1384:
397:
374:
296:
271:
648:
403:
302:
40:
1970:
On the other hand, the study of the dynamics of replication and repair in living cells can be done by introducing translational fusions of PCNA. To eliminate the need for transfection and bypass the problem of difficult to transfect and/or short lived cells, cell permeable replication and/or repair
4802:
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P,
1531:
pathway, which is carried out by specialised DNA polymerases that are able to incorporate damaged DNA bases into their active sites (unlike the normal replicative polymerase, which stall), and hence bypass the damage, and (2) a proposed "template switch" pathway that is thought to involve damage
1428:
Many proteins interact with PCNA via the two known PCNA-interacting motifs PCNA-interacting peptide (PIP) box and AlkB homologue 2 PCNA interacting motif (APIM). Proteins binding to PCNA via the PIP-box are mainly involved in DNA replication whereas proteins binding to PCNA via APIM are mainly
66:
1437:
The protein encoded by this gene is found in the nucleus and is a cofactor of DNA polymerase delta. The encoded protein acts as a homotrimer and helps increase the processivity of leading strand synthesis during DNA replication. In response to DNA damage, this protein is
1536:. Mono-ubiquitin of lysine number 164 on PCNA activates the translesion synthesis pathway. Extension of this mono-ubiquitin by a non-canonical lysine-63-linked poly-ubiquitin chain on PCNA is thought to activate the template switch pathway. Furthermore, sumoylation (by
5750:
Rodríguez-López AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS (February 2003). "Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA".
5892:
Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, et al. (January 1999). "Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen".
5391:
van der Kuip H, Carius B, Haque SJ, Williams BR, Huber C, Fischer T (April 1999). "The DNA-binding subunit p140 of replication factor C is upregulated in cycling cells and associates with G1 phase cell cycle regulatory proteins".
4803:
Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network".
1532:
bypass by recruitment of the homologous recombination machinery. PCNA is pivotal to the activation of these pathways and the choice as to which pathway is utilised by the cell. PCNA becomes post-translationally modified by
410:
309:
1768:
4536:
Matheos D, Ruiz MT, Price GB, Zannis-Hadjopoulos M (October 2002). "Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication".
3025:"A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2-5 complex as a novel PCNA-binding protein"
3962:
Dianova II, Bohr VA, Dianov GL (October 2001). "Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair".
5036:"Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta"
3864:"The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21"
5255:
Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai K, Koiwai O (July 2002). "Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme".
6371:
Chen IT, Smith ML, O'Connor PM, Fornace AJ (November 1995). "Direct interaction of Gadd45 with PCNA and evidence for competitive interaction of Gadd45 and p21Waf1/Cip1 with PCNA".
4090:
Chen IT, Smith ML, O'Connor PM, Fornace AJ (November 1995). "Direct interaction of Gadd45 with PCNA and evidence for competitive interaction of Gadd45 and p21Waf1/Cip1 with PCNA".
6881:
Prelich G, Kostura M, Marshak DR, Mathews MB, Stillman B (1987). "The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro".
4390:
Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K (October 2001). "UV-induced binding of ING1 to PCNA regulates the induction of apoptosis".
7084:"A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen"
4427:"A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen"
4119:"Characterization of MyD118, Gadd45, and proliferating cell nuclear antigen (PCNA) interacting domains. PCNA impedes MyD118 AND Gadd45-mediated negative growth control"
1421:
and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and
6590:
Ku DH, Travali S, Calabretta B, Huebner K, Baserga R (July 1989). "Human gene for proliferating cell nuclear antigen has pseudogenes and localizes to chromosome 20".
5542:"Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment"
6530:
Kemeny MM, Alava G, Oliver JM (November 1992). "Improving responses in hepatomas with circadian-patterned hepatic artery infusions of recombinant interleukin-2".
2798:
Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (September 2002). "RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO".
5442:"A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme"
4755:"A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells"
7125:
Webb G, Parsons P, Chenevix-Trench G (November 1990). "Localization of the gene for human proliferating nuclear antigen/cyclin by in situ hybridization".
