
9267:
8534:
5985:"Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation"
7864:"Therapeutic Potential of Wnt and Notch Signaling and Epigenetic Regulation in Mammalian Sensory Hair Cell Regeneration"
253:
mediate binary cell fate decisions, and lateral inhibition involves feedback mechanisms to amplify initial differences.
9112:
8840:
8502:
598:
influencing of binary fate decisions of cells that must choose between the secretory and absorptive lineages in the gut
2382:"lin-12, a nematode homeotic gene, is homologous to a set of mammalian proteins that includes epidermal growth factor"
8812:
8280:
7913:"Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling"
7169:"Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells"
7084:"FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development"
5851:"Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2"
3193:
1094:
cells use the Notch signaling pathway to coordinate cellular behaviors during the blood vessel sprouting that occurs
1005:
986:
Notch signaling can regulate the atrioventricular boundary formation between the AV canal and the chamber myocardium.
9069:
8855:
8850:
8665:
3878:"Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells"
784:
Notch-Delta lateral inhibition in neural stem cells, resulting in the generation of neuronal and glial progenitors.
5498:"Targeted deletion of numb and numblike in sensory neurons reveals their essential functions in axon arborization"
4810:"Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters"
4183:"Signaling axis involving Hedgehog, Notch, and Scl promotes the embryonic endothelial-to-hematopoietic transition"
1647:
Recent extensions of this model incorporate long-range signaling, acknowledging the role of cell protrusions like
467:
sugar inhibits signaling through one ligand, and potentiates signaling through another is not clearly understood.
7455:
Ristic Petrovic A, Stokanović D, Stojnev S, Potić Floranović M, Krstić M, Djordjević I, et al. (July 2022).
6745:"Signaling transduction mechanisms mediating biological actions of the vascular endothelial growth factor family"
6442:"An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation"
5802:"Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors"
4765:
Wahi K, Bochter MS, Cole SE (January 2016). "The many roles of Notch signaling during vertebrate somitogenesis".
4540:
Lambie EJ, Kimble J (May 1991). "Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions".
1198:
to activate anabolic pathways such as ribosome and protein biosynthesis thereby promoting leukemia cell growth.
367:
may be added between the second and third conserved cysteines. These sugars are added by an as-yet-unidentified
3371:
1288:
854:
in the proliferative state, amounting to a mechanism regulating cortical surface area growth and, potentially,
9246:
7457:"The association between NOTCH3 expression and the clinical outcome in the urothelial bladder cancer patients"
6100:
Croquelois A, Domenighetti AA, Nemir M, Lepore M, Rosenblatt-Velin N, Radtke F, Pedrazzini T (December 2008).
5944:"VE-cadherin is not required for the formation of nascent blood vessels but acts to prevent their disassembly"
9097:
4603:
Conlon RA, Reaume AG, Rossant J (May 1995). "Notch1 is required for the coordinate segmentation of somites".
1891:"A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE"
9321:
8529:
8273:
5351:
Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, et al. (August 2006).
1716:
1590:
1584:
806:
was associated with Notch dysfunction indicated that Notch mutations can lead to the failure of neural and
5895:"Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation"
2130:
9064:
7912:
7682:"γ-Secretase inhibition affects viability, apoptosis, and the stem cell phenotype of endometriotic cells"
7680:
Ramirez Williams L, Brüggemann K, Hubert M, Achmad N, Kiesel L, Schäfer SD, et al. (December 2019).
6198:
Campa VM, Gutiérrez-Lanza R, Cerignoli F, Díaz-Trelles R, Nelson B, Tsuji T, et al. (October 2008).
5623:
Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, et al. (April 2004).
5545:
Woo HN, Park JS, Gwon AR, Arumugam TV, Jo DG (December 2009). "Alzheimer's disease and Notch signaling".
2907:"Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes"
1052:
9256:
9122:
8954:
8348:
8331:
8314:
7083:
613:
6581:
Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ (February 2007).
5893:
Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, et al. (January 2004).
1971:
Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. (November 2009).
1759:
biology and offering potential therapeutic pathways in diseases related to Notch-Delta dysregulation.
109:
to promote neural differentiation. It plays a major role in the regulation of embryonic development.
8949:
8544:
8539:
8353:
3977:
Grego-Bessa J, Luna-Zurita L, del Monte G, Bolós V, Melgar P, Arandilla A, et al. (March 2007).
1067:
931:
756:
748:
147:
7314:
6920:
Lammert E, Brown J, Melton DA (June 2000). "Notch gene expression during pancreatic organogenesis".
2752:"Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila"
1112:
signaling, only a restricted number of them initiate the angiogenic process. Vegf is able to induce
8636:
8326:
8259:
1678:
1216:
843:
5255:"FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis"
4978:"rax, Hes1, and notch1 promote the formation of Müller glia by postnatal retinal progenitor cells"
900:, Notch appears to have an instructive role that can directly promote the differentiation of many
9190:
9102:
8985:
8905:
8700:
8464:
8436:
8363:
4867:"A Presenilin-2-ARF4 trafficking axis modulates Notch signaling during epidermal differentiation"
4497:
Levin M (January 2005). "Left-right asymmetry in embryonic development: a comprehensive review".
2094:
1748:
1055:
in mammals and zebrafish. A regulatory correspondence likely exists between Notch signaling and
602:
339:
298:
134:
6786:"Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function"
6247:
Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, MacRae CA, Lee RT, et al. (January 2014).
3176:
Lu L, Stanley P (2006). "Roles of O-Fucose Glycans in Notch Signaling Revealed by Mutant Mice".
3106:
Ma B, Simala-Grant JL, Taylor DE (December 2006). "Fucosylation in prokaryotes and eukaryotes".
1171:. Notch1 is expressed in the mesenchymal condensation area and subsequently in the hypertrophic
609:
lineage, suggesting a potential therapeutic role for notch in bone regeneration and osteoporosis
9175:
9074:
9059:
9028:
8929:
8476:
8449:
8128:
Binshtok U, Sprinzak D (2018). "Modeling the Notch Response". In Borggrefe T, Giaimo B (eds.).
7811:
Erni ST, Gill JC, Palaferri C, Fernandes G, Buri M, Lazarides K, et al. (13 August 2021).
7309:
7126:
Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J (March 2005).
6583:"Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting"
3738:
Gaiano N, Fishell G (2002). "The role of notch in promoting glial and neural stem cell fates".
1658:
1095:
971:
967:
955:
927:
789:
708:
519:
Notch signaling is initiated when Notch receptors on the cell surface engage ligands presented
485:
323:
156:
8065:
700:. Also, Notch signaling is required during left-right asymmetry determination in vertebrates.
9286:
9180:
9107:
9006:
8944:
8939:
8870:
8805:
8732:
8657:
8602:
8564:
8444:
8390:
8368:
8321:
7721:
Schüring AN, Dahlhues B, Korte A, Kiesel L, Titze U, Heitkötter B, et al. (March 2018).
6332:
Wang J, Sridurongrit S, Dudas M, Thomas P, Nagy A, Schneider MD, et al. (October 2005).
2421:
Oswald F, Täuber B, Dobner T, Bourteele S, Kostezka U, Adler G, et al. (November 2001).
1843:
1641:
1056:
858:. In this way, Notch signaling controls NPC self-renewal as well as cell fate specification.
677:
283:
to activate transcription of target genes. The structure of the complex has been determined.
261:
205:
75:
24:
1291:
to describe the feedback loop between adjacent cells. The model is defined by the equations:
904:
subtypes. For example, activation of Notch signaling in the retina favors the generation of
9195:
9038:
9033:
8915:
8910:
8897:
8573:
8569:
8521:
8255:
7924:
7366:
7301:
7167:
Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, et al. (March 2003).
7128:"Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine"
6878:
6649:
6594:
6498:
6394:
6334:"Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart"
6260:
6249:"Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration"
6061:"Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling"
5364:
5125:
4927:
4821:
4647:
4402:
4291:"Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells"
4194:
4050:
3832:
3751:
3555:
3482:
3299:
van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M (November 2009).
3016:
2818:
2763:
2656:
2041:
1984:
1805:
1495:
1468:
966:
Notch signal pathway plays a crucial role in at least three cardiac development processes:
780:
497:
7813:"Hair Cell Generation in Cochlear Culture Models Mediated by Novel γ-Secretase Inhibitors"
5942:
Crosby CV, Fleming PA, Argraves WS, Corada M, Zanetta L, Dejana E, Drake CJ (April 2005).
5408:"Notch3 is necessary for neuronal differentiation and maturation in the adult spinal cord"
865:) has been shown to regulate the number of NPCs in culture and in the adult rodent brain.
8:
9291:
8688:
8631:
8614:
8300:
7353:
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. (November 2006).
6381:
Xin M, Small EM, van Rooij E, Qi X, Richardson JA, Srivastava D, et al. (May 2007).
6308:"An Evolving Role for Notch Signaling in Heart Regeneration of the Zebrafish Danio rerio"
6102:"Control of the adaptive response of the heart to stress via the Notch1 receptor pathway"
5983:
Noseda M, McLean G, Niessen K, Chang L, Pollet I, Montpetit R, et al. (April 2004).
862:
807:
432:
428:
338:
The notch extracellular domain is composed primarily of small cystine-rich motifs called
117:
7928:
7532:
7505:
7370:
7305:
6882:
6653:
6598:
6540:
Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J (March 2007).
6502:
6398:
6264:
5368:
5204:
Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, et al. (December 2003).
5129:
4931:
4825:
4651:
4406:
4340:
Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, Rajagopal J (May 2009).
4198:
4140:
Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, et al. (April 2005).
4054:
3876:
Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, et al. (April 2002).
3836:
3559:
3486:
3020:
2822:
2767:
2660:
2045:
1988:
1889:
Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, et al. (February 2000).
1809:
132:
In 1914, John S. Dexter noticed the appearance of a notch in the wings of the fruit fly
8980:
8373:
8265:
8205:
8180:
8156:
8105:
8080:
7888:
7863:
7839:
7812:
7793:
7750:
7632:
7607:
7583:
7481:
7456:
7437:
7389:
7354:
7335:
7233:
7023:
6945:
6902:
6810:
6785:
6720:
6695:
6673:
6617:
6582:
6522:
6466:
6441:
6440:
High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA (February 2007).
6417:
6382:
6358:
6333:
6283:
6248:
6224:
6199:
6175:
6150:
6126:
6101:
6077:
6060:
6001:
5984:
5826:
5801:
5654:
5605:
5432:
5407:
5388:
5328:
5303:
5279:
5254:
5235:
5151:
5091:
5007:
4950:
4915:
4891:
4866:
4847:
4790:
4671:
4522:
4474:
4449:
4425:
4390:
4366:
4341:
4317:
4290:
4266:
4241:
4217:
4182:
4003:
3978:
3853:
3821:"Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal"
3820:
3715:
3690:
3666:
3641:
3576:
3543:
3506:
3454:
3327:
3300:
3088:
3042:
2985:
2936:
2883:
2727:
2702:
2680:
2629:
2580:
2536:
2362:
2319:
2271:
2111:
2085:
2067:
2005:
1972:
1871:
1623:
1566:
1546:
1526:
1039:
for proper trabeculae development subsequent to myocardial specification by regulating
659:
589:
488:) cleaves the remaining part of the notch protein just inside the inner leaflet of the
412:-glucose on notch can be further elongated to a trisaccharide with the addition of two
143:
44:
8240:
8179:
Hunter GL, Hadjivasiliou Z, Bonin H, He L, Perrimon N, Charras G, Baum B (July 2016).
7723:"The endometrial stem cell markers notch-1 and numb are associated with endometriosis"
7656:"Notch inhibitors could help overcome therapy resistance in ER-positive breast cancer"
6976:
6933:
6761:
6744:
5919:
5894:
5641:
5624:
5522:
5497:
5222:
5205:
5138:
5113:
4994:
4977:
4834:
4809:
4073:
4038:
3951:
3926:
3902:
3877:
3185:
3029:
3004:
2870:
2853:
2616:
2599:
2223:
2198:
1907:
1890:
751:
has been proposed in order to spatially determine the location and boundaries between
585:, as well as implications in other human disorders involving the cardiovascular system
9241:
9021:
9016:
8925:
8481:
8210:
8161:
8141:
8110:
8052:
8019:
7984:
7940:
7893:
7844:
7797:
7785:
7742:
7703:
7637:
7623:
7588:
7568:
7537:
7486:
7429:
7394:
7327:
7274:
7225:
7190:
7149:
7105:
7064:
7015:
6980:
6937:
6894:
6851:
6815:
6766:
6725:
6665:
6622:
6563:
6514:
6471:
6422:
6363:
6307:
6288:
6229:
6180:
6131:
6082:
6041:
6006:
5965:
5924:
5872:
5831:
5782:
5736:
5726:
5695:
5646:
5597:
5593:
5562:
5527:
5478:
5437:
5380:
5333:
5284:
5227:
5186:
5143:
5083:
5045:
4999:
4955:
4896:
4839:
4782:
4747:
4706:
4663:
4620:
4580:
4557:
4514:
4479:
4430:
4371:
4342:"Notch signaling promotes airway mucous metaplasia and inhibits alveolar development"
4322:
4271:
4222:
4163:
4119:
4078:
4008:
3956:
3907:
3858:
3801:
3755:
3720:
3671:
3622:
3581:
3498:
3494:
3446:
3442:
3410:
3367:
3332:
3281:
3240:
3199:
3189:
3158:
3123:
3080:
3063:
Lai EC (March 2004). "Notch signaling: control of cell communication and cell fate".
3034:
2977:
2928:
2875:
2834:
2791:
2786:
2751:
2732:
2672:
2621:
2572:
2568:
2528:
2524:
2493:
2452:
2447:
2438:
2422:
2403:
2398:
2381:
2354:
2350:
2311:
2307:
2263:
2259:
2228:
2179:
2174:
2157:
2136:
2071:
2059:
2010:
1953:
1912:
1863:
1821:
1768:
1014:
744:
593:
573:
237:
gene. Compelling evidence for this model was provided in 1998 by in vivo analysis in
146:. Its molecular analysis and sequencing was independently undertaken in the 1980s by
121:
9225:
7339:
7027:
5658:
5155:
5095:
5011:
4794:
4526:
4181:
Kim PG, Albacker CE, Lu YF, Jang IH, Lim Y, Heffner GC, et al. (January 2013).
3510:
3458:
3046:
2940:
2887:
2633:
2584:
2540:
2323:
2275:
2115:
1875:
942:
The Notch signaling pathway is a critical component of cardiovascular formation and
9251:
9220:
9215:
9170:
8798:
8607:
8578:
8507:
8403:
8336:
8200:
8192:
8151:
8133:
8100:
8092:
8038:
8011:
7974:
7932:
7883:
7875:
7834:
7824:
7777:
7754:
7734:
7693:
7627:
7619:
7578:
7560:
7527:
7517:
7476:
7468:
7441:
7421:
7384:
7374:
7319:
7264:
7237:
7217:
7180:
7139:
7095:
7054:
7007:
6972:
6949:
6929:
6906:
6886:
6843:
6805:
6797:
6756:
6715:
6707:
6677:
6657:
6612:
6602:
6553:
6526:
6506:
6461:
6453:
6412:
6402:
6353:
6345:
6278:
6268:
6219:
6211:
6170:
6162:
6121:
6113:
6072:
6033:
5996:
5955:
5914:
5906:
5862:
5821:
5813:
5772:
5718:
5685:
5636:
5609:
5589:
5554:
5517:
5509:
5496:
Huang EJ, Li H, Tang AA, Wiggins AK, Neve RL, Zhong W, et al. (January 2005).