6830:"The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis"
5483:"The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis"
1544:
the template switch pathway. This antagonistic effect occurs because sumoylated PCNA recruits a DNA helicase called Srs2, which has a role in disrupting
1116:
1097:
232:
5216:"Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis"
6629:"Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen"
3591:
Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF (September 1997). "Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1".
6560:
5077:"Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen"
3626:
Hasan S, Hassa PO, Imhof R, Hottiger MO (March 2001). "Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis".
2849:
Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S (July 2005). "SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase".
2085:
3442:"Suppression of cell transformation by the cyclin-dependent kinase inhibitor p57KIP2 requires binding to proliferating cell nuclear antigen"
3386:
Serrano M, Hannon GJ, Beach D (December 1993). "A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4".
2067:
1527:
PCNA is also involved in the DNA damage tolerance pathway known as post-replication repair (PRR). In PRR, there are two sub-pathways: (1) a
7004:"Association of proliferating cell nuclear antigen with cyclin-dependent kinases and cyclins in normal and transformed human T lymphocytes"
1442:
and is involved in the RAD6-dependent DNA repair pathway. Two transcript variants encoding the same protein have been found for this gene.
6480:
Hall PA, Kearsey JM, Coates PJ, Norman DG, Warbrick E, Cox LS (June 1995). "Characterisation of the interaction between PCNA and Gadd45".
4158:
Hall PA, Kearsey JM, Coates PJ, Norman DG, Warbrick E, Cox LS (June 1995). "Characterisation of the interaction between PCNA and Gadd45".
3499:
Rountree MR, Bachman KE, Baylin SB (July 2000). "DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci".
4310:"Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification"
5583:"Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks"
2165:
1331:
8073:
5922:"Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA"
4711:"Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair"
1324:
1959:
value. Imaging of the nuclear distribution of PCNA (via antibody labeling) can be used to distinguish between early, mid and late
7228:
6501:
Kato S, Sekine S, Oh SW, Kim NS, Umezawa Y, Abe N, et al. (December 1994). "Construction of a human full-length cDNA bank".
4946:"A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome"
4753:
Fotedar R, Mossi R, Fitzgerald P, Rousselle T, Maga G, Brickner H, Messier H, Kasibhatla S, Hübscher U, Fotedar A (August 1996).
3815:"Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen"
2282:
8637:
8600:
8178:
6926:
Prosperi E (1998). "Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair and cell cycle control".
1903:
1773:
8815:
6943:
6570:
2451:
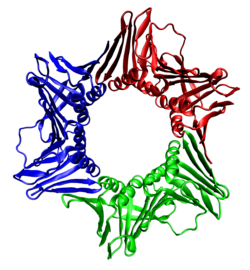
Bowman GD, O'Donnell M, Kuriyan J (June 2004). "Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex".
8572:
3905:"p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex"
2050:
4856:"Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1. A dynamic exchange of partners"
4269:"A novel oncostatin M-inducible gene OIG37 forms a gene family with MyD118 and GADD45 and negatively regulates cell growth"
1983:
1753:
63:
5298:
Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K (September 2002).
4489:"Chromatin-bound PCNA complex formation triggered by DNA damage occurs independent of the ATM gene product in human cells"
1383:
7179:
2029:
2239:
Mailand N, Gibbs-Seymour I, Bekker-Jensen S (May 2013). "Regulation of PCNA-protein interactions for genome stability".
1920:
Proteins interacting with PCNA via APIM include human AlkB homologue 2, TFIIS-L, TFII-I, Rad51B, XPA, ZRANB3, and FBH1.
6394:"Sequence and expression in Escherichia coli of the 40-kDa subunit of activator 1 (replication factor C) of HeLa cells"
5214:
Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hübscher U (December 2002).
3679:"Multivalent interaction of ESCO2 with replication machinery is required for sister chromatid cohesion in vertebrates"
8729:
8295:
4574:"Regulation of apoptosis and cell cycle progression by MCL1. Differential role of proliferating cell nuclear antigen"
2659:
Shivji KK, Kenny MK, Wood RD (April 1992). "Proliferating cell nuclear antigen is required for DNA excision repair".
8443:
8111:
396:
8782:
7667:
7606:
7846:
7438:
2054:
2033:
1979:
373:
5300:"A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase"
8820:
8642:
7742:
5981:"Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress"
2563:"Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development"
1161:
6447:"Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen"
6028:
Bacquin A, Pouvelle C, Siaud N, Perderiset M, Salomé-Desnoulez S, Tellier-Lebegue C, et al. (July 2013).
3771:"Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300"
2612:"HER2 Signaling Drives DNA Anabolism and Proliferation through SRC-3 Phosphorylation and E2F1-Regulated Genes"
8452:
8302:
8066:
7836:
7506:
2504:"Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen"
1537:
295:
270:
8765:
3730:"Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation"
1142:
8099:
7789:
7720:
7692:
7465:
6795:"Analysis of the proliferating cell nuclear antigen promoter and its response to adenovirus early region 1"
4228:"Interaction of CR6 (GADD45gamma ) with proliferating cell nuclear antigen impedes negative growth control"
1577:
212:
4043:
Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O'Connor PM, Fornace AJ (November 1994).
2702:
Essers J, Theil AF, Baldeyron C, van
Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W (November 2005).
8086:
7221:
4995:"Direct interaction of proliferating cell nuclear antigen with the small subunit of DNA polymerase delta"
3164:"Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function"
2951:"Proliferating cell nuclear antigen acts as a cytoplasmic platform controlling human neutrophil survival"
1710:
1517:
8431:
8051:
3066:"A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks"
2296:
Madru C, Henneke G, Raia P, Hugonneau-Beaufet I, Pehau-Arnaudet G, England P, et al. (March 2020).
8548:
8417:
8249:
8196:
8090:
7831:
1677:
1671:
409:
308:
5642:"Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination"
5034:
Ducoux M, Urbach S, Baldacci G, Hübscher U, Koundrioukoff S, Christensen J, Hughes P (December 2001).
8626:
7826:
6742:"Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis"
6574:
4670:"Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes"
1982:
isoform of PCNA common in cancer cells, is a potential therapeutic target in cancer therapy. In 2023
1748:
1636:
402:
301:
199:
8825:
8787:
8059:
7621:
7596:
7568:
7496:
7326:
7317:
7173:
6030:"The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells"
2949:
Witko-Sarsat V, Mocek J, Bouayad D, Tamassia N, Ribeil JA, Candalh C, et al. (November 2010).
2355:"PCNA and Ki67 expression in breast carcinoma: correlations with clinical and biological variables"
2005:
220:
3298:"Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation"
3162:
Rolef Ben-Shahar T, Castillo AG, Osborne MJ, Borden KL, Kornblatt J, Verreault A (December 2009).
2298:"Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA"
8189:
8120:
4854:
Frouin I, Maga G, Denegri M, Riva F, Savio M, Spadari S, Prosperi E, Scovassi AI (October 2003).
3769:
Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hübscher U, Hottiger MO (June 2001).
2752:
1975:
in living tissue and even distinguish cells undergoing replication from cells undergoing repair.
1913:
4349:
Komatsu K, Wharton W, Hang H, Wu C, Singh S, Lieberman HB, Pledger WJ, Wang HG (November 2000).
2561:
Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L (December 2002).
1307:
1286:
1282:
1256:
1235:
1231:
8686:
8617:
7889:
7700:
7214:
6699:"Detection of chromatin-bound PCNA in mammalian cells and its use to study DNA excision repair"
5352:"Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen"
6320:"Cloning and sequence of the human nuclear protein cyclin: homology with DNA-binding proteins"
2610:
Nikolai BC, Lanz RB, York B, Dasgupta S, Mitsiades N, Creighton CJ, et al. (March 2016).
1561:
8612:
8381:
7734:
7601:
6565:
5165:
Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S (February 2002).