5468:
5427:
5419:
5392:
5372:
5323:
5319:
5315:
5274:
5270:
5266:
5239:
5217:
5178:
5133:
5075:
5037:
4989:
4945:
4935:
4916:"Chromosomal Deficiencies and the Embryonic Development of Drosophila Melanogaster"
4886:
4878:
4851:
4829:
4774:
4737:
4698:
4675:
4655:
4612:
4549:
4506:
4469:
4461:
4420:
4410:
4361:
4353:
4312:
4302:
4261:
4253:
4212:
4202:
4153:
4109:
4068:
4058:
3998:
3990:
3946:
3938:
3897:
3889:
3848:
3840:
3791:
3747:
3710:
3702:
3661:
3653:
3612:
3571:
3563:
3490:
3438:
3400:
3359:
3322:
3312:
3271:
3230:
3181:
3150:
3115:
3092:
3072:
3024:
2989:
2967:
2918:
2865:
2826:
2781:
2771:
2722:
2714:
2684:
2664:
2611:
2564:
2520:
2483:
2442:
2434:
2393:
2366:
2346:
2303:
2255:
2218:
2210:
2169:
2103:
2049:
2000:
1992:
1943:
1902:
1855:
1813:
151:
7559:. Advances in Experimental Medicine and Biology. Vol. 727. pp. 305–319.
7555:
Purow B (2012). "Notch Inhibition as a Promising New Approach to Cancer Therapy".
5777:
5760:
5717:. Advances in Experimental Medicine and Biology. Vol. 727. pp. 210–222.
9261:
9165:
8975:
8486:
8250:
7564:
7522:
7269:
7252:
5722:
4778:
4742:
4725:
4415:
4389:
Bhandari DR, Seo KW, Roh KH, Jung JW, Kang SK, Kang KS (May 2010). Pera M (ed.).
3994:
3657:
1859:
1817:
1655:) that reach non-neighboring cells. One extended model, often referred to as the
1251:
1233:
946:
in both development and disease. It is required for the selection of endothelial
923:
851:
818:
643:
612:
expansion of the hemogenic endothelial cells along with signaling axis involving
481:
471:
315:
213:
79:
8137:
7879:
7454:
7100:
6542:"Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis"
6349:
6200:"Notch activates cell cycle reentry and progression in quiescent cardiomyocytes"
5800:
Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M (November 2006).
5473:
5456:
5304:"Cortical gyrification induced by fibroblast growth factor 2 in the mouse brain"
3942:
3925:
Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, et al. (January 2003).
2718:
8790:
8619:
8426:
8296:
7738:
7359:
Proceedings of the National Academy of Sciences of the United States of America
7251:
Göthert JR, Brake RL, Smeets M, Dührsen U, Begley CG, Izon DJ (November 2007).
7185:
7168:
6801:
6587:
Proceedings of the National Academy of Sciences of the United States of America
6387:
Proceedings of the National Academy of Sciences of the United States of America
6253:
Proceedings of the National Academy of Sciences of the United States of America
5960:
5943:
5690:
5673:
5558:
4920:
Proceedings of the National Academy of Sciences of the United States of America
4598:
4596:
4187:
Proceedings of the National Academy of Sciences of the United States of America
4114:
4097:
4043:
Proceedings of the National Academy of Sciences of the United States of America
3642:"Combinatorial Antigen Targeting: Ideal T-Cell Sensing and Anti-Tumor Response"
3542:
Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC (February 2015).
2972:
2955:
2923:
2906:
2756:
Proceedings of the National Academy of Sciences of the United States of America
1520:
1238:
The involvement of Notch signaling in many cancers has led to investigation of
375:
303:
55:
47:
8043:
8015:
7979:
7962:
7829:
7221:
4510:
4289:
Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS (2004).
4257:
3706:
1163:
studies have found the Notch signaling pathway functions as down-regulator in
9315:
9229:
9143:
9011:
8845:
8822:
8421:
7472:
7125:
2809:
Artavanis-Tsakonas S, Matsuno K, Fortini ME (April 1995). "Notch signaling".
1844:"Notch Antagonists: Potential Modulators of Cancer and Inflammatory Diseases"
1752:
1255:
1080:
943:
740:
733:
629:
579:
489:
460:
421:
391:
368:
361:
354:
201:
197:
191:
95:
83:
7379:
7323:
6847:
6607:
6407:
6273:
6037:
5182:
4593:
4207:
4063:
3567:
3544:"Structural biology. Structural basis for Notch1 engagement of Delta-like 4"
3363:
3317:
3154:
3119:
2956:"Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA"
2830:
802:
experiments. For example, the finding that an embryonic lethal phenotype in
470:
Once the notch extracellular domain interacts with a ligand, an ADAM-family
360:
sugar may be added between the first and second conserved cysteines, and an
9054:
8875:
8865:
8712:
8646:
8411:
8407:
8214:
8165:
8114:
8096:
8023:
7988:
7936:
7897:
7848:
7789:
7746:
7707:
7641:
7592:
7541:
7490:
7433:
7398:
7331:
7278:
7229:
7194:
7153:
7109:
7068:
7019:
6984:
6941:
6898:
6855:
6819:
6770:
6729:
6669:
6626:
6567:
6518:
6475:
6426:
6367:
6292:
6233:
6184:
6135:
6086:
6045:
6010:
5969:
5928:
5876:
5835:
5786:
5740:
5699:
5650:
5601:
5566:
5531:
5482:
5441:
5384:
5350:
5337:
5288:
5231:
5147:
5087:
5049:
5041:
5003:
4959:
4900:
4843:
4786:
4751:
4616:
4518:
4483:
4434:
4375:
4326:
4275:
4226:
4167:
4158:
4141:
4123:
4082:
4012:
3960:
3911:
3862:
3805:
3759:
3724:
3675:
3626:
3585:
3414:
3336:
3285:
3276:
3259:
3244:
3203:
3162:
3127:
3084:
3038:
2981:
2932:
2795:
2736:
2456:
2063:
2014:
1957:
1916:
1867:
1825:
1271:
1172:
855:
639:
605:
compartment during bone development and participation in commitment to the
559:
493:
436:
401:
is absolutely necessary for notch function, and, without the enzyme to add
209:
102:
91:
7944:
6215:
6166:
5190:
4940:
4882:
4710:
4702:
4667:
4624:
4561:
4553:
3691:"Chimeric antigen receptors: driving immunology towards synthetic biology"
3502:
3450:
3405:
3388:
2879:
2838:
2776:
2676:
2625:
2576:
2532:
2497:
2488:
2471:
2407:
2358:
2315:
2267:
2232:
2183:
1620:
denotes the average level of Delta activity in the cells adjacent to cell
775:
8990:
8970:
8641:
8454:
6711:
6197:
6117:
5713:
Kume T (2012). "Ligand-Dependent Notch Signaling in Vascular Formation".
4026:
The notch signaling pathway in cardiac development and tissue homeostasis
3796:
3779:
2214:
1091:
1036:
1018:
951:
905:
897:
839:
799:
582:
565:
476:
452:
448:
106:
7253:"NOTCH1 pathway activation is an early hallmark of SCL T leukemogenesis"
6661:
6510:
5513:
5376:
3844:
3298:
1996:
1948:
1931:
1587:, reflecting the regulatory dynamics of the signaling process. The term
9234:
8934:
8887:
8594:
8196:
7698:
7681:
7059:
7042:
6558:
6541:
5910:
5423:
4465:
4357:
4039:"Notch signaling controls multiple steps of pancreatic differentiation"
3893:
3617:
3600:
3524:"Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1".
1168:
1164:
1009:
918:
901:
822:
817:
The Notch signaling pathway was found to be critical mainly for neural
794:
606:
569:
142:
of the gene were identified in 1917 by American evolutionary biologist
7781:
7679:
7144:
7127:
6151:"Notch1 signaling stimulates proliferation of immature cardiomyocytes"
5867:
5850:
5817:
4447:
3235:
3218:
3076:
219:
The cleavage model was first proposed in 1993 based on work done with
9185:
9153:
8920:
8826:
8624:
8358:
6457:
4659:
2156:
Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S (December 1985).
1713:
to balance juxtacrine and long-range signaling. The interaction term
1648:
1263:
947:
847:
444:
440:
7722:
7425:
6539:
5352:
4448:
Feller J, Schneider A, Schuster-Gossler K, Gossler A (August 2008).
3976:
3389:"Fine-tuning of the intracellular canonical Notch signaling pathway"
2089:
2054:
2029:
427:
can be elongated to a tetrasaccharide by the ordered addition of an
9148:
9117:
8416:
5353:"Notch signalling regulates stem cell numbers in vitro and in vivo"
4307:
2155:
2107:
1652:
1275:
1130:
1126:
826:
704:
646:
7011:
6890:
6784:
Williams CK, Li JL, Murga M, Harris AL, Tosato G (February 2006).
5079:
3979:"Notch signaling is essential for ventricular chamber development"
2668:
2423:"p300 acts as a transcriptional coactivator for mammalian Notch-1"
31:
8880:
8774:
8769:
8764:
8398:
6099:
5457:"Murine numb regulates granule cell maturation in the cerebellum"
4976:
Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL (May 2000).
3601:"Protein binders and their applications in developmental biology"
1841:
1773:
1267:
1032:
881:
752:
697:
347:
139:
87:
67:
7411:
6246:
2808:
1970:
511:
8860:
6058:
5892:
5849:
Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y (February 2007).
5848:
4637:
3301:"Metalloprotease ADAM10 is required for Notch1 site 2 cleavage"
2470:
Lieber T, Kidd S, Alcamo E, Corbin V, Young MW (October 1993).
1063:
1048:
622:
552:
417:
413:
71:
63:
59:
51:
8687:
4288:
1842:
Kumar R, Juillerat-Jeanneret L, Golshayan D (September 2016).
1776:– A curated resource of signal transduction pathways in humans
1262:, are being studied for their potential ability to regenerate
7207:
5302:
Rash BG, Tomasi S, Lim HD, Suh CY, Vaccarino FM (June 2013).
4723:
4037:
Murtaugh LC, Stanger BZ, Kwan KM, Melton DA (December 2003).
3875:
1973:"Direct inhibition of the NOTCH transcription factor complex"
1220:
1191:
1040:
768:
636:
635:
possibly some non-nuclear mechanisms, such as control of the
280:
233:, informed by the first oncogenic mutation affecting a human
6488:
6148:
6023:
5027:
4098:"Expression of notch receptors and ligands in the adult gut"
9158:
8748:
8743:
8722:
8717:
7720:
7166:
6580:
6331:
6149:
Collesi C, Zentilin L, Sinagra G, Giacca M (October 2008).
6059:
Nemir M, Croquelois A, Pedrazzini T, Radtke F (June 2006).
5941:
5622:
4808:
Lowell S, Jones P, Le Roux I, Dunne J, Watt FM (May 2000).
4688:
3777:
2851:
2420:
2135:(revised ed.). Yale University Press. pp. 77–81.
1113:
1109:
1102:
1044:
993:
989:
673:
663:
617:
500:
276:
19:
8178:
7810:
7767:
5799:
5253:
Rash BG, Lim HD, Breunig JJ, Vaccarino FM (October 2011).
4975:
4724:
Huppert SS, Ilagan MX, De Strooper B, Kopan R (May 2005).
3180:. Methods in Enzymology. Vol. 417. pp. 127–136.
1795:
977:
496:, where it can regulate gene expression by activating the
7911:
Collier JR, Monk NA, Maini PK, Lewis JH (December 1996).
7861:
7250:
6997:
4036:
3140:
2510:
2245:
1195:
163:
genes were identified based on developmental phenotypes:
8295:
5982:
5065:
4139:
3471:
3427:
2646:
1073:
1004:
Notch signaling is also important for the process of AV
564:
regulation of crucial cell communication events between
101:
Notch signaling promotes proliferative signaling during
6868:
5455:
Klein AL, Zilian O, Suter U, Taylor V (February 2004).
5252:
5168:
4807:
776:
Role in central nervous system development and function
747:
cells dictates the precise rate of somite formation. A
275:
The intracellular domain of Notch forms a complex with
7503:
5454:
2469:
2336:
572:
during both the formation of the valve primordial and
7910:
7291:
6962:
6783:
6696:"Notch signalling and the regulation of angiogenesis"
4339:
3541:
3349:
2553:
1929:
1888:
1719:
1681:
1661:
1626:
1593:
1569:
1549:
1529:
1498:
1471:
1297:
937:
676:
has inhibitory effects on the expression of notch in
7352:
7081:
6439:
5203:
3105:
762:
8001:
7956:
7954:
7043:"Gene regulatory factors in pancreatic development"
6832:
6380:
5674:"Lessons from a failed γ-secretase Alzheimer trial"
5547:
Biochemical and Biophysical Research Communications
5544:
5114:"Learning and memory deficits in Notch mutant mice"
4864:
4602:
4242:"Notch signaling in CD4 and CD8 T cell development"
3972:
3970:
3778:Bolós V, Grego-Bessa J, de la Pompa JL (May 2007).
926:inhibitors that underwent human clinical trials in
8078:
7082:Norgaard GA, Jensen JN, Jensen J (December 2003).
6919:
5495:
4388:
3924:
2852:Singson A, Mercer KB, L'Hernault SW (April 1998).
2030:"Chemical biology: A Notch above other inhibitors"
1738:
1705:
1667:
1632:
1612:
1575:
1555:
1535:
1511:
1484:
1455:
651:Regulation of the mitotic/meiotic decision in the
625:lineage commitment from common lymphoid precursor
7862:Samarajeewa A, Jacques BE, Dabdoub A (May 2019).
7855:
7121:
7119:
6689:
6687:
5579:
4180:
3818:
1930:Sharma VM, Draheim KM, Kelliher MA (April 2007).
1675:-Collier model, introduces a weighting parameter
1051:B2 expression. Notch signaling sustains immature
9313:
8820:
8127:
7963:"Coupling dynamics of 2D Notch-Delta signalling"
7951:
5301:
3967:
3598:
3350:Desbordes S, López-Schier H (2006). "Drosophila
2947:
2900:
2894:
2743:
2691:
1120:
1117:patent connection to the original blood vessel.
5111:
4764:
4135:
4133:
4102:The Journal of Histochemistry and Cytochemistry
3819:Aguirre A, Rubio ME, Gallo V (September 2010).
3773:
3771:
3769:
3639:
3386:
3216:
2749:
2696:
2694:
2640:
2591:
2547:
2504:
2463:
2373:
2330:
2196:
974:, and cardiac outflow tract (OFT) development.
558:stabilization of arterial endothelial fate and
7960:
7116:
6693:
6684:
6639:
2289:
2287:
2285:
2197:Kidd S, Kelley MR, Young MW (September 1986).
1791:
1789:
832:
8806:
8673:
8281:
8172:
7686:Acta Obstetricia et Gynecologica Scandinavica
7605:
7075:
7034:
6991:
6956:
6913:
6862:
6777:
6736:
6633:
6533:
5888:
5886:
5758:
4865:Ezratty EJ, Pasolli HA, Fuchs E (July 2016).
3737:
1932:"The Notch1/c-Myc pathway in T cell leukemia"
829:development, as well as learning and memory.
302:(the fruit fly), there are two ligands named
6742:
5616:
5448:
5405:
4767:Seminars in Cell & Developmental Biology
4539:
4282:
4174:
4130:
4095:
4089:
4030:
3918:
3869:
3812:
3766:
3731:
3537:
3535:
3380:
3292:
3257:
3251:
3210:
3169:
3134:
3099:
3002:
2996:
2953:
2600:"Nuclear access and action of notch in vivo"
2597:
2414:
2293:
1837:
1835:
1733:
1720:
1607:
1594:
1353:
1340:
1201:
531:
7817:Frontiers in Cell and Developmental Biology
5671:
5112:Costa RM, Honjo T, Silva AJ (August 2003).
5107:
5105:
4239:
3780:"Notch signaling in development and cancer"
2845:
2802:
2282:
2190:
2027:
1923:
1882:
1786:
792:(CNS) development were performed mainly in
204:, with part of it inside and part outside.
8813:
8799:
8680:
8666:
8288:
8274:
8079:Cohen M, Baum B, Miodownik M (June 2011).