3337:
Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, et al. (July 1999).
3113:
Moggs JG, Grandi P, Quivy JP, Jónsson ZO, Hübscher U, Becker PB, Almouzni G (February 2000).
1603:
1528:
1007:
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest
284:
5116:
Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S (November 2001).
8659:
8632:
8583:
7360:
6974:
6890:
6841:
6753:
6710:
6405:
6331:
6264:"Small molecule targeting of transcription-replication conflict for selective chemotherapy"
5933:
5653:
5594:
5494:
4812:
4438:
4185:
Yang Q, Manicone A, Coursen JD, Linke SP, Nagashima M, Forgues M, Wang XW (November 2000).
4056:
3916:
3635:
3453:
3395:
2858:
2807:
2515:
2460:
2404:"Molecular cloning of cDNA coding for rat proliferating cell nuclear antigen (PCNA)/cyclin"
2309:
1588:
1582:
1571:
1505:
1494:
1407:
17:
2353:
Leonardi E, Girlando S, Serio G, Mauri FA, Perrone G, Scampini S, et al. (May 1992).
240:
8:
8830:
8389:
7681:
6963:"Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen"
4045:"Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen"
3544:"PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA"
2190:
Gilljam KM, Feyzi E, Aas PA, Sousa MM, Müller R, Vågbø CB, et al. (September 2009).
1932:
1649:
1644:
6978:
6894:
6845:
6757:
6714:
6409:
6335:
6290:
6263:
5937:
5657:
5598:
5498:
4816:
4442:
4060:
3920:
3639:
3457:
3399:
2862:
2811:
2519:
2464:
2313:
1450:
7150:
7113:
6914:
6776:
6741:
6615:
6239:
6214:
6178:
Wang SC (April 2014). "PCNA: a silent housekeeper or a potential therapeutic target?".
6155:
6130:
6106:
6078:
6054:
6029:
6005:
5980:
5956:
5921:
5776:
5727:
5700:
5676:
5641:
5617:
5582:
5417:
5280:
4926:
4836:
4770:
3839:
3814:
3705:
3678:
3659:
3573:
3524:
3419:
3363:
3338:
3188:
3163:
3090:
3065:
2975:
2950:
2931:
2882:
2831:
2728:
2703:
2684:
2636:
2611:
2484:
2419:
2330:
2297:
2264:
2216:
2191:
2134:
1971:
markers can be used. These peptides offer the distinct advantage that they can be used
1692:
1477:. Part of the protein was sequenced and that sequence was used to allow isolation of a
244:
7100:
7083:
7055:
7038:
6811:
6794:
6680:
6663:
6354:
6319:
5869:
5844:
5820:
5795:
5764:
5324:
5299:
5191:
5166:
4970:
4945:
4913:
4896:
4779:
4754:
4642:
4617:
4550:
4351:"PCNA interacts with hHus1/hRad9 in response to DNA damage and replication inhibition"
3787:
3770:
3440:
Watanabe H, Pan ZQ, Schreiber-Agus N, DePinho RA, Hurwitz J, Xiong Y (February 1998).
3270:
3253:
2587:
2562:
2428:
2403:
2379:
2354:
2192:"Identification of a novel, widespread, and functionally important PCNA-binding motif"
1031:
1026:
1021:
1016:
1011:
1006:
1001:
996:
991:
986:
981:
976:
971:
966:
961:
956:
951:
946:
941:
936:
931:
926:
921:
916:
911:
906:
901:
896:
891:
886:
881:
876:
871:
866:
861:
856:
851:
846:
841:
825:
820:
815:
810:
805:
800:
795:
790:
785:
780:
775:
770:
754:
749:
744:
739:
734:
729:
724:
719:
714:
709:
704:
699:
694:
689:
8835:
8595:
8412:
7794:
7781:
7706:
7629:
7583:
7555:
7391:
7195:
7186:
7142:
7105:
7070:
7025:
6990:
6949:
6939:
6906:
6869:
6864:
6829:
6816:
6781:
6728:
6685:
6650:
6607:
6547:
6543:
6518:
6514:
6489:
6468:
6433:
6428:
6393:
6380:
6359:
6295:
6244:
6195:
6160:
6111:
6059:
6010:
5961:
5902:
5874:
5825:
5768:
5732:
5681:
5622:
5563:
5522:
5517:
5482:
5463:
5409:
5373:
5329:
5315:
5272:
5268:
5237:
5196:
5147:
5142:
5133:
5117:
5098:
5057:
5016:
4975:
4918:
4877:
4828:
4784:
4732:
4691:
4647:
4595:
4554:
4518:
4513:
4488:
4466:
4461:
4426:
4407:
4372:
4331:
4290:
4249:
4208:
4167:
4140:
4099:
4072:
4025:
4004:"p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues"
3979:
3944:
3939:
3904:
3885:
3844:
3792:
3751:
3710:
3651:
3608:
3565:
3560:
3543:
3542:
Iida T, Suetake I, Tajima S, Morioka H, Ohta S, Obuse C, Tsurimoto T (October 2002).
3516:
3481:
3476:
3441:
3411:
3368:
3319:
3275:
3234:
3193:
3144:
3139:
3114:
3095:
3046:
2980:
2923:
2874:
2835:
2823:
2775:
2733:
2719:
2676:
2672:
2641:
2592:
2543:
2538:
2503:
2476:
2433:
2384:
2335:
2256:
2221:
2169:
2126:
1626:
1621:
192:
56:
7154:
6619:
5979:
Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, et al. (August 2012).
5780:
5421:
5284:
4930:
3577:
3528:
3130:
2688:
2268:
8552:
8401:
8396:
8288:
8028:
7337:
7278:
7134:
7117:
7095:
7060:
7050:
7020:
7015:
7003:
6982:
6961:
Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, et al. (November 1994).
6931:
6918:
6898:
6859:
6849:
6806:
6771:
6761:
6718:
6675:
6640:
6599:
6539:
6510:
6458:
6423:
6413:
6349:
6339:
6285:
6280:
6275:
6234:
6226:
6213:
Gu L, Lingeman R, Yakushijin F, Sun E, Cui Q, Chao J, et al. (December 2018).
6187:
6150:
6142:
6101:
6091:
6049:
6041:
6000:
5992:
5951:
5941:
5864:
5856:
5815:
5807:
5760:
5722:
5712:
5671:
5661:
5640:
Unk I, Hajdú I, Fátyol K, Hurwitz J, Yoon JH, Prakash L, et al. (March 2008).
5612:
5602:
5553:
5512:
5502:
5453:
5401:
5363:
5319:
5311:
5264:
5227:
5186:
5178:
5137:
5129:
5088:
5047:
5006:
4965:
4957:
4908:
4867:
4840:
4820:
4774:
4766:
4722:
4681:
4637:
4629:
4585:
4546:
4508:
4500:
4456:
4446:
4399:
4362:
4321:
4280:
4239:
4198:
4130:
4064:
4015:
3971:
3934:
3924:
3875:
3834:
3826:
3782:
3741:
3700:
3690:
3677:
Bender D, De Silva E, Chen J, Poss A, Gawey L, Rulon Z, Rankin, S (December 2019).
3663:
3643:
3600:
3555:
3508:
3471:
3461:
3423:
3403:
3358:
3350:
3309:
3265:
3224:
3183:
3175:
3134:
3126:
3085:
3077:
3036:
2970:
2962:
2935:
2913:
2886:
2866:
2815:
2767:
2723:
2715:
2668:
2631:
2623:
2582:
2574:
2533:
2523:
2488:
2468:
2423:
2415:
2374:
2366:
2325:
2317:
2248:
2211:
2203:
2161:
2138:
2116:
1940:
1660:
489:
420:
364:
319:
6230:
5440:
Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O'Donnell M, Hurwitz J (July 1997).