7804:
7548:
5883:
5412:Journal of Cellular and Molecular Medicine
5061:
5059:
4574:
2149:
1026:
1001:AV epithelial-mesenchymal transition (EMT)
838:Modulators of the Notch signal, e.g., the
588:timely cell lineage specification of both
310:. In mammals, the corresponding names are
8258:at the U.S. National Library of Medicine
8204:
8155:
8104:
8042:
7978:
7887:
7838:
7828:
7697:
7631:
7582:
7531:
7521:
7480:
7461:Bosnian Journal of Basic Medical Sciences
7388:
7378:
7313:
7268:
7184:
7143:
7099:
7058:
6809:
6760:
6719:
6616:
6606:
6557:
6465:
6416:
6406:
6357:
6282:
6272:
6223:
6174:
6125:
6076:
6000:
5959:
5918:
5866:
5825:
5776:
5689:
5640:
5521:
5472:
5431:
5327:
5278:
5221:
5137:
4993:
4949:
4939:
4890:
4833:
4741:
4473:
4424:
4414:
4365:
4316:
4306:
4265:
4216:
4206:
4157:
4113:
4072:
4062:
4002:
3950:
3901:
3852:
3795:
3714:
3665:
3616:
3575:
3532:
3404:
3326:
3316:
3275:
3234:
3217:Thomas GB, van Meyel DJ (February 2007).
3175:
3028:
2971:
2922:
2869:
2785:
2775:
2726:
2703:"Notch and the awesome power of genetics"
2700:
2615:
2487:
2446:
2397:
2379:
2222:
2173:
2078:
2053:
2004:
1947:
1906:
1832:
1145:
1136:
846:pathway promotes Notch signaling to keep
7557:Notch Signaling in Embryology and Cancer
5761:"Notch signaling in cardiac development"
5715:Notch Signaling in Embryology and Cancer
5102:
3688:
3592:
1281:
779:
725:
683:
510:
327:, two genes encode homologous proteins,
30:
18:
8130:Molecular Mechanisms of Notch Signaling
8085:Journal of the Royal Society, Interface
7608:"Notch inhibitors for cancer treatment"
7599:
5754:
5752:
5750:
5056:
5023:
5021:
4971:
4969:
4913:
4579:(11th ed.). Sinauer. p. 272.
3599:Harmansa S, Affolter M (January 2018).
3258:LaVoie MJ, Selkoe DJ (September 2003).
1219:, for instance, it was suggesting that
978:Atrioventricular (AV) canal development
911:
9314:
9303:Index of evolutionary biology articles
7210:Journal of Bone and Mineral Metabolism
7040:
3752:10.1146/annurev.neuro.25.030702.130823
3387:Borggrefe T, Liefke R (January 2012).
3058:
3056:
2750:Struhl G, Greenwald I (January 2001).
2128:
2084:
1017:2 (MMP2) expression, or by inhibiting
961:
880:studies show that Notch can influence
872:
632:at several distinct development stages
506:
171:. The cloning and partial sequence of
74:. The notch receptor is a single-pass
8794:
8661:
8269:
8036:
7554:
6446:The Journal of Clinical Investigation
4496:
2028:Arora PS, Ansari AZ (November 2009).
1739:{\displaystyle \langle d_{i}\rangle }
1613:{\displaystyle \langle d_{i}\rangle }
1270:, which could lead to treatments for
1074:Ventricular outflow tract development
788:Early findings on Notch signaling in
6743:Zachary I, Gliki G (February 2001).
6106:The Journal of Experimental Medicine
5747:
5712:
5018:
4966:
3640:Themeli M, Sadelain M (April 2016).
628:regulation of cell-fate decision in
8535:Transcription preinitiation complex
7961:Berkemeier F, Page KM (June 2023).
7648:
7606:Espinoza I, Miele L (August 2013).
5406:Rusanescu G, Mao J (October 2014).
4146:The Journal of Biological Chemistry
4096:Sander GR, Powell BC (April 2004).
3305:The Journal of Biological Chemistry
3264:The Journal of Biological Chemistry
3062:
3053:
2954:Wilson JJ, Kovall RA (March 2006).
1227:
1154:
105:, and its activity is inhibited by
13:
9113:Evolutionary developmental biology
8503:Signal transducing adaptor protein
6078:10.1161/01.RES.0000226497.52052.2a
6002:10.1161/01.RES.0000124300.76171.C9
1019:vascular endothelial (VE)-cadherin
938:Role in cardiovascular development
691:
14:
9333:
8224:
6305:
4240:Laky K, Fowlkes BJ (April 2008).
2901:Nam Y, Sliz P, Song L, Aster JC,
1179:
1066:and the endocardial cells of the
763:Role in epidermal differentiation
390:), respectively. The addition of
175:was reported at the same time as
54:. Mammals possess four different
9070:Evolution of sexual reproduction
8121:
8072:
8030:
7995:
7904:
7761:
7714:
7673:
7624:10.1016/j.pharmthera.2013.02.003
7497:
7448:
7405:
7346:
7285:
7244:
7201:
7160:
6826:
6574:
6482:
6433:
6374:
6325:
6299:
5759:Niessen K, Karsan A (May 2008).
5594:10.1097/00001756-200110290-00035
3003:Munro S, Freeman M (July 2000).
2439:10.1128/MCB.21.22.7761-7774.2001
7727:Reproductive Biomedicine Online
7612:Pharmacology & Therapeutics
6694:Siekmann AF, Lawson ND (2007).
6240:
6191:
6142:
6093:
6052:
6017:
5976:
5935:
5842:
5793:
5706:
5672:De Strooper B (November 2014).
5665:
5573:
5538:
5489:
5399:
5344:
5295:
5246:
5197:
5162:
4907:
4858:
4801:
4758:
4717:
4682:
4631:
4568:
4533:
4490:
4441:
4382:
4333:
4233:
4019:
3682:
3633:
3526:Journal of Biological Chemistry
3517:
3465:
3421:
3343:
2598:Struhl G, Adachi A (May 1998).
2239:
1289:ordinary differential equations
1210:
1125:During development, definitive
1086:
576:development and differentiation
433:N-Acetylglucosaminyltransferase
86:portion, which associates in a
8841:Genotype–phenotype distinction
7917:Journal of Theoretical Biology
7823:. Frontiers Media SA: 710159.
5320:10.1523/JNEUROSCI.3621-12.2013
5271:10.1523/JNEUROSCI.4439-11.2011
3931:Molecular and Cellular Biology
2427:Molecular and Cellular Biology
2203:Molecular and Cellular Biology
2122:
2021:
1964:
1848:Journal of Medicinal Chemistry
1700:
1688:
1446:
1430:
1417:
1411:
1356:
1337:
891:
732:Notch signaling is central to
680:, preventing differentiation.
1:
9098:Regulation of gene expression
6977:10.1016/S0012-1606(03)00308-7
6934:10.1016/S0925-4773(00)00317-8
6762:10.1016/S0008-6363(00)00268-6
6700:Cell Adhesion & Migration
5778:10.1161/CIRCRESAHA.108.174318
5642:10.1016/S0896-6273(04)00182-5
5223:10.1016/S0896-6273(03)00755-4
5139:10.1016/S0960-9822(03)00492-5
4995:10.1016/S0896-6273(00)81171-X
4835:10.1016/s0960-9822(00)00451-6
4246:Current Opinion in Immunology
3740:Annual Review of Neuroscience
3695:Current Opinion in Immunology
3356:Encyclopedia of Life Sciences
3354:: Delta-Notch Interactions".
3186:10.1016/S0076-6879(06)17010-X
3030:10.1016/S0960-9822(00)00578-9
2871:10.1016/S0092-8674(00)81147-2
2617:10.1016/S0092-8674(00)81193-9
2380:Greenwald I (December 1985).
1908:10.1016/S1097-2765(00)80417-7
1780:
1706:{\displaystyle \epsilon \in }
1121:Role in endocrine development
863:STAT3-Ser/Hes3 Signaling Axis
9268:Endless Forms Most Beautiful
9048:Evolution of genetic systems
8856:Gene–environment correlation
8851:Gene–environment interaction
7565:10.1007/978-1-4614-0899-4_23
7523:10.1371/journal.pone.0304607
7270:10.1182/blood-2006-12-063644
5723:10.1007/978-1-4614-0899-4_16
4779:10.1016/j.semcdb.2014.11.010
4743:10.1016/j.devcel.2005.02.019
4416:10.1371/journal.pone.0010493
3995:10.1016/j.devcel.2006.12.011
3658:10.1016/j.molmed.2016.02.009
3646:Trends in Molecular Medicine
3495:10.1016/0092-8674(91)90064-6
3443:10.1016/0092-8674(91)90064-6
2569:10.1016/0092-8674(91)90111-B
2525:10.1016/0092-8674(93)90424-O
2399:10.1016/0092-8674(85)90230-2
2351:10.1016/0092-8674(87)90129-2
2308:10.1016/0092-8674(87)90128-0
2260:10.1016/0092-8674(83)90377-x
2175:10.1016/0092-8674(85)90229-6
1860:10.1021/acs.jmedchem.5b01516
1818:10.1126/science.284.5415.770
743:, a molecular oscillator in
672:It has also been found that
260:consists of Notch and Notch
185:
127:
7:
9247:Christiane Nüsslein-Volhard
8232:notch signaling pathway in
8138:10.1007/978-3-319-89512-3_5
7880:10.1016/j.ymthe.2019.03.017
7874:(5). Elsevier BV: 904–911.
7101:10.1016/j.ydbio.2003.08.013
6350:10.1016/j.ydbio.2005.07.035
6204:The Journal of Cell Biology
6155:The Journal of Cell Biology
5474:10.1016/j.ydbio.2003.10.017
5308:The Journal of Neuroscience
5259:The Journal of Neuroscience
4871:The Journal of Cell Biology
3943:10.1128/MCB.23.1.14-25.2003
2719:10.1534/genetics.112.141812
1762:
1523:and Delta activity in cell
1184:
1053:cardiomyocyte proliferation
833:Neuron cell differentiation
540:
16:Series of molecular signals
10:
9338:
9123:Hedgehog signaling pathway
9000:Developmental architecture
8758:Receptor on receiving cell
7739:10.1016/j.rbmo.2017.11.010
7186:10.1182/blood-2002-06-1740
6802:10.1182/blood-2005-03-1000
5961:10.1182/blood-2004-06-2244
5691:10.1016/j.cell.2014.10.016
5559:10.1016/j.bbrc.2009.10.093
4115:10.1177/002215540405200409
3689:Sadelain M (August 2016).
2973:10.1016/j.cell.2006.01.035
2924:10.1016/j.cell.2005.12.037
1543:, respectively. Functions
1244:gamma-secretase inhibitors
1231:
810:cell segregation in early
286:
189:
9300:
9279:
9208:
9136:
9090:
9083:
9047:
8999:
8963:
8950:Transgressive segregation
8896:
8833:
8757:
8731:
8699:
8587:
8557:
8520:
8495:
8472:
8463:
8435:
8389:
8382:
8307:
8044:10.3410/f.4361976.4187082
8016:10.1016/j.mbs.2015.12.008
7980:10.1016/j.mbs.2023.109012
7830:10.3389/fcell.2021.710159
7222:10.1007/s00774-003-0428-4
7041:Jensen J (January 2004).
6922:Mechanisms of Development
4914:Poulson DF (March 1937).
4511:10.1016/j.mod.2004.08.006
4499:Mechanisms of Development
4258:10.1016/j.coi.2008.03.004
3707:10.1016/j.coi.2016.06.004
2701:Greenwald I (July 2012).
1668:{\displaystyle \epsilon }
1202:Urothelial bladder cancer
757:spondylocostal dysostosis
749:clock and wavefront model
532:Synthetic Notch signaling
148:Spyros Artavanis-Tsakonas
8615:Neuroendocrine signaling
8260:Medical Subject Headings
8004:Mathematical Biosciences
7967:Mathematical Biosciences
7770:The Journal of Pathology
7473:10.17305/bjbms.2021.6767
2090:"The theory of the gene"
1519:represent the levels of
1217:hepatocellular carcinoma
844:fibroblast growth factor
555:function and development
484:(which is implicated in
447:, and the addition of a
9128:Notch signaling pathway
9103:Gene regulatory network
8986:Dual inheritance theory
8693:notch signaling pathway
7380:10.1073/pnas.0606108103
7324:10.1126/science.1102160
6848:10.1242/dev.128.19.3675
6749:Cardiovascular Research
6608:10.1073/pnas.0611206104
6408:10.1073/pnas.0702447104
6274:10.1073/pnas.1311705111
6038:10.1242/dev.127.17.3865
5899:Genes & Development
5502:Genes & Development
5183:10.1242/dev.124.10.1887
4454:Genes & Development
4208:10.1073/pnas.1214361110
4064:10.1073/pnas.2436557100
3882:Genes & Development
3568:10.1126/science.1261093
3364:10.1038/npg.els.0004194
3318:10.1074/jbc.M109.006775
2831:10.1126/science.7716513
2476:Genes & Development
2095:The American Naturalist
1027:Ventricular development
603:hematopoietic stem cell
299:Drosophila melanogaster
135:Drosophila melanogaster
50:system present in most
41:Notch signaling pathway
9176:cis-regulatory element
9084:Control of development
8964:Non-genetic influences
8930:evolutionary landscape
8477:cAMP-dependent pathway
8097:10.1098/rsif.2010.0488
7937:10.1006/jtbi.1996.0233
7658:. 2015. Archived from
7047:Developmental Dynamics
5042:10.1242/dev.128.7.1099
4617:10.1242/dev.121.5.1533
4295:Breast Cancer Research
4159:10.1074/jbc.M412891200
3277:10.1074/jbc.M302659200
2132:The theory of the gene
1740:
1707:
1669:
1634:
1614:
1577:
1557:
1537:
1513:
1486:
1457:
1146:Intestinal development
1137:Pancreatic development
1096:sprouting angiogenesis
972:myocardial development
968:Atrioventricular canal
956:sprouting angiogenesis
790:central nervous system
785:
678:mesenchymal stem cells
516:
353:at specific sites. An
76:transmembrane receptor
36:
28:
27:between adjacent cells
9287:Nature versus nurture
9191:Cell surface receptor
9108:Evo-devo gene toolkit
9007:Developmental biology
8945:Polygenic inheritance
8871:Quantitative genetics
8603:Synaptic transmission
8599:Neurocrine signaling
8522:Transcription factors
8369:PI3K/AKT/mTOR pathway
7088:Developmental Biology
6965:Developmental Biology
6338:Developmental Biology
6216:10.1083/jcb.200806104
6167:10.1083/jcb.200806091
5461:Developmental Biology
4941:10.1073/pnas.23.3.133
4883:10.1083/jcb.201508082
4703:10.1242/dev.123.1.153
4577:Developmental biology
4554:10.1242/dev.112.1.231
3406:10.4161/cc.11.2.18995
3155:10.1093/glycob/cwf085
3120:10.1093/glycob/cwl040
2777:10.1073/pnas.98.1.229
2489:10.1101/gad.7.10.1949
1741:
1708:
1670:
1635:
1615:
1578:
1558:
1538:
1514:
1512:{\displaystyle d_{i}}
1487:
1485:{\displaystyle n_{i}}
1458:
1282:Mathematical modeling
983:AV boundary formation
783:
726:Role in somitogenesis
703:Early studies in the
684:Role in embryogenesis
514:
445:galactosyltransferase
431:(GlcNAc) sugar by an
384:-fucosyltransferase 1
190:Further information:
154:. Alleles of the two
35:Notch signaling steps
34:
22:
9196:Transcription factor
8911:Genetic assimilation
8898:Genetic architecture
6712:10.4161/cam.1.2.4488
6118:10.1084/jem.20081427
6065:Circulation Research
5989:Circulation Research
5765:Circulation Research
3797:10.1210/er.2006-0046
3178:Functional Glycomics
2215:10.1128/mcb.6.9.3094
1717:
1679:
1659:
1642:juxtacrine signaling
1624:
1591:
1567:
1547:
1527:
1496:
1469:
1295:
912:Adult brain function
498:transcription factor
439:, the addition of a
372:-glucosyltransferase
82:composed of a large
9322:Signal transduction
9292:Morphogenetic field
9209:Influential figures
8632:Mechanotransduction
8620:Exocrine signalling
8427:Signaling molecules
8301:Signal transduction
8241:Notch signaling in
7929:1996JThBi.183..429C
7371:2006PNAS..10318261P
7365:(48): 18261–18266.
7306:2004Sci...306..269W
6883:1999Natur.400..877A
6662:10.1038/nature05577
6654:2007Natur.445..781S
6599:2007PNAS..104.3219L
6511:10.1038/nature05571
6503:2007Natur.445..776H
6399:2007PNAS..104.7975X
6265:2014PNAS..111.1403Z
5514:10.1101/gad.1246005
5377:10.1038/nature04940
5369:2006Natur.442..823A
5314:(26): 10802–10814.
5265:(43): 15604–15617.
5130:2003CBio...13.1348C
5068:Nature Neuroscience
4932:1937PNAS...23..133P
4826:2000CBio...10..491L
4652:1997Natur.386..717D
4575:Gilbert SF (2016).
4407:2010PLoSO...510493B
4199:2013PNAS..110E.141K
4152:(16): 15842–15848.
4055:2003PNAS..10014920M
4049:(25): 14920–14925.
3845:10.1038/nature09347
3837:2010Natur.467..323A
3560:2015Sci...347..847L
3487:2012Sci...338.1229Y
3311:(45): 31018–31027.
3270:(36): 34427–34437.
3021:2000CBio...10..813M
2823:1995Sci...268..225A
2768:2001PNAS...98..229S
2661:1998Natur.393..382S
2392:(3 Pt 2): 583–590.
2168:(3 Pt 2): 567–581.
2046:2009Natur.462..171A
1997:10.1038/nature08543
1989:2009Natur.462..182M
1949:10.4161/cc.6.8.4134
1810:1999Sci...284..770A
1103:Delta-like ligand 4
962:Cardiac development
928:Alzheimer's disease
873:Neurite development
507:Ligand interactions
486:Alzheimer's disease
429:N-acetylglucosamine
418:xylosyltransferases
380:GDP-fucose Protein
118:Tetralogy of Fallot
8981:Genomic imprinting
8642:Ion channel gating
8374:Integrin receptors
8308:Signaling pathways
8249:2017-11-09 at the
8197:10.1242/dev.134213
7699:10.1111/aogs.13707
7060:10.1002/dvdy.10460
6559:10.1242/dev.003244
5911:10.1101/gad.276304
5424:10.1111/jcmm.12362
4730:Developmental Cell
4466:10.1101/gad.480408
4358:10.1242/dev.029249
3983:Developmental Cell
3894:10.1101/gad.975202
3618:10.1242/dev.148874
2129:Morgan TH (1928).
1736:
1703:
1665:
1630:
1610:
1573:
1553:
1533:
1509:
1482:
1453:
1451:
1169:osteoblastogenesis
1165:osteoclastogenesis
786:
614:Hedgehog signaling
517:
182:by Iva Greenwald.
144:Thomas Hunt Morgan
37:
29:
9309:
9308:
9242:Eric F. Wieschaus
9204:
9203:
9022:Pattern formation
8926:Fitness landscape
8788:
8787:
8703:on signaling cell
8689:Signaling pathway
8655:
8654:
8637:Phototransduction
8595:Intracrine action
8553:
8552:
8516:
8515:
8404:Neurotransmitters
8191:(13): 2305–2310.
8147:978-3-319-89512-3
8060:Missing or empty
7868:Molecular Therapy
7782:10.1002/path.2364
7692:(12): 1565–1574.
7574:978-1-4614-0898-7
7420:(10): 1199–1205.
7300:(5694): 269–271.
7263:(10): 3753–3762.
7145:10.1242/dev.01644
6877:(6747): 877–881.
6842:(19): 3675–3683.
6648:(7129): 781–784.
6497:(7129): 776–780.
6393:(19): 7975–7980.
6112:(13): 3173–3185.
6071:(12): 1471–1478.
6032:(17): 3865–3876.
5868:10.1242/dev.02777
5818:10.1242/dev.02607
5812:(21): 4381–4390.
5771:(10): 1169–1181.
5732:978-1-4614-0898-7
5588:(15): 3321–3325.
5418:(10): 2103–2116.
5363:(7104): 823–826.
5177:(10): 1887–1897.
5124:(15): 1348–1354.
4646:(6626): 717–721.
4586:978-1-60535-470-5
4460:(16): 2166–2171.
4352:(10): 1751–1759.
3831:(7313): 323–327.
3784:Endocrine Reviews
3554:(6224): 847–853.
3236:10.1242/dev.02754
3114:(12): 158R–184R.
3077:10.1242/dev.01074
2817:(5208): 225–232.
2655:(6683): 382–386.
2482:(10): 1949–1965.
2433:(22): 7761–7774.
2142:978-0-8240-1384-4
2040:(7270): 171–173.
1983:(7270): 182–188.
1854:(17): 7719–7737.
1804:(5415): 770–776.
1769:Alagille syndrome
1633:{\displaystyle i}
1576:{\displaystyle g}
1556:{\displaystyle f}
1536:{\displaystyle i}
1389:
1315:
1015:metalloproteinase
745:paraxial mesoderm
601:expansion of the
594:exocrine pancreas
523:on opposing cells
453:sialyltransferase
122:Alagille syndrome
78:protein. It is a
58:, referred to as
25:juxtacrine signal
9329:
9252:William McGinnis
9221:Richard Lewontin
9216:C. H. Waddington
9088:
9087:
9065:Neutral networks
8815:
8808:
8801:
8792:
8791:
8682:
8675:
8668:
8659:
8658:
8608:Chemical synapse
8508:Scaffold protein
8470:
8469:
8465:Second messenger
8391:Receptor ligands
8387:
8386:
8290:
8283:
8276:
8267:
8266:
8219:
8218:
8208:
8176:
8170:
8169:
8159:
8125:
8119:
8118:
8108:
8076:
8070:
8069:
8063:
8058:
8056:
8048:
8046:
8034:
8028:
8027:
7999:
7993:
7992:
7982:
7958:
7949:
7948:
7908:
7902:
7901:
7891:
7859:
7853:
7852:
7842:
7832:
7808:
7802:
7801:
7765:
7759:
7758:
7718:
7712:
7711:
7701:
7677:
7671:
7670:
7668:
7667:
7652:
7646:
7645:
7635:
7603:
7597:
7596:
7586:
7552:
7546:
7545:
7535:
7525:
7501:
7495:
7494:
7484:
7452:
7446:
7445:
7409:
7403:
7402:
7392:
7382:
7350:
7344:
7343:
7317:
7289:
7283:
7282:
7272:
7248:
7242:
7241:
7205:
7199:
7198:
7188:
7179:(6): 2227–2234.
7164:
7158:
7157:
7147:
7138:(5): 1093–1104.
7123:
7114:
7113:
7103:
7079:
7073:
7072:
7062:
7038:
7032:
7031:
6995:
6989:
6988:
6960:
6954:
6953:
6928:(1–2): 199–203.
6917:
6911:
6910:
6866:
6860:
6859:
6830:
6824:
6823:
6813:
6781:
6775:
6774:
6764:
6740:
6734:
6733:
6723:
6691:
6682:
6681:
6637:
6631:
6630:
6620:
6610:
6593:(9): 3219–3224.
6578:
6572:
6571:
6561:
6537:
6531:
6530:
6486:
6480:
6479:
6469:
6458:10.1172/JCI30070
6437:
6431:
6430:
6420:
6410:
6378:
6372:
6371:
6361:
6329:
6323:
6322:
6320:
6318:
6312:Researchgate.com
6303:
6297:
6296:
6286:
6276:
6259:(4): 1403–1408.
6244:
6238:
6237:
6227:
6195:
6189:
6188:
6178:
6146:
6140:
6139:
6129:
6097:
6091:
6090:
6080:
6056:
6050:
6049:
6021:
6015:
6014:
6004:
5980:
5974:
5973:
5963:
5954:(7): 2771–2776.
5939:
5933:
5932:
5922:
5890:
5881:
5880:
5870:
5846:
5840:
5839:
5829:
5797:
5791:
5790:
5780:
5756:
5745:
5744:
5710:
5704:
5703:
5693:
5669:
5663:
5662:
5644:
5620:
5614:
5613:
5577:
5571:
5570:
5553:(4): 1093–1097.
5542:
5536:
5535:
5525:
5493:
5487:
5486:
5476:
5452:
5446:
5445:
5435:
5403:
5397:
5396:
5348:
5342:
5341:
5331:
5299:
5293:
5292:
5282:
5250:
5244:
5243:
5225:
5216:(6): 1105–1118.
5201:
5195:
5194:
5166:
5160:
5159:
5141:
5109:
5100:
5099:
5063:
5054:
5053:
5036:(7): 1099–1107.
5025:
5016:
5015:
4997:
4973:
4964:
4963:
4953:
4943:
4911:
4905:
4904:
4894:
4862:
4856:
4855:
4837:
4805:
4799:
4798:
4762:
4756:
4755:
4745:
4721:
4715:
4714:
4686:
4680:
4679:
4660:10.1038/386717a0
4635:
4629:
4628:
4611:(5): 1533–1545.
4600:
4591:
4590:
4572:
4566:
4565:
4537:
4531:
4530:
4494:
4488:
4487:
4477:
4445:
4439:
4438:
4428:
4418:
4386:
4380:
4379:
4369:
4337:
4331:
4330:
4320:
4310:
4301:(6): R605–R615.
4286:
4280:
4279:
4269:
4237:
4231:
4230:
4220:
4210:
4193:(2): E141–E150.
4178:
4172:
4171:
4161:
4137:
4128:
4127:
4117:
4093:
4087:
4086:
4076:
4066:
4034:
4028:
4023:
4017:
4016:
4006:
3974:
3965:
3964:
3954:
3922:
3916:
3915:
3905:
3873:
3867:
3866:
3856:
3816:
3810:
3809:
3799:
3775:
3764:
3763:
3735:
3729:
3728:
3718:
3686:
3680:
3679:
3669:
3637:
3631:
3630:
3620:
3611:(2): dev148874.
3596:
3590:
3589:
3579:
3539:
3530:
3529:
3521:
3515:
3514:
3469:
3463:
3462:
3425:
3419:
3418:
3408:
3384:
3378:
3377:
3347:
3341:
3340:
3330:
3320:
3296:
3290:
3289:
3279:
3255:
3249:
3248:
3238:
3214:
3208:
3207:
3173:
3167:
3166:
3138:
3132:
3131:
3103:
3097:
3096:
3060:
3051:
3050:
3032:
3000:
2994:
2993:
2975:
2951:
2945:
2944:
2926:
2898:
2892:
2891:
2873:
2849:
2843:
2842:
2806:
2800:
2799:
2789:
2779:
2747:
2741:
2740:
2730:
2698:
2689:
2688:
2644:
2638:
2637:
2619:
2595:
2589:
2588:
2551:
2545:
2544:
2508:
2502:
2501:
2491:
2467:
2461:
2460:
2450:
2418:
2412:
2411:
2401:
2377:
2371:
2370:
2334:
2328:
2327:
2291:
2280:
2279:
2243:
2237:
2236:
2226:
2209:(9): 3094–3108.
2194:
2188:
2187:
2177:
2153:
2147:
2146:
2126:
2120:
2119:
2102:(609): 513–544.
2082:
2076:
2075:
2057:
2025:
2019:
2018:
2008:
1968:
1962:
1961:
1951:
1927:
1921:
1920:
1910:
1886:
1880:
1879:
1839:
1830:
1829:
1793:
1745:
1743:
1742:
1737:
1732:
1731:
1712:
1710:
1709:
1704:
1674:
1672:
1671:
1666:
1639:
1637:
1636:
1631:
1619:
1617:
1616:
1611:
1606:
1605:
1582:
1580:
1579:
1574:
1562:
1560:
1559:
1554:
1542:
1540:
1539:
1534:
1518:
1516:
1515:
1510:
1508:
1507:
1491:
1489:
1488:
1483:
1481:
1480:
1462:
1460:
1459:
1454:
1452:
1445:
1444:
1429:
1428:
1400:
1399:
1390:
1388:
1377:
1371:
1370:
1352:
1351:
1326:
1325:
1316:
1314:
1303:
1240:notch inhibitors
1228:Notch inhibitors
1155:Bone development
1068:cardiac cushions
1031:Some studies in
321:In the nematode
152:Michael W. Young
9337:
9336:
9332:
9331:
9330:
9328:
9327:
9326:
9312:
9311:
9310:
9305:
9296:
9275:
9262:Sean B. Carroll
9200:
9132:
9079:
9043:
8995:
8976:Maternal effect
8959:
8892:
8829:
8819:
8789:
8784:
8753:
8727:
8695:
8686:
8656:
8651:
8583:
8549:
8512:
8491:
8487:Lipid signaling
8459:
8431:
8378:
8303:
8294:
8256:Notch+Receptors
8251:Wayback Machine
8227:
8222:
8177:
8173:
8148:
8126:
8122:
8091:(59): 787–798.
8077:
8073:
8061:
8059:
8050:
8049:
8035:
8031:
8000:
7996:
7959:
7952:
7909:
7905:
7860:
7856:
7809:
7805:
7766:
7762:
7719:
7715:
7678:
7674:
7665:
7663:
7654:
7653:
7649:
7604:
7600:
7575:
7553:
7549:
7516:(6): e0304607.
7502:
7498:
7453:
7449:
7426:10.1038/nm.3678
7414:Nature Medicine
7410:
7406:
7351:
7347:
7315:10.1.1.459.5126
7290:
7286:
7249:
7245:
7206:
7202:
7165:
7161:
7124:
7117:
7080:
7076:
7039:
7035:
7000:Nature Genetics
6996:
6992:
6961:
6957:
6918:
6914:
6867:
6863:
6831:
6827:
6782:
6778:
6741:
6737:
6692:
6685:
6638:
6634:
6579:
6575:
6538:
6534:
6487:
6483:
6438:
6434:
6379:
6375:
6330:
6326:
6316:
6314:
6304:
6300:
6245:
6241:
6196:
6192:
6147:
6143:
6098:
6094:
6057:
6053:
6022:
6018:
5981:
5977:
5940:
5936:
5891:
5884:
5847:
5843:
5798:
5794:
5757:
5748:
5733:
5711:
5707:
5670:
5666:
5621:
5617:
5578:
5574:
5543:
5539:
5494:
5490:
5453:
5449:
5404:
5400:
5349:
5345:
5300:
5296:
5251:
5247:
5202:
5198:
5167:
5163:
5118:Current Biology
5110:
5103:
5064:
5057:
5026:
5019:
4974:
4967:
4912:
4908:
4863:
4859:
4814:Current Biology
4806:
4802:
4763:
4759:
4722:
4718:
4687:
4683:
4636:
4632:
4601:
4594:
4587:
4573:
4569:
4538:
4534:
4495:
4491:
4446:
4442:
4387:
4383:
4338:
4334:
4287:
4283:
4238:
4234:
4179:
4175:
4138:
4131:
4094:
4090:
4035:
4031:
4024:
4020:
3975:
3968:
3923:
3919:
3874:
3870:
3817:
3813:
3776:
3767:
3736:
3732:
3687:
3683:
3638:
3634:
3597:
3593:
3540:
3533:
3523:
3522:
3518:
3470:
3466:
3426:
3422:
3385:
3381:
3374:
3348:
3344:
3297:
3293:
3256:
3252:
3215:
3211:
3196:
3174:
3170:
3149:(11): 763–770.
3139:
3135:
3104:
3100:
3061:
3054:
3015:(14): 813–820.
3009:Current Biology
3001:
2997:
2952:
2948:
2899:
2895:
2850:
2846:
2807:
2803:
2748:
2744:
2699:
2692:
2645:
2641:
2596:
2592:
2552:
2548:
2509:
2505:
2468:
2464:
2419:
2415:
2378:
2374:
2335:
2331:
2292:
2283:
2244:
2240:
2195:
2191:
2154:
2150:
2143:
2127:
2123:
2083:
2079:
2055:10.1038/462171a
2026:
2022:
1969:
1965:
1928:
1924:
1887:
1883:
1840:
1833:
1794:
1787:
1783:
1765:
1727:
1723:
1718:
1715:
1714:
1680:
1677:
1676:
1660:
1657:
1656:
1625:
1622:
1621:
1601:
1597:
1592:
1589:
1588:
1568:
1565:
1564:
1548:
1545:
1544:
1528:
1525:
1524:
1503:
1499:
1497:
1494:
1493:
1476:
1472:
1470:
1467:
1466:
1450:
1449:
1440:
1436:
1424:
1420:
1401:
1395:
1391:
1381:
1376:
1373:
1372:
1366:
1362:
1347:
1343:
1327:
1321:
1317:
1307:
1302:
1298:
1296:
1293:
1292:
1284:
1252:gamma-secretase
1236:
1234:DAPT (chemical)
1230:
1213:
1204:
1187:
1182:
1157:
1148:
1139:
1123:
1089:
1076:
1029:
987:
980:
964:
940:
924:gamma secretase
914:
894:
875:
852:cerebral cortex
835:
825:specification,
819:progenitor cell
778:
765:
728:
707:model organism
694:
692:Embryo polarity
686:
658:development of
644:tyrosine kinase
543:
534:
509:
472:metalloprotease
351:-linked glycans
342:-like repeats.