5182:
3604:
3115:"A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage"
2627:
224:
8557:
8149:
8082:
7881:
7818:
7756:
7714:
7521:
7345:
6935:
6445:
Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T (September 1995).
6262:
Gu L, Li M, Li CM, Haratipour P, Lingeman R, Jossart J, et al. (July 2023).
6129:
Herce HD, Rajan M, Lättig-Tünnemann G, Fillies M, Cardoso MC (3 September 2014).
5996:
5946:
2771:
1999:
1936:
1687:
1654:
1608:
464:
248:
7169:
6740:
Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K (September 2005).
4897:"Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA"
2090:
National Center for
Biotechnology Information, U.S. National Library of Medicine
2072:
National Center for
Biotechnology Information, U.S. National Library of Medicine
1990:
that appears to suppress tumor growth without causing discernable side effects.
1548:
nucleoprotein filaments fundamental for initiation of homologous recombination.
8405:
8385:
7899:
7658:
7373:
7304:
6834:
Proceedings of the
National Academy of Sciences of the United States of America
6746:
Proceedings of the
National Academy of Sciences of the United States of America
6398:
Proceedings of the
National Academy of Sciences of the United States of America
6324:
Proceedings of the
National Academy of Sciences of the United States of America
6318:
Almendral JM, Huebsch D, Blundell PA, Macdonald-Bravo H, Bravo R (March 1987).
6191:
6083:
5646:
Proceedings of the
National Academy of Sciences of the United States of America
5587:
Proceedings of the
National Academy of Sciences of the United States of America
5487:
Proceedings of the
National Academy of Sciences of the United States of America
4961:
4618:"hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci"
3909:
Proceedings of the National Academy of Sciences of the United States of America
3446:
Proceedings of the National Academy of Sciences of the United States of America
3354:
2918:
2901:
2508:
Proceedings of the National Academy of Sciences of the United States of America
2321:
2121:
2104:
1482:
1414:
565:
6096:
5581:
Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, et al. (August 2008).
4944:
Touitou R, Richardson J, Bose S, Nakanishi M, Rivett J, Allday MJ (May 2001).
3830:
8809:
8607:
8534:
8144:
7922:
7383:
7368:
7312:
7270:
6645:
6628:
6463:
6446:
5811:
5458:
5441:
5167:"Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA"
4285:
4268:
4135:
4118:
3929:
3880:
3863:
1697:
897:
regulation of transcription involved in G1/S transition of mitotic cell cycle
676:
6986:
6766:
5666:
5607:
5118:"Physical and functional interactions of human DNA polymerase eta with PCNA"
4504:
4403:
4187:"Identification of a functional domain in a GADD45-mediated G2/M checkpoint"
4068:
4002:
Yu P, Huang B, Shen M, Lau C, Chan E, Michel J, et al. (January 2001).
3695:
625:
503:
8746:
7748:
7728:
7460:
7448:
7237:
7180:
ANA: Cell cycle related (Mitotic): PCNA type 1 and type 2 Antibody Patterns
7037:
Travali S, Ku DH, Rizzo MG, Ottavio L, Baserga R, Calabretta B (May 1989).
6785:
6732:
6418:
6344:
6299:
6248:
6215:"The Anticancer Activity of a First-in-class Small-molecule Targeting PCNA"
6199:
6164:
6115:
6063:
6014:
5965:
5878:
5829:
5772:
5736:
5717:
5685:
5626:
5567:
5558:
5541:
5413:
5368:
5351:
5333:
5276:
5241:
5232:
5215:
5200:
5151:
5102:
5093:
5076:
5061:
5052:
5035:
5020:
5011:
4994:
4979:
4881:
4872:
4855:
4832:
4736:
4727:
4710:
4695:
4686:
4669:
4651:
4599:
4590:
4573:
4558:
4522:
4470:
4451:
4411:
4376:
4367:
4350:
4335:
4326:
4309:
4294:
4253:
4244:
4227:
4212:
4203:
4186:
4144:
4029:
4020:
4003:
3983:
3796:
3755:
3746:
3729:
3714:
3655:
3569:
3520:
3466:
3372:
3238:
3229:
3212:
3197:
3148:
3099:
3050:
3041:
3024:
2984:
2927:
2878:
2827:
2779:
2737:
2645:
2596:
2547:
2528:
2480:
2339:
2260:
2225:
2130:
1614:
1466:
1403:
647:
482:
261:
7146:
7109:
7074:
7029:
6994:
6953:
6910:
6873:
6820:
6689:
6654:
6611:
6551:
6522:
6493:
6472:
6437:
6384:
6363:
5906:
5526:
5467:
5405:
5377:
4922:
4788:
4171:
4103:
4076:
3948:
3889:
3848:
3612:
3485:
3415:
3323:
3279:
3064:
Zhang K, Gao Y, Li J, Burgess R, Han J, Liang H, et al. (June 2016).
2680:
2437:
2388:
2207:
2173:
1446:
of this gene have been described on chromosome 4 and on the X chromosome.
1371:
1366:
8691:
8081:
8007:
7945:
7534:
7525:
7488:
7430:
7412:
6045:
5860:
5699:
Brun J, Chiu R, Lockhart K, Xiao W, Wouters BG, Gray DA (February 2008).
4993:
Lu X, Tan CK, Zhou JQ, You M, Carastro LM, Downey KM, So AG (July 2002).
3314:
3297:
3179:
3081:
2966:
2370:
2166:
10.1002/(sici)1521-1878(199803)20:3<195::aid-bies2>3.0.co;2-r
1948:
1898:
1682:
1520:
epsilon is involved in resynthesis of excised damaged DNA strands during
1422:
1206:
1187:
7065:
7039:"Structure of the human gene for the proliferating cell nuclear antigen"
6854:
6076:
5507:
4824:
4226:
Azam N, Vairapandi M, Zhang W, Hoffman B, Liebermann DA (January 2001).
4117:
Vairapandi M, Azam N, Balliet AG, Hoffman B, Liebermann DA (June 2000).
3862:
Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS (September 1997).
3211:
Kawabe T, Suganuma M, Ando T, Kimura M, Hori H, Okamoto T (March 2002).
2870:
2819:
2472:
8777:
8751:
8539:
8524:
7998:
7321:
7260:
7138:
6603:
6317:
6146:
4895:
Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J (October 1996).
4633:
2578:
1718:
1598:
1521:
1474:
1443:
1418:
1411:
381:
278:
228:
6077:
Schönenberger F, Deutzmann A, Ferrando-May E, Merhof D (29 May 2015).
5794:
Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J (June 2000).
4668:
Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA (November 2000).
3975:
169:
165:
161:
157:
153:
149:
145:
141:
137:
133:
129:
125:
121:
117:
113:
109:
105:
101:
97:
93:
89:
85:
8715:
8529:
8519:
7994:
7501:
7350:
6902:
6723:
6698:
6664:"D-type cyclin-binding regions of proliferating cell nuclear antigen"
5350:
Mossi R, Jónsson ZO, Allen BL, Hardin SH, Hübscher U (January 1997).
5075:
Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY (March 2003).
3647:
3407:
3254:"D-type cyclin-binding regions of proliferating cell nuclear antigen"
3161:
1956:
1952:
1944:
1738:
1533:
1453:, the single multi-functional DNA polymerase in this domain of life.
1439:
1399:
1061:
448:
435:
347:
334:
236:
6962:
4709:
Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL (February 2001).
4044:
2252:
8792:
8773:
8345:
8336:
8134:
7984:
7950:
7935:
7452:
7252:
4801:
3023:
Ohta S, Shiomi Y, Sugimoto K, Obuse C, Tsurimoto T (October 2002).
2152:
Warbrick E (March 1998). "PCNA binding through a conserved motif".
1964:
1928:
1808:
1743:
1631:
1355:
6627:
Li X, Li J, Harrington J, Lieber MR, Burgers PM (September 1995).