289:
214:gene expression
194:
188:
130:
80:hetero-oligomer
56:notch receptors
23:Notch-mediated
17:
12:
11:
5:
9335:
9325:
9324:
9307:
9306:
9301:
9298:
9297:
9295:
9294:
9289:
9283:
9281:
9277:
9276:
9274:
9273:
9272:
9271:
9259:
9254:
9249:
9244:
9239:
9238:
9237:
9226:François Jacob
9223:
9218:
9212:
9210:
9206:
9205:
9202:
9201:
9199:
9198:
9193:
9188:
9183:
9178:
9173:
9168:
9163:
9162:
9161:
9151:
9146:
9140:
9138:
9134:
9133:
9131:
9130:
9125:
9120:
9115:
9110:
9105:
9100:
9094:
9092:
9085:
9081:
9080:
9078:
9077:
9072:
9067:
9062:
9057:
9051:
9049:
9045:
9044:
9042:
9041:
9036:
9031:
9026:
9025:
9024:
9019:
9009:
9003:
9001:
8997:
8996:
8994:
8993:
8988:
8983:
8978:
8973:
8967:
8965:
8961:
8960:
8958:
8957:
8955:Sequence space
8952:
8947:
8942:
8937:
8932:
8923:
8918:
8913:
8908:
8902:
8900:
8894:
8893:
8891:
8890:
8885:
8884:
8883:
8873:
8868:
8863:
8858:
8853:
8848:
8843:
8837:
8835:
8831:
8830:
8818:
8817:
8810:
8803:
8795:
8786:
8785:
8783:
8782:
8777:
8772:
8767:
8761:
8759:
8755:
8754:
8752:
8751:
8746:
8741:
8737:
8735:
8729:
8728:
8726:
8725:
8720:
8715:
8710:
8706:
8704:
8697:
8696:
8685:
8684:
8677:
8670:
8662:
8653:
8652:
8650:
8649:
8644:
8639:
8634:
8629:
8628:
8627:
8617:
8612:
8611:
8610:
8605:
8597:
8591:
8589:
8588:Other concepts
8585:
8584:
8582:
8581:
8576:
8567:
8561:
8559:
8555:
8554:
8551:
8550:
8548:
8547:
8542:
8537:
8532:
8526:
8524:
8518:
8517:
8514:
8513:
8511:
8510:
8505:
8499:
8497:
8493:
8492:
8490:
8489:
8484:
8479:
8473:
8467:
8461:
8460:
8458:
8457:
8452:
8447:
8441:
8439:
8433:
8432:
8430:
8429:
8424:
8422:Growth factors
8419:
8414:
8401:
8395:
8393:
8384:
8380:
8379:
8377:
8376:
8371:
8366:
8361:
8356:
8351:
8346:
8341:
8340:
8339:
8334:
8324:
8319:
8317:
8311:
8309:
8305:
8304:
8297:Cell signaling
8293:
8292:
8285:
8278:
8270:
8264:
8263:
8253:
8237:
8226:
8225:External links
8223:
8221:
8220:
8171:
8146:
8120:
8071:
8029:
7994:
7950:
7923:(4): 429–446.
7903:
7854:
7803:
7776:(3): 317–329.
7760:
7733:(3): 294–301.
7713:
7672:
7647:
7598:
7573:
7547:
7496:
7467:(4): 523–530.
7447:
7404:
7345:
7284:
7243:
7216:(6): 344–352.
7200:
7159:
7115:
7094:(2): 323–338.
7074:
7053:(1): 176–200.
7033:
6990:
6971:(1): 197–208.
6955:
6912:
6861:
6825:
6796:(3): 931–939.
6776:
6755:(3): 568–581.
6735:
6706:(2): 104–106.
6683:
6632:
6573:
6552:(5): 839–844.
6532:
6481:
6452:(2): 353–363.
6432:
6373:
6344:(1): 299–310.
6324:
6298:
6239:
6210:(1): 129–141.
6190:
6161:(1): 117–128.
6141:
6092:
6051:
6016:
5995:(7): 910–917.
5975:
5934:
5882:
5861:(4): 747–755.
5841:
5792:
5746:
5731:
5705:
5684:(4): 721–726.
5664:
5615:
5572:
5537:
5508:(1): 138–151.
5488:
5467:(1): 161–177.
5447:
5398:
5343:
5294:
5245:
5196:
5161:
5101:
5055:
5017:
4988:(2): 383–394.
4965:
4926:(3): 133–137.
4906:
4857:
4820:(9): 491–500.
4800:
4757:
4736:(5): 677–688.
4716:
4681:
4630:
4592:
4585:
4567:
4548:(1): 231–240.
4532:
4489:
4440:
4381:
4332:
4308:10.1186/bcr920
4281:
4252:(2): 197–202.
4232:
4173:
4129:
4108:(4): 509–516.
4088:
4029:
4018:
3989:(3): 415–429.
3966:
3917:
3888:(7): 846–858.
3868:
3811:
3790:(3): 339–363.
3765:
3746:(1): 471–490.
3730:
3681:
3652:(4): 271–273.
3632:
3591:
3531:
3516:
3481:(4): 687–699.
3464:
3437:(4): 687–699.
3420:
3399:(2): 264–276.
3379:
3372:
3342:
3291:
3250:
3229:(3): 591–600.
3209:
3194:
3168:
3133:
3098:
3071:(5): 965–973.
3052:
2995:
2966:(5): 985–996.
2946:
2917:(5): 973–983.
2905:(March 2006).
2893:
2844:
2801:
2762:(1): 229–234.
2742:
2713:(3): 655–669.
2690:
2639:
2610:(4): 649–660.
2590:
2563:(4): 649–661.
2546:
2519:(2): 331–345.
2503:
2462:
2413:
2372:
2345:(4): 601–611.
2329:
2302:(4): 589–599.
2281:
2254:(2): 435–444.
2238:
2189:
2148:
2141:
2121:
2108:10.1086/279629
2077:
2020:
1963:
1942:(8): 927–930.
1922:
1901:(2): 207–216.
1895:Molecular Cell
1881:
1831:
1784:
1782:
1779:
1778:
1777:
1771:
1764:
1761:
1735:
1730:
1726:
1722:
1702:
1699:
1696:
1693:
1690:
1687:
1684:
1664:
1640:, integrating
1629:
1609:
1604:
1600:
1596:
1585:Hill functions
1583:are typically
1572:
1552:
1532:
1506:
1502:
1479:
1475:
1448:
1443:
1439:
1435:
1432:
1427:
1423:
1419:
1416:
1413:
1410:
1407:
1404:
1402:
1398:
1394:
1387:
1384:
1380:
1375:
1374:
1369:
1365:
1361:
1358:
1355:
1350:
1346:
1342:
1339:
1336:
1333:
1330:
1328:
1324:
1320:
1313:
1310:
1306:
1301:
1300:
1283:
1280:
1254:inhibitors in
1229:
1226:
1212:
1209:
1203:
1200:
1186:
1183:
1181:
1180:Role in cancer
1178:
1176:regeneration.
1156:
1153:
1147:
1144:
1138:
1135:
1122:
1119:
1088:
1085:
1075:
1072:
1028:
1025:
1024:
1023:
1002:
998:
997:
984:
979:
976:
963:
960:
939:
936:
913:
910:
893:
890:
874:
871:
834:
831:
777:
774:
764:
761:
727:
724:
693:
690:
685:
682:
670:
669:
668:
667:
656:
649:
633:
630:mammary glands
626:
620:
610:
599:
596:
586:
577:
562:
556:
542:
539:
533:
530:
508:
505:
288:
285:
187:
184:
129:
126:
48:cell signaling
15:
9:
6:
4:
3:
2:
9334:
9323:
9320:
9319:
9317:
9304:
9299:
9293:
9290:
9288:
9285:
9284:
9282:
9278:
9270:
9269:
9265:
9264:
9263:
9260:
9258:
9255:
9253:
9250:
9248:
9245:
9243:
9240:
9236:
9233:
9232:
9231:
9230:Jacques Monod
9227:
9224:
9222:
9219:
9217:
9214:
9213:
9211:
9207:
9197:
9194:
9192:
9189:
9187:
9184:
9182:
9179:
9177:
9174:
9172:
9169:
9167:
9164:
9160:
9157:
9156:
9155:
9152:
9150:
9147:
9145:
9144:Homeotic gene
9142:
9141:
9139:
9135:
9129:
9126:
9124:
9121:
9119:
9116:
9114:
9111:
9109:
9106:
9104:
9101:
9099:
9096:
9095:
9093:
9089:
9086:
9082:
9076:
9073:
9071:
9068:
9066:
9063:
9061:
9058:
9056:
9053:
9052:
9050:
9046:
9040:
9037:
9035:
9032:
9030:
9027:
9023:
9020:
9018:
9015:
9014:
9013:
9012:Morphogenesis
9010:
9008:
9005:
9004:
9002:
8998:
8992:
8989:
8987:
8984:
8982:
8979:
8977:
8974:
8972:
8969:
8968:
8966:
8962:
8956:
8953:
8951:
8948:
8946:
8943:
8941:
8938:
8936:
8933:
8931:
8927:
8924:
8922:
8919:
8917:
8914:
8912:
8909:
8907:
8904:
8903:
8901:
8899:
8895:
8889:
8886:
8882:
8879:
8878:
8877:
8874:
8872:
8869:
8867:
8864:
8862:
8859:
8857:
8854:
8852:
8849:
8847:
8846:Reaction norm
8844:
8842:
8839:
8838:
8836:
8832:
8828:
8824:
8816:
8811:
8809:
8804:
8802:
8797:
8796:
8793:
8781:
8778:
8776:
8773:
8771:
8768:
8766:
8763:
8762:
8760:
8756:
8750:
8747:
8745:
8742:
8739:
8738:
8736:
8734:
8730:
8724:
8721:
8719:
8716:
8714:
8711:
8708:
8707:
8705:
8702:
8698:
8694:
8690:
8683:
8678:
8676:
8671:
8669:
8664:
8663:
8660:
8648:
8645:
8643:
8640:
8638:
8635:
8633:
8630:
8626:
8623:
8622:
8621:
8618:
8616:
8613:
8609:
8606:
8604:
8601:
8600:
8598:
8596:
8593:
8592:
8590:
8586:
8580:
8577:
8575:
8571:
8568:
8566:
8563:
8562:
8560:
8556:
8546:
8543:
8541:
8538:
8536:
8533:
8531:
8528:
8527:
8525:
8523:
8519:
8509:
8506:
8504:
8501:
8500:
8498:
8494:
8488:
8485:
8483:
8480:
8478:
8475:
8474:
8471:
8468:
8466:
8462:
8456:
8453:
8451:
8450:Intracellular
8448:
8446:
8443:
8442:
8440:
8438:
8434:
8428:
8425:
8423:
8420:
8418:
8415:
8413:
8412:Neurohormones
8409:
8408:Neuropeptides
8405:
8402:
8400:
8397:
8396:
8394:
8392:
8388:
8385:
8381:
8375:
8372:
8370:
8367:
8365:
8362:
8360:
8359:Fas apoptosis
8357:
8355:
8352:
8350:
8347:
8345:
8342:
8338:
8335:
8333:
8330:
8329:
8328:
8325:
8323:
8320:
8318:
8316:
8313:
8312:
8310:
8306:
8302:
8298:
8291:
8286:
8284:
8279:
8277:
8272:
8271:
8268:
8261:
8257:
8254:
8252:
8248:
8245:
8244:
8238:
8236:
8235:
8229:
8228:
8216:
8212:
8207:
8202:
8198:
8194:
8190:
8186:
8182:
8175:
8167:
8163:
8158:
8153:
8149:
8143:
8139:
8135:
8131:
8124:
8116:
8112:
8107:
8102:
8098:
8094:
8090:
8086:
8082:
8075:
8067:
8054:
8045:
8040:
8033:
8025:
8021:
8017:
8013:
8009:
8005:
7998:
7990:
7986:
7981:
7976:
7972:
7968:
7964:
7957:
7955:
7946:
7942:
7938:
7934:
7930:
7926:
7922:
7918:
7914:
7907:
7899:
7895:
7890:
7885:
7881:
7877:
7873:
7869:
7865:
7858:
7850:
7846:
7841:
7836:
7831:
7826:
7822:
7818:
7814:
7807:
7799:
7795:
7791:
7787:
7783:
7779:
7775:
7771:
7764:
7756:
7752:
7748:
7744:
7740:
7736:
7732:
7728:
7724:
7717:
7709:
7705:
7700:
7695:
7691:
7687:
7683:
7676:
7662:on 2015-02-05
7661:
7657:
7651:
7643:
7639:
7634:
7629:
7625:
7621:
7618:(2): 95–110.
7617:
7613:
7609:
7602:
7594:
7590:
7585:
7580:
7576:
7570:
7566:
7562:
7558:
7551:
7543:
7539:
7534:
7529:
7524:
7519:
7515:
7511:
7507:
7500:
7492:
7488:
7483:
7478:
7474:
7470:
7466:
7462:
7458:
7451:
7443:
7439:
7435:
7431:
7427:
7423:
7419:
7415:
7408:
7400:
7396:
7391:
7386:
7381:
7376:
7372:
7368:
7364:
7360:
7356:
7349:
7341:
7337:
7333:
7329:
7325:
7321:
7316:
7311:
7307:
7303:
7299:
7295:
7288:
7280:
7276:
7271:
7266:
7262:
7258:
7254:
7247:
7239:
7235:
7231:
7227:
7223:
7219:
7215:
7211:
7204:
7196:
7192:
7187:
7182:
7178:
7174:
7170:
7163:
7155:
7151:
7146:
7141:
7137:
7133:
7129:
7122:
7120:
7111:
7107:
7102:
7097:
7093:
7089:
7085:
7078:
7070:
7066:
7061:
7056:
7052:
7048:
7044:
7037:
7029:
7025:
7021:
7017:
7013:
7012:10.1038/71657
7009:
7005:
7001:
6994:
6986:
6982:
6978:
6974:
6970:
6966:
6959:
6951:
6947:
6943:
6939:
6935:
6931:
6927:
6923:
6916:
6908:
6904:
6900:
6896:
6892:
6891:10.1038/23716
6888:
6884:
6880:
6876:
6872:
6865:
6857:
6853:
6849:
6845:
6841:
6837:
6829:
6821:
6817:
6812:
6807:
6803:
6799:
6795:
6791:
6787:
6780:
6772:
6768:
6763:
6758:
6754:
6750:
6746:
6739:
6731:
6727:
6722:
6717:
6713:
6709:
6705:
6701:
6697:
6690:
6688:
6679:
6675:
6671:
6667:
6663:
6659:
6655:
6651:
6647:
6643:
6636:
6628:
6624:
6619:
6614:
6609:
6604:
6600:
6596:
6592:
6588:
6584:
6577:
6569:
6565:
6560:
6555:
6551:
6547:
6543:
6536:
6528:
6524:
6520:
6516:
6512:
6508:
6504:
6500:
6496:
6492:
6485:
6477:
6473:
6468:
6463:
6459:
6455:
6451:
6447:
6443:
6436:
6428:
6424:
6419:
6414:
6409:
6404:
6400:
6396:
6392:
6388:
6384:
6377:
6369:
6365:
6360:
6355:
6351:
6347:
6343:
6339:
6335:
6328:
6313:
6309:
6302:
6294:
6290:
6285:
6280:
6275:
6270:
6266:
6262:
6258:
6254:
6250:
6243:
6235:
6231:
6226:
6221:
6217:
6213:
6209:
6205:
6201:
6194:
6186:
6182:
6177:
6172:
6168:
6164:
6160:
6156:
6152:
6145:
6137:
6133:
6128:
6123:
6119:
6115:
6111:
6107:
6103:
6096:
6088:
6084:
6079:
6074:
6070:
6066:
6062:
6055:
6047:
6043:
6039:
6035:
6031:
6027:
6020:
6012:
6008:
6003:
5998:
5994:
5990:
5986:
5979:
5971:
5967:
5962:
5957:
5953:
5949:
5945:
5938:
5930:
5926:
5921:
5916:
5912:
5908:
5905:(1): 99–115.
5904:
5900:
5896:
5889:
5887:
5878:
5874:
5869:
5864:
5860:
5856:
5852:
5845:
5837:
5833:
5828:
5823:
5819:
5815:
5811:
5807:
5803:
5796:
5788:
5784:
5779:
5774:
5770:
5766:
5762:
5755:
5753:
5751:
5742:
5738:
5734:
5728:
5724:
5720:
5716:
5709:
5701:
5697:
5692:
5687:
5683:
5679:
5675:
5668:
5660:
5656:
5652:
5648:
5643:
5638:
5634:
5630:
5626:
5619:
5611:
5607:
5603:
5599:
5595:
5591:
5587:
5583:
5576:
5568:
5564:
5560:
5556:
5552:
5548:
5541:
5533:
5529:
5524:
5519:
5515:
5511:
5507:
5503:
5499:
5492:
5484:
5480:
5475:
5470:
5466:
5462:
5458:
5451:
5443:
5439:
5434:
5429:
5425:
5421:
5417:
5413:
5409:
5402:
5394:
5390:
5386:
5382:
5378:
5374:
5370:
5366:
5362:
5358:
5354:
5347:
5339:
5335:
5330:
5325:
5321:
5317:
5313:
5309:
5305:
5298:
5290:
5286:
5281:
5276:
5272:
5268:
5264:
5260:
5256:
5249:
5241:
5237:
5233:
5229:
5224:
5219:
5215:
5211:
5207:
5200:
5192:
5188:
5184:
5180:
5176:
5172:
5165:
5157:
5153:
5149:
5145:
5140:
5135:
5131:
5127:
5123:
5119:
5115:
5108:
5106:
5097:
5093:
5089:
5085:
5081:
5080:10.1038/71104
5077:
5073:
5069:
5062:
5060:
5051:
5047:
5043:
5039:
5035:
5031:
5024:
5022:
5013:
5009:
5005:
5001:
4996:
4991:
4987:
4983:
4979:
4972:
4970:
4961:
4957:
4952:
4947:
4942:
4937:
4933:
4929:
4925:
4921:
4917:
4910:
4902:
4898:
4893:
4888:
4884:
4880:
4877:(1): 89–101.
4876:
4872:
4868:
4861:
4853:
4849:
4845:
4841:
4836:
4831:
4827:
4823:
4819:
4815:
4811:
4804:
4796:
4792:
4788:
4784:
4780:
4776:
4772:
4768:
4761:
4753:
4749:
4744:
4739:
4735:
4731:
4727:
4720:
4712:
4708:
4704:
4700:
4696:
4692:
4685:
4677:
4673:
4669:
4665:
4661:
4657:
4653:
4649:
4645:
4641:
4634:
4626:
4622:
4618:
4614:
4610:
4606:
4599:
4597:
4588:
4582:
4578:
4571:
4563:
4559:
4555:
4551:
4547:
4543:
4536:
4528:
4524:
4520:
4516:
4512:
4508:
4504:
4500:
4493:
4485:
4481:
4476:
4471:
4467:
4463:
4459:
4455:
4451:
4444:
4436:
4432:
4427:
4422:
4417:
4412:
4408:
4404:
4401:(5): e10493.
4400:
4396:
4392:
4385:
4377:
4373:
4368:
4363:
4359:
4355:
4351:
4347:
4343:
4336:
4328:
4324:
4319:
4314:
4309:
4304:
4300:
4296:
4292:
4285:
4277:
4273:
4268:
4263:
4259:
4255:
4251:
4247:
4243:
4236:
4228:
4224:
4219:
4214:
4209:
4204:
4200:
4196:
4192:
4188:
4184:
4177:
4169:
4165:
4160:
4155:
4151:
4147:
4143:
4136:
4134:
4125:
4121:
4116:
4111:
4107:
4103:
4099:
4092:
4084:
4080:
4075:
4070:
4065:
4060:
4056:
4052:
4048:
4044:
4040:
4033:
4027:
4022:
4014:
4010:
4005:
4000:
3996:
3992:
3988:
3984:
3980:
3973:
3971:
3962:
3958:
3953:
3948:
3944:
3940:
3936:
3932:
3928:
3921:
3913:
3909:
3904:
3899:
3895:
3891:
3887:
3883:
3879:
3872:
3864:
3860:
3855:
3850:
3846:
3842:
3838:
3834:
3830:
3826:
3822:
3815:
3807:
3803:
3798:
3793:
3789:
3785:
3781:
3774:
3772:
3770:
3761:
3757:
3753:
3749:
3745:
3741:
3734:
3726:
3722:
3717:
3712:
3708:
3704:
3700:
3696:
3692:
3685:
3677:
3673:
3668:
3663:
3659:
3655:
3651:
3647:
3643:
3636:
3628:
3624:
3619:
3614:
3610:
3606:
3602:
3595:
3587:
3583:
3578:
3573:
3569:
3565:
3561:
3557:
3553:
3549:
3545:
3538:
3536:
3527:
3520:
3512:
3508:
3504:
3500:
3496:
3492:
3488:
3484:
3480:
3476:
3468:
3460:
3456:
3452:
3448:
3444:
3440:
3436:
3432:
3424:
3416:
3412:
3407:
3402:
3398:
3394:
3390:
3383:
3375:
3369:
3365:
3361:
3357:
3353:
3346:
3338:
3334:
3329:
3324:
3319:
3314:
3310:
3306:
3302:
3295:
3287:
3283:
3278:
3273:
3269:
3265:
3261:
3254:
3246:
3242:
3237:
3232:
3228:
3224:
3220:
3213:
3205:
3201:
3197:
3195:9780121828226
3191:
3187:
3183:
3179:
3172:
3164:
3160:
3156:
3152:
3148:
3144:
3137:
3129:
3125:
3121:
3117:
3113:
3109:
3102:
3094:
3090:
3086:
3082:
3078:
3074:
3070:
3066:
3059:
3057:
3048:
3044:
3040:
3036:
3031:
3026:
3022:
3018:
3014:
3010:
3006:
2999:
2991:
2987:
2983:
2979:
2974:
2969:
2965:
2961:
2957:
2950:
2942:
2938:
2934:
2930:
2925:
2920:
2916:
2912:
2908:
2904:
2897:
2889:
2885:
2881:
2877:
2872:
2867:
2863:
2859:
2855:
2848:
2840:
2836:
2832:
2828:
2824:
2820:
2816:
2812:
2805:
2797:
2793:
2788:
2783:
2778:
2773:
2769:
2765:
2761:
2757:
2753:
2746:
2738:
2734:
2729:
2724:
2720:
2716:
2712:
2708:
2704:
2697:
2695:
2686:
2682:
2678:
2674:
2670:
2669:10.1038/30756
2666:
2662:
2658:
2654:
2650:
2643:
2635:
2631:
2627:
2623:
2618:
2613:
2609:
2605:
2601:
2594:
2586:
2582:
2578:
2574:
2570:
2566:
2562:
2558:
2550:
2542:
2538:
2534:
2530:
2526:
2522:
2518:
2514:
2507:
2499:
2495:
2490:
2485:
2481:
2477:
2473:
2466:
2458:
2454:
2449:
2444:
2440:
2436:
2432:
2428:
2424:
2417:
2409:
2405:
2400:
2395:
2391:
2387:
2383:
2376:
2368:
2364:
2360:
2356:
2352:
2348:
2344:
2340:
2333:
2325:
2321:
2317:
2313:
2309:
2305:
2301:
2297:
2290:
2288:
2286:
2277:
2273:
2269:
2265:
2261:
2257:
2253:
2249:
2242:
2234:
2230:
2225:
2220:
2216:
2212:
2208:
2204:
2200:
2193:
2185:
2181:
2176:
2171:
2167:
2163:
2159:
2152:
2144:
2138:
2134:
2133:
2125:
2117:
2113:
2109:
2105:
2101:
2097:
2096:
2091:
2087:
2081:
2073:
2069:
2065:
2061:
2056:
2051:
2047:
2043:
2039:
2035:
2031:
2024:
2016:
2012:
2007:
2002:
1998:
1994:
1990:
1986:
1982:
1978:
1974:
1967:
1959:
1955:
1950:
1945:
1941:
1937:
1933:
1926:
1918:
1914:
1909:
1904:
1900:
1896:
1892:
1885:
1877:
1873:
1869:
1865:
1861:
1857:
1853:
1849:
1845:
1838:
1836:
1827:
1823:
1819:
1815:
1811:
1807:
1803:
1799:
1792:
1790:
1785:
1775:
1772:
1770:
1767:
1766:
1760:
1756:
1754:
1751:'s notum and
1750:
1728:
1724:
1697:
1694:
1691:
1685:
1682:
1662:
1654:
1650:
1645:
1643:
1627:
1602:
1598:
1586:
1570:
1550:
1530:
1522:
1504:
1500:
1477:
1473:
1463:
1441:
1437:
1433:
1425:
1421:
1414:
1408:
1405:
1403:
1396:
1392:
1385:
1382:
1378:
1367:
1363:
1359:
1348:
1344:
1334:
1331:
1329:
1322:
1318:
1311:
1308:
1304:
1290:
1279:
1277:
1273:
1269:
1265:
1261:
1257:
1256:endometriosis
1253:
1249:
1245:
1241:
1235:
1225:
1222:
1218:
1208:
1199:
1197:
1193:
1177:
1174:
1170:
1166:
1162:
1152:
1143:
1134:
1132:
1128:
1118:
1115:
1111:
1106:
1104:
1099:
1097:
1093:
1084:
1082:
1081:smooth muscle
1071:
1069:
1065:
1060:
1058:
1057:Wnt signaling
1054:
1050:
1046:
1042:
1038:
1034:
1020:
1016:
1011:
1007:
1003:
1000:
999:
995:
991:
985:
982:
981:
975:
973:
970:development,
969:
959:
957:
953:
949:
945:
944:morphogenesis
935:
933:
929:
925:
920:
909:
907:
903:
899:
889:
887:
884:development.
883:
879:
870:
866:
864:
859:
857:
853:
849:
845:
841:
830:
828:
824:
820:
815:
813:
809:
805:
801:
797:
796:
791:
782:
773:
770:
760:
758:
754:
750:
746:
742:
741:somitogenesis
737:
735:
734:somitogenesis
730:
723:
721:
717:
712:
711:
706:
701:
699:
689:
681:
679:
675:
665:
661:
657:
654:
650:
648:
645:
641:
638:
634:
631:
627:
624:
621:
619:
615:
611:
608:
604:
600:
597:
595:
591:
587:
584:
581:
580:cardiac valve
578:
575:
571:
567:
563:
561:
557:
554:
551:
550:
549:
548:
547:
538:
529:
526:
522:
513:
504:
502:
499:
495:
491:
490:cell membrane
487:
483:
478:
473:
468:
465:
463:
456:
454:
450:
446:
442:
438:
434:
430:
426:
424:
419:
415:
411:
406:
404:
400:
396:
394:
389:
385:
383:
377:
373:
371:
366:
364:
359:
357:
352:
350:
343:
341:
336:
334:
330:
326:
325:
319:
317:
313:
309:
305:
301:
300:
296:proteins. In
293:
284:
282:
278:
273:
271:
267:
263:
259:
258:Notch cascade
254:
252:
248:
242:
240:
236:
232:
229:
225:
222:
217:
215:
211:
207:
203:
202:cell membrane
199:
198:Notch protein
193:
192:Notch protein
183:
181:
178:
174:
170:
166:
162:
159:
158:
153:
149:
145:
141:
137:
136:
125:
123:
119:
115:
110:
108:
104:
99:
97:
96:intracellular
93:
89:
85:
84:extracellular
81:
77:
73:
69:
65:
61:
57:
53:
49:
46:
42:
33:
26:
21:
9266:
9159:eyeless gene
9127:
9055:Evolvability
9029:Segmentation
8906:Canalisation
8876:Heterochrony
8866:Heritability
8834:Key concepts
8692:
8647:Gap junction
8482:Ca signaling
8445:Cell surface
8343:
8242:
8234:Homo sapiens
8233:
8188:
8184:
8174:
8129:
8123:
8088:
8084:
8074:
8032:
8007:
8003:
7997:
7970:
7966:
7920:
7916:
7906:
7871:
7867:
7857:
7820:
7816:
7806:
7773:
7769:
7763:
7730:
7726:
7716:
7689:
7685:
7675:
7664:. Retrieved
7660:the original
7650:
7615:
7611:
7601:
7556:
7550:
7513:
7509:
7499:
7464:
7460:
7450:
7417:
7413:
7407:
7362:
7358:
7348:
7297:
7293:
7287:
7260:
7256:
7246:
7213:
7209:
7203:
7176:
7172:
7162:
7135:
7131:
7091:
7087:
7077:
7050:
7046:
7036:
7006:(1): 36–44.
7003:
6999:
6993:
6968:
6964:
6958:
6925:
6921:
6915:
6874:
6870:
6864:
6839:
6835:
6828:
6793:
6789:
6779:
6752:
6748:
6738:
6703:
6699:
6645:
6641:
6635:
6590:
6586:
6576:
6549:
6545:
6535:
6494:
6490:
6484:
6449:
6445:
6435:
6390:
6386:
6376:
6341:
6337:
6327:
6315:. Retrieved
6311:
6301:
6256:
6252:
6242:
6207:
6203:
6193:
6158:
6154:
6144:
6109:
6105:
6095:
6068:
6064:
6054:
6029:
6025:
6019:
5992:
5988:
5978:
5951:
5947:
5937:
5902:
5898:
5858:
5854:
5844:
5809:
5805:
5795:
5768:
5764:
5714:
5708:
5681:
5677:
5667:
5635:(1): 23–36.
5632:
5628:
5618:
5585:
5581:
5575:
5550:
5546:
5540:
5505:
5501:
5491:
5464:
5460:
5450:
5415:
5411:
5401:
5360:
5356:
5346:
5311:
5307:
5297:
5262:
5258:
5248:
5213:
5209:
5199:
5174:
5170:
5164:
5121:
5117:
5074:(1): 30–40.
5071:
5067:
5033:
5029:
4985:
4981:
4923:
4919:
4909:
4874:
4870:
4860:
4817:
4813:
4803:
4770:
4766:
4760:
4733:
4729:
4719:
4694:
4690:
4684:
4643:
4639:
4633:
4608:
4604:
4576:
4570:
4545:
4541:
4535:
4502:
4498:
4492:
4457:
4453:
4443:
4398:
4394:
4384:
4349:
4345:
4335:
4298:
4294:
4284:
4249:
4245:
4235:
4190:
4186:
4176:
4149:
4145:
4105:
4101:
4091:
4046:
4042:
4032:
4021:
3986:
3982:
3937:(1): 14–25.
3934:
3930:
3920:
3885:
3881:
3871:
3828:
3824:
3814:
3787:
3783:
3743:
3739:
3733:
3698:
3694:
3684:
3649:
3645:
3635:
3608:
3604:
3594:
3551:
3547:
3525:
3519:
3478:
3474:
3467:
3434:
3430:
3423:
3396:
3392:
3382:
3355:
3351:
3345:
3308:
3304:
3294:
3267:
3263:
3253:
3226:
3222:
3212:
3177:
3171:
3146:
3143:Glycobiology
3142:
3136:
3111:
3108:Glycobiology
3107:
3101:
3068:
3064:
3012:
3008:
2998:
2963:
2959:
2949:
2914:
2910:
2896:
2864:(1): 71–79.
2861:
2857:
2847:
2814:
2810:
2804:
2759:
2755:
2745:
2710:
2706:
2652:
2648:
2642:
2607:
2603:
2593:
2560:
2556:
2549:
2516:
2512:
2506:
2479:
2475:
2465:
2430:
2426:
2416:
2389:
2385:
2375:
2342:
2338:
2332:
2299:
2295:
2251:
2247:
2241:
2206:
2202:
2192:
2165:
2161:
2151:
2131:
2124:
2099:
2093:
2080:
2037:
2033:
2023:
1980:
1976:
1966:
1939:
1935:
1925:
1898:
1894:
1884:
1851:
1847:
1801:
1797:
1757:
1646:
1464:
1285:
1272:hearing loss
1242:(especially
1237:
1214:
1211:Liver cancer
1205:
1188:
1173:chondrocytes
1160:
1158:
1149:
1140:
1124:
1107:
1100:
1090:
1087:Angiogenesis
1077:
1061:
1030:
965:
941:
915:
895:
885:
877:
876:
867:
860:
856:gyrification
836:
816:
811:
803:
793:
787:
769:presenilin-2
766:
738:
731:
729:
719:
715:
709:
702:
695:
687:
671:
652:
642:through the
640:cytoskeleton
607:osteoblastic
560:angiogenesis
544:
535:
524:
520:
518:
469:
461:
457:
422:
409:
407:
402:
392:
381:
374:(except for
369:
362:
355:
348:
344:
337:
332:
328:
322:
320:
297:
294:
290:
274:
269:
265:
257:
255:
250:
246:
243:
238:
234:
230:
227:
223:
220:
218:
210:cell nucleus
195:
179:
176:
172:
168:
164:
160:
155:
133:
131:
111:
103:neurogenesis
100:
92:non-covalent
90:-dependent,
43:is a highly
40:
38:
9257:Mike Levine
9166:Distal-less
8991:Polyphenism
8971:Epigenetics
8823:development
8558:By distance
8496:Assistants:
8455:Co-receptor
8185:Development
7132:Development
6836:Development
6546:Development
6026:Development
5855:Development
5806:Development
5582:NeuroReport
5171:Development
5030:Development
4697:: 153–164.
4691:Development
4605:Development
4542:Development
4505:(1): 3–25.
4346:Development
3605:Development
3223:Development
3065:Development
2903:Blacklow SC
1753:wing margin
1092:Endothelial
1037:endocardium
952:stalk cells
919:presenilins
906:Muller glia
898:gliogenesis
892:Gliogenesis
800:mutagenesis
583:homeostasis
574:ventricular
566:endocardium
482:γ-secretase
477:endocytosed
449:sialic acid
9235:Lac operon
9060:Robustness
9039:Modularity
9034:Metamerism
8940:Plasticity
8935:Pleiotropy
8888:Heterotopy
8625:Pheromones
8565:Juxtacrine
8243:Drosophila
8062:|url=
7973:: 109012.
7666:2015-02-05
6306:Teske CM.
3393:Cell Cycle
3373:0470016175
3352:Patterning
1936:Cell Cycle
1781:References
1749:Drosophila
1264:hair cells
1232:See also:
1022:signaling.
1010:myocardium
902:glial cell
848:stem cells
823:glial cell
812:Drosophila
804:Drosophila
795:Drosophila
710:C. elegans
653:C. elegans
570:myocardium
420:, and the
416:sugars by
324:C. elegans
312:Delta-like
281:Mastermind
270:C. elegans
266:Drosophila
239:Drosophila
228:C. elegans
221:Drosophila
212:to modify
200:spans the
177:Drosophila
157:C. elegans
9186:Morphogen
9171:Engrailed
9154:Pax genes
9075:Tinkering
8921:Epistasis
8916:Dominance
8827:phenotype
8579:Endocrine
8574:Paracrine
8570:Autocrine
8437:Receptors
8417:Cytokines
8239:Diagram:
8230:Diagram:
7798:206323361
7310:CiteSeerX
6317:4 October
4773:: 68–75.
3701:: 68–76.
2086:Morgan TH
2072:205050842
1734:⟩
1721:⟨
1686:∈
1683:ϵ
1663:ϵ
1653:cytonemes
1649:filopodia
1644:effects.
1608:⟩
1595:⟨
1434:−
1409:ν
1360:−
1354:⟩
1341:⟨
1260:LY3056480
808:Epidermal
590:endocrine
441:galactose
186:Mechanism
128:Discovery
45:conserved
9316:Category
9149:Hox gene
9137:Elements
9118:Homeobox
8701:Receptor
8399:Hormones
8349:JAK-STAT
8337:MAPK/ERK
8332:TGF beta
8247:Archived
8215:27226324
8166:30030823
8115:21084342
8053:cite web
8024:26748293
8010:: 1–15.
7989:37142213
7898:30982678
7849:34485296
7790:18473332
7747:29398419
7708:31424097
7642:23458608
7593:22399357
7542:38848383
7533:11161089
7510:PLOS ONE
7491:35073251
7434:25194568
7399:17114293
7340:24049536
7332:15472075
7279:17698635
7230:14586790
7195:12411305
7154:15689380
7110:14651921
7069:14699589
7028:52872659
7020:10615124
6985:12941629
6942:10842072
6899:10476967
6856:11585794
6820:16219802
6771:11166270
6730:19329884
6670:17259972
6627:17296940
6568:17251261
6519:17259973
6476:17273555
6427:17468400
6368:16140292
6293:24474765
6234:18838555
6185:18824567
6136:19064701
6087:16690879
6046:10934030
6011:14988227
5970:15604224
5929:14701881
5877:17259303
5836:17021042
5787:18497317
5741:22399350
5700:25417150
5659:17550860
5651:15066262
5602:11711879
5567:19853579
5532:15598981
5483:14729486
5442:25164209
5385:16799564
5338:23804101
5289:22031906
5232:14687546
5156:15150614
5148:12906797
5096:14774606
5088:10607392
5050:11245575
5012:16444353
5004:10839357
4960:16588136
4901:27354375
4844:10801437
4795:10822545
4787:25483003
4752:15866159
4527:15211728
4519:15582774
4484:18708576
4435:20463961
4395:PLOS ONE
4376:19369400
4327:15535842
4276:18434124
4227:23236128
4168:15695512
4124:15034002
4083:14657333
4013:17336907
3961:12482957
3912:11937492
3863:20844536
3806:17409286
3760:12052917
3725:27372731
3676:26971630
3627:29374062
3586:25700513
3511:12643727
3459:12643727
3415:22223095
3337:19726682
3286:12826675
3245:17215308
3204:17132502
3163:12460944
3128:16973733
3085:14973298
3047:13909969
3039:10899003
2982:16530045
2941:17809522
2933:16530044
2888:17455442
2796:11134525
2737:22785620
2707:Genetics
2634:10828910
2585:45604279
2541:27966283
2457:11604511
2324:31484517
2276:40668388
2116:84050307
2088:(1917).
2064:19907487
2015:19907488
1958:17404512
1917:10882063
1876:43654713
1868:27045975
1826:10221902
1763:See also
1276:tinnitus
1185:Leukemia
1161:in vitro
1131:ectoderm
1127:endoderm
878:In vitro
827:neurites
705:nematode
655:germline
553:neuronal
541:Function
521:in trans
358:-glucose
98:region.
9280:Debates
9091:Systems
9017:Eyespot
8881:Neoteny
8780:Notch 4
8775:Notch 3
8770:Notch 2
8765:Notch 1
8530:General
8354:Akt/PKB
8206:4958321
8157:6879322
8106:3104346
7945:9015458
7925:Bibcode
7889:6520458
7840:8414802
7755:4912558
7633:3732476
7584:3361718
7482:9392971
7442:5390234
7390:1838740
7367:Bibcode
7302:Bibcode
7294:Science
7238:1881239
6950:9931966
6907:4338027
6879:Bibcode
6811:1895896
6721:2633979
6678:4349541
6650:Bibcode
6618:1805530
6595:Bibcode
6527:4407198
6499:Bibcode
6467:1783803
6418:1876557
6395:Bibcode
6359:1361261
6284:3910613
6261:Bibcode
6225:2557048
6176:2557047
6127:2605223
5827:3619037
5610:8329715
5433:4244024
5393:4372065
5365:Bibcode
5329:3693057
5280:3235689
5240:6525042
5191:9169836
5126:Bibcode
4951:1076884
4928:Bibcode
4892:4932368
4852:8927528
4822:Bibcode
4711:9007237
4676:4331445
4668:9109488
4648:Bibcode
4625:7789282
4562:1769331
4475:2518812
4426:2864743
4403:Bibcode
4367:2673763
4318:1064073
4267:2475578
4218:3545793
4195:Bibcode
4051:Bibcode
4004:2746361
3854:2941915
3833:Bibcode
3716:5520666
3667:4994806
3577:4445638
3556:Bibcode
3548:Science
3528:. 2013.
3503:1657403
3483:Bibcode
3451:1657403
3328:2781502
3093:6930563
3017:Bibcode
2990:9224353
2880:9546393
2839:7716513
2819:Bibcode
2811:Science
2764:Bibcode
2728:3389966
2685:4431882
2677:9620803
2657:Bibcode
2626:9604939
2577:1831692
2533:8343960
2498:8406001
2408:3000611
2367:6282210
2359:3677169
2316:3677168
2268:6616618
2233:3097517
2184:3935325
2042:Bibcode
2006:2951323
1985:Bibcode
1806:Bibcode
1798:Science
1774:Netpath
1268:cochlea
1266:in the
1248:MK-0752
1033:Xenopus
954:during
886:In vivo
882:neurite
850:of the
753:somites
739:During
698:somites
662:in the
660:alveoli
494:nucleus
464:-fucose
435:called
425:-fucose
395:-fucose
378:), and
365:-fucose
308:Serrate
287:Pathway
262:ligands
140:alleles
88:calcium
52:animals
9181:Ligand
8861:Operon
8740:Jagged
8733:Ligand
8383:Agents
8262:(MeSH)
8213:
8203:
8164:
8154:
8144:
8113:
8103:
8022:
7987:
7943:
7896:
7886:
7847:
7837:
7796:
7788:
7753:
7745:
7706:
7640:
7630:
7591:
7581:
7571:
7540:
7530:
7489:
7479:
7440:
7432:
7397:
7387:
7338:
7330:
7312:
7277:
7236:
7228:
7193:
7152:
7108:
7067:
7026:
7018:
6983:
6948:
6940:
6905:
6897:
6871:Nature
6854:
6818:
6808:
6769:
6728:
6718:
6676:
6668:
6642:Nature
6625:
6615:
6566:
6525:
6517:
6491:Nature
6474:
6464:
6425:
6415:
6366:
6356:
6291:
6281:
6232:
6222:
6183:
6173:
6134:
6124:
6085:
6044:
6009:
5968:
5927:
5920:314285
5917:
5875:
5834:
5824:
5785:
5739:
5729:
5698:
5657:
5649:
5629:Neuron
5608:
5600:
5565:
5530:
5523:540232
5520:
5481:
5440:
5430:
5391:
5383:
5357:Nature
5336:
5326:
5287:
5277:
5238:
5230:
5210:Neuron
5189:
5154:
5146:
5094:
5086:
5048:
5010:
5002:
4982:Neuron
4958:
4948:
4899:
4889:
4850:
4842:
4793:
4785:
4750:
4709:
4674:
4666:
4640:Nature
4623:
4583:
4560:
4525:
4517:
4482:
4472:
4433:
4423:
4374:
4364:
4325:
4315:
4274:
4264:
4225:
4215:
4166:
4122:
4081:
4074:299853
4071:
4011:
4001:
3959:
3952:140667
3949:
3910:
3903:186324
3900:
3861:
3851:
3825:Nature
3804:
3758:
3723:
3713:
3674:
3664:
3625:
3584:
3574:
3509:
3501:
3457:
3449:
3413:
3370:
3335:
3325:
3284:
3243:
3202:
3192:
3161:
3126:
3091:
3083:
3045:
3037:
2988:
2980:
2939:
2931:
2886:
2878:
2837:
2794:
2784:
2735:
2725:
2683:
2675:
2649:Nature
2632:
2624:
2583:
2575:
2539:
2531:
2496:
2455:
2445:
2406:
2365:
2357:
2322:
2314:
2274:
2266:
2231:
2224:367044
2221:
2182:
2139:
2114:
2070:
2062:
2034:Nature
2013:
2003:
1977:Nature
1956:
1915:
1874:
1866:
1824:
1465:where
1159:Early
1064:septum
1049:Ephrin
1047:, and
720:lin-12
623:T cell
437:Fringe
414:xylose
399:POFUT1
388:POFUT1
333:lin-12
316:Jagged
247:lin-12
231:lin-12
206:Ligand
173:lin-12
165:lin-12
138:. The
120:, and
72:NOTCH4
70:, and
68:NOTCH3
64:NOTCH2
60:NOTCH1
8709:Delta
8545:TFIIH
8540:TFIID
8364:Hippo
8344:Notch
7794:S2CID
7751:S2CID
7438:S2CID
7336:S2CID
7257:Blood
7234:S2CID
7173:Blood
7024:S2CID
6946:S2CID
6903:S2CID
6790:Blood
6674:S2CID
6523:S2CID
5948:Blood
5655:S2CID
5606:S2CID
5389:S2CID
5236:S2CID
5152:S2CID
5092:S2CID
5008:S2CID
4848:S2CID
4791:S2CID
4672:S2CID
4523:S2CID
3507:S2CID
3455:S2CID
3089:S2CID
3043:S2CID
2986:S2CID
2937:S2CID
2884:S2CID
2787:14573
2681:S2CID
2630:S2CID
2581:S2CID
2537:S2CID
2448:99946
2363:S2CID
2320:S2CID
2272:S2CID
2112:S2CID
2068:S2CID
1872:S2CID
1521:Notch
1221:AXIN1
1196:c-MYC
1192:FBXW7
1041:BMP10
798:with
716:glp-1
637:actin
451:by a
443:by a
329:glp-1
304:Delta
251:Notch
235:Notch
224:Notch
180:Notch
169:glp-1
161:Notch
114:T-ALL
8821:The
8749:JAG2
8744:JAG1
8723:DLL4
8718:DLL3
8713:DLL1
8315:GPCR
8211:PMID
8162:PMID
8142:ISBN
8111:PMID
8066:help
8020:PMID
7985:PMID
7941:PMID
7894:PMID
7845:PMID
7786:PMID
7743:PMID
7704:PMID
7638:PMID
7589:PMID
7569:ISBN
7538:PMID
7487:PMID
7430:PMID
7395:PMID
7328:PMID
7275:PMID
7226:PMID
7191:PMID
7150:PMID
7106:PMID
7065:PMID
7016:PMID
6981:PMID
6938:PMID
6895:PMID
6852:PMID
6816:PMID
6767:PMID
6726:PMID
6666:PMID
6623:PMID
6564:PMID
6515:PMID
6472:PMID
6423:PMID
6364:PMID
6319:2022
6289:PMID
6230:PMID
6181:PMID
6132:PMID
6083:PMID
6042:PMID
6007:PMID
5966:PMID
5925:PMID
5873:PMID
5832:PMID
5783:PMID
5737:PMID
5727:ISBN
5696:PMID
5678:Cell
5647:PMID
5598:PMID
5563:PMID
5528:PMID
5479:PMID
5438:PMID
5381:PMID
5334:PMID
5285:PMID
5228:PMID
5187:PMID
5144:PMID
5084:PMID
5046:PMID
5000:PMID
4956:PMID
4897:PMID
4840:PMID
4783:PMID
4748:PMID
4707:PMID
4664:PMID
4621:PMID
4581:ISBN
4558:PMID
4515:PMID
4480:PMID
4431:PMID
4372:PMID
4323:PMID
4272:PMID
4223:PMID
4164:PMID
4120:PMID
4079:PMID
4009:PMID
3957:PMID
3908:PMID
3859:PMID
3802:PMID
3756:PMID
3721:PMID
3672:PMID
3623:PMID
3582:PMID
3499:PMID
3475:Cell
3447:PMID
3431:Cell
3411:PMID
3368:ISBN
3333:PMID
3282:PMID
3241:PMID
3200:PMID
3190:ISBN
3159:PMID
3124:PMID
3081:PMID
3035:PMID
2978:PMID
2960:Cell
2929:PMID
2911:Cell
2876:PMID
2858:Cell
2835:PMID
2792:PMID
2733:PMID
2673:PMID
2622:PMID
2604:Cell
2573:PMID
2557:Cell
2529:PMID
2513:Cell
2494:PMID
2453:PMID
2404:PMID
2386:Cell
2355:PMID
2339:Cell
2312:PMID
2296:Cell
2264:PMID
2248:Cell
2229:PMID
2180:PMID
2162:Cell
2137:ISBN
2060:PMID
2011:PMID
1954:PMID
1913:PMID
1864:PMID
1822:PMID
1563:and
1492:and
1274:and
1167:and
1129:and
1114:DLL4
1110:VEGF
1045:NRG1
994:HEY2
992:and
990:HEY1
950:and
930:and
840:Numb
718:and
674:Rex1
664:lung
616:and
592:and
568:and
408:The
376:Rumi
331:and
314:and
306:and
279:and
277:CBF1
268:and
256:The
249:and
226:and
196:The
167:and
150:and
107:Numb
39:The
8825:of
8327:RTK
8322:Wnt
8201:PMC
8193:doi
8189:143
8152:PMC
8134:doi
8101:PMC
8093:doi
8039:doi
8012:doi
8008:273
7975:doi
7971:360
7933:doi
7921:183
7884:PMC
7876:doi
7835:PMC
7825:doi
7778:doi
7774:215
7735:doi
7694:doi
7628:PMC
7620:doi
7616:139
7579:PMC
7561:doi
7528:PMC
7518:doi
7477:PMC
7469:doi
7422:doi
7385:PMC
7375:doi
7363:103
7320:doi
7298:306
7265:doi
7261:110
7218:doi
7181:doi
7177:101
7140:doi
7136:132
7096:doi
7092:264
7055:doi
7051:229
7008:doi
6973:doi
6969:261
6930:doi
6887:doi
6875:400
6844:doi
6840:128
6806:PMC
6798:doi
6794:107
6757:doi
6716:PMC
6708:doi
6658:doi
6646:445
6613:PMC
6603:doi
6591:104
6554:doi
6550:134
6507:doi
6495:445
6462:PMC
6454:doi
6450:117
6413:PMC
6403:doi
6391:104
6354:PMC
6346:doi
6342:286
6279:PMC
6269:doi
6257:111
6220:PMC
6212:doi
6208:183
6171:PMC
6163:doi
6159:183
6122:PMC
6114:doi
6110:205
6073:doi
6034:doi
6030:127
5997:doi
5956:doi
5952:105
5915:PMC
5907:doi
5863:doi
5859:134
5822:PMC
5814:doi
5810:133
5773:doi
5769:102
5719:doi
5686:doi
5682:159
5637:doi
5590:doi
5555:doi
5551:390
5518:PMC
5510:doi
5469:doi
5465:266
5428:PMC
5420:doi
5373:doi
5361:442
5324:PMC
5316:doi
5275:PMC
5267:doi
5218:doi
5179:doi
5175:124
5134:doi
5076:doi
5038:doi
5034:128
4990:doi
4946:PMC
4936:doi
4887:PMC
4879:doi
4875:214
4830:doi
4775:doi
4738:doi
4699:doi
4695:123
4656:doi
4644:386
4613:doi
4609:121
4550:doi
4546:112
4507:doi
4503:122
4470:PMC
4462:doi
4421:PMC
4411:doi
4362:PMC
4354:doi
4350:136
4313:PMC
4303:doi
4262:PMC
4254:doi
4213:PMC
4203:doi
4191:110
4154:doi
4150:280
4110:doi
4069:PMC
4059:doi
4047:100
3999:PMC
3991:doi
3947:PMC
3939:doi
3898:PMC
3890:doi
3849:PMC
3841:doi
3829:467
3792:doi
3748:doi
3711:PMC
3703:doi
3662:PMC
3654:doi
3613:doi
3609:145
3572:PMC
3564:doi
3552:347
3491:doi
3439:doi
3401:doi
3360:doi
3323:PMC
3313:doi
3309:284
3272:doi
3268:278
3231:doi
3227:134
3182:doi
3151:doi
3116:doi
3073:doi
3069:131
3025:doi
2968:doi
2964:124
2919:doi
2915:124
2866:doi
2827:doi
2815:268
2782:PMC
2772:doi
2723:PMC
2715:doi
2711:191
2665:doi
2653:393
2612:doi
2565:doi
2521:doi
2484:doi
2443:PMC
2435:doi
2394:doi
2347:doi
2304:doi
2256:doi
2219:PMC
2211:doi
2170:doi
2104:doi
2050:doi
2038:462
2001:PMC
1993:doi
1981:462
1944:doi
1903:doi
1856:doi
1814:doi
1802:284
1215:In
1006:EMT
948:tip
932:MCI
896:In
647:Abl
618:Scl
501:CSL
397:by
340:EGF
9318::
9228:+
8691::
8572:/
8299:/
8209:.
8199:.
8187:.
8183:.
8160:.
8150:.
8140:.
8109:.
8099:.
8087:.
8083:.
8057::
8055:}}
8051:{{
8018:.
8006:.
7983:.
7969:.
7965:.
7953:^
7939:.
7931:.
7919:.
7915:.
7892:.
7882:.
7872:27
7870:.
7866:.
7843:.
7833:.
7819:.
7815:.
7792:.
7784:.
7772:.
7749:.
7741:.
7731:36
7729:.
7725:.
7702:.
7690:98
7688:.
7684:.
7636:.
7626:.
7614:.
7610:.
7587:.
7577:.
7567:.
7536:.
7526:.
7514:19
7512:.
7508:.
7485:.
7475:.
7465:22
7463:.
7459:.
7436:.
7428:.
7418:20
7416:.
7393:.
7383:.
7373:.
7361:.
7357:.
7334:.
7326:.
7318:.
7308:.
7296:.
7273:.
7259:.
7255:.
7232:.
7224:.
7214:21
7212:.
7189:.
7175:.
7171:.
7148:.
7134:.
7130:.
7118:^
7104:.
7090:.
7086:.
7063:.
7049:.
7045:.
7022:.
7014:.
7004:24
7002:.
6979:.
6967:.
6944:.
6936:.
6926:94
6924:.
6901:.
6893:.
6885:.
6873:.
6850:.
6838:.
6814:.
6804:.
6792:.
6788:.
6765:.
6753:49
6751:.
6747:.
6724:.
6714:.
6702:.
6698:.
6686:^
6672:.
6664:.
6656:.
6644:.
6621:.
6611:.
6601:.
6589:.
6585:.
6562:.
6548:.
6544:.
6521:.
6513:.
6505:.
6493:.
6470:.
6460:.
6448:.
6444:.
6421:.
6411:.
6401:.
6389:.
6385:.
6362:.
6352:.
6340:.
6336:.
6310:.
6287:.
6277:.
6267:.
6255:.
6251:.
6228:.
6218:.
6206:.
6202:.
6179:.
6169:.
6157:.
6153:.
6130:.
6120:.
6108:.
6104:.
6081:.
6069:98
6067:.
6063:.
6040:.
6028:.
6005:.
5993:94
5991:.
5987:.
5964:.
5950:.
5946:.
5923:.
5913:.
5903:18
5901:.
5897:.
5885:^
5871:.
5857:.
5853:.
5830:.
5820:.
5808:.
5804:.
5781:.
5767:.
5763:.
5749:^
5735:.
5725:.
5694:.
5680:.
5676:.
5653:.
5645:.
5633:42
5631:.
5627:.
5604:.
5596:.
5586:12
5584:.
5561:.
5549:.
5526:.
5516:.
5506:19
5504:.
5500:.
5477:.
5463:.
5459:.
5436:.
5426:.
5416:18
5414:.
5410:.
5387:.
5379:.
5371:.
5359:.
5355:.
5332:.
5322:.
5312:33
5310:.
5306:.
5283:.
5273:.
5263:31
5261:.
5257:.
5234:.
5226:.
5214:40
5212:.
5208:.
5185:.
5173:.
5150:.
5142:.
5132:.
5122:13
5120:.
5116:.
5104:^
5090:.
5082:.
5070:.
5058:^
5044:.
5032:.
5020:^
5006:.
4998:.
4986:26
4984:.
4980:.
4968:^
4954:.
4944:.
4934:.
4924:23
4922:.
4918:.
4895:.
4885:.
4873:.
4869:.
4846:.
4838:.
4828:.
4818:10
4816:.
4812:.
4789:.
4781:.
4771:49
4769:.
4746:.
4732:.
4728:.
4705:.
4693:.
4670:.
4662:.
4654:.
4642:.
4619:.
4607:.
4595:^
4556:.
4544:.
4521:.
4513:.
4501:.
4478:.
4468:.
4458:22
4456:.
4452:.
4429:.
4419:.
4409:.
4397:.
4393:.
4370:.
4360:.
4348:.
4344:.
4321:.
4311:.
4297:.
4293:.
4270:.
4260:.
4250:20
4248:.
4244:.
4221:.
4211:.
4201:.
4189:.
4185:.
4162:.
4148:.
4144:.
4132:^
4118:.
4106:52
4104:.
4100:.
4077:.
4067:.
4057:.
4045:.
4041:.
4007:.
3997:.
3987:12
3985:.
3981:.
3969:^
3955:.
3945:.
3935:23
3933:.
3929:.
3906:.
3896:.
3886:16
3884:.
3880:.
3857:.
3847:.
3839:.
3827:.
3823:.
3800:.
3788:28
3786:.
3782:.
3768:^
3754:.
3744:25
3742:.
3719:.
3709:.
3699:41
3697:.
3693:.
3670:.
3660:.
3650:22
3648:.
3644:.
3621:.
3607:.
3603:.
3580:.
3570:.
3562:.
3550:.
3546:.
3534:^
3505:.
3497:.
3489:.
3479:67
3477:.
3453:.
3445:.
3435:67
3433:.
3409:.
3397:11
3395:.
3391:.
3366:.
3358:.
3331:.
3321:.
3307:.
3303:.
3280:.
3266:.
3262:.
3239:.
3225:.
3221:.
3198:.
3188:.
3157:.
3147:12
3145:.
3122:.
3112:16
3110:.
3087:.
3079:.
3067:.
3055:^
3041:.
3033:.
3023:.
3013:10
3011:.
3007:.
2984:.
2976:.
2962:.
2958:.
2935:.
2927:.
2913:.
2909:.
2882:.
2874:.
2862:93
2860:.
2856:.
2833:.
2825:.
2813:.
2790:.
2780:.
2770:.
2760:98
2758:.
2754:.
2731:.
2721:.
2709:.
2705:.
2693:^
2679:.
2671:.
2663:.
2651:.
2628:.
2620:.
2608:93
2606:.
2602:.
2579:.
2571:.
2561:66
2559:.
2535:.
2527:.
2517:74
2515:.
2492:.
2478:.
2474:.
2451:.
2441:.
2431:21
2429:.
2425:.
2402:.
2390:43
2388:.
2384:.
2361:.
2353:.
2343:51
2341:.
2318:.
2310:.
2300:51
2298:.
2284:^
2270:.
2262:.
2252:34
2250:.
2227:.
2217:.
2205:.
2201:.
2178:.
2166:43
2164:.
2160:.
2110:.
2100:51
2098:.
2092:.
2066:.
2058:.
2048:.
2036:.
2032:.
2009:.
1999:.
1991:.
1979:.
1975:.
1952:.
1938:.
1934:.
1911:.
1897:.
1893:.
1870:.
1862:.
1852:59
1850:.
1846:.
1834:^
1820:.
1812:.
1800:.
1788:^
1755:.
1278:.
1098:.
1043:,
958:.
455:.
272:.
216:.
66:,
62:,
8928:/
8814:e
8807:t
8800:v
8681:e
8674:t
8667:v
8410:/
8406:/
8289:e
8282:t
8275:v
8217:.
8195::
8168:.
8136::
8117:.
8095::
8089:8
8068:)
8064:(
8047:.
8041::
8026:.
8014::
7991:.
7977::
7947:.
7935::
7927::
7900:.
7878::
7851:.
7827::
7821:9
7800:.
7780::
7757:.
7737::
7710:.
7696::
7669:.
7644:.
7622::
7595:.
7563::
7544:.
7520::
7493:.
7471::
7444:.
7424::
7401:.
7377::
7369::
7342:.
7322::
7304::
7281:.
7267::
7240:.
7220::
7197:.
7183::
7156:.
7142::
7112:.
7098::
7071:.
7057::
7030:.
7010::
6987:.
6975::
6952:.
6932::
6909:.
6889::
6881::
6858:.
6846::
6822:.
6800::
6773:.
6759::
6732:.
6710::
6704:1
6680:.
6660::
6652::
6629:.
6605::
6597::
6570:.
6556::
6529:.
6509::
6501::
6478:.
6456::
6429:.
6405::
6397::
6370:.
6348::
6321:.
6295:.
6271::
6263::
6236:.
6214::
6187:.
6165::
6138:.
6116::
6089:.
6075::
6048:.
6036::
6013:.
5999::
5972:.
5958::
5931:.
5909::
5879:.
5865::
5838:.
5816::
5789:.
5775::
5743:.
5721::
5702:.
5688::
5661:.
5639::
5612:.
5592::
5569:.
5557::
5534:.
5512::
5485:.
5471::
5444:.
5422::
5395:.
5375::
5367::
5340:.
5318::
5291:.
5269::
5242:.
5220::
5193:.
5181::
5158:.
5136::
5128::
5098:.
5078::
5072:3
5052:.
5040::
5014:.
4992::
4962:.
4938::
4930::
4903:.
4881::
4854:.
4832::
4824::
4797:.
4777::
4754:.
4740::
4734:8
4713:.
4701::
4678:.
4658::
4650::
4627:.
4615::
4589:.
4564:.
4552::
4529:.
4509::
4486:.
4464::
4437:.
4413::
4405::
4399:5
4378:.
4356::
4329:.
4305::
4299:6
4278:.
4256::
4229:.
4205::
4197::
4170:.
4156::
4126:.
4112::
4085:.
4061::
4053::
4015:.
3993::
3963:.
3941::
3914:.
3892::
3865:.
3843::
3835::
3808:.
3794::
3762:.
3750::
3727:.
3705::
3678:.
3656::
3629:.
3615::
3588:.
3566::
3558::
3513:.
3493::
3485::
3461:.
3441::
3417:.
3403::
3376:.
3362::
3339:.
3315::
3288:.
3274::
3247:.
3233::
3206:.
3184::
3165:.
3153::
3130:.
3118::
3095:.
3075::
3049:.
3027::
3019::
2992:.
2970::
2943:.
2921::
2890:.
2868::
2841:.
2829::
2821::
2798:.
2774::
2766::
2739:.
2717::
2687:.
2667::
2659::
2636:.
2614::
2587:.
2567::
2543:.
2523::
2500:.
2486::
2480:7
2459:.
2437::
2410:.
2396::
2369:.
2349::
2326:.
2306::
2278:.
2258::
2235:.
2213::
2207:6
2186:.
2172::
2145:.
2118:.
2106::
2074:.
2052::
2044::
2017:.
1995::
1987::
1960:.
1946::
1940:6
1919:.
1905::
1899:5
1878:.
1858::
1828:.
1816::
1808::
1729:i
1725:d
1701:]
1698:1
1695:,
1692:0
1689:[
1651:(
1628:i
1603:i
1599:d
1571:g
1551:f
1531:i
1505:i
1501:d
1478:i
1474:n
1447:)
1442:i
1438:d
1431:)
1426:i
1422:n
1418:(
1415:g
1412:(
1406:=
1397:i
1393:d
1386:t
1383:d
1379:d
1368:i
1364:n
1357:)
1349:i
1345:d
1338:(
1335:f
1332:=
1323:i
1319:n
1312:t
1309:d
1305:d
666:.
525:.
462:O
423:O
410:O
403:O
393:O
386:(
382:O
370:O
363:O
356:O
349:O
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.