5749:
4535:
3512:
8356:
8331:
8326:
8158:
7976:
7930:
7866:
7762:
7200:
7190:
6128:
6079:"Discrimination of cell cycle phases in PCNA-immunolabeled cells"
2701:
2502:
Zhang G, Gibbs E, Kelman Z, O'Donnell M, Hurwitz J (March 1999).
2295:
2238:
1987:
1960:
1858:
1783:
1778:
1641:
1593:
1566:
1470:
1462:
1456:
1173:
1128:
1046:
1042:
7206:
5701:"hMMS2 serves a redundant role in human PCNA polyubiquitination"
4894:
4267:
Nakayama K, Hara T, Hibi M, Hirano T, Miyajima A (August 1999).
3439:
2948:
1986:
published preclinical research on a targeted chemotherapy using
1540:, SUMO) of PCNA lysine-164 (and to a lesser extent, lysine-127)
216:
39:
8036:
7851:
7798:
7673:
7633:
7591:
7563:
5796:"Characterization of the human and mouse WRN 3'→5' exonuclease"
5540:
Merkle CJ, Karnitz LM, Henry-Sánchez JT, Chen J (August 2003).
5390:
5074:
4752:
4615:
2797:
2560:
1967:
is that cells need to be fixed leading to potential artifacts.
1803:
1733:
1728:
1387:
Cryo-EM structure of the DNA-bound PolD–PCNA processive complex
1339:
1083:
656:
6027:
5539:
4943:
927:
mitotic telomere maintenance via semi-conservative replication
8706:
8701:
8696:
8679:
8674:
8669:
8664:
8652:
8647:
8365:
8360:
8317:
8312:
8307:
7955:
7940:
7912:
7907:
7871:
7856:
7841:
7808:
7770:
7611:
7573:
7511:
7283:
5254:
4616:
Kleczkowska HE, Marra G, Lettieri T, Jiricny J (March 2001).
2753:"Gaps and forks in DNA replication: Rediscovering old models"
2501:
1908:
1853:
1848:
1838:
1788:
1763:
1758:
1723:
1665:
1545:
1501:
class of ATPases. Expression of PCNA is under the control of
1498:
750:
purine-specific mismatch base pair DNA N-glycosylase activity
7185:
Overview of all the structural information available in the
6880:
6370:
5919:
5843:
Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM (2004).
5033:
4116:
4089:
3727:
3336:
8588:
8512:
8507:
8502:
8497:
8492:
8487:
8482:
8477:
8472:
8467:
8462:
8457:
8349:
8279:
8274:
8269:
8264:
8259:
8254:
8242:
8235:
8226:
8221:
8216:
8211:
8206:
8201:
8163:
8139:
8127:
7861:
7803:
7687:
4389:
3861:
3339:"Post-replicative base excision repair in replication foci"
2401:
1893:
1888:
1883:
1878:
1873:
1868:
1863:
1833:
1828:
1823:
1818:
1813:
1798:
1793:
1524:, PCNA is important for both DNA synthesis and DNA repair.
1478:
1449:
PCNA is also found in archaea, as a processivity factor of
7124:
6661:
6589:
6131:"A novel cell permeable DNA replication and repair marker"
5213:
5164:
5115:
4225:
3251:
2848:
2450:
2352:
6739:
4667:
2402:
Matsumoto K, Moriuchi T, Koji T, Nakane PK (March 1987).
1843:
1502:
1490:
6444:
5842:
4184:
3541:
3210:
2704:"Nuclear dynamics of PCNA in DNA replication and repair"
1963:
of the cell cycle. However, an important limitation of
472:
6479:
6212:
5845:"XRCC1 co-localizes and physically interacts with PCNA"
5439:
5349:
5297:
4157:
3625:
3112:
3022:
2609:
5891:
4266:
3812:
3676:
2899:
2283:"Entrez Gene: PCNA proliferating cell nuclear antigen"
2102:
1027:
telomere maintenance via semi-conservative replication
977:
nucleotide-excision repair, DNA incision, 5'-to lesion
7082:
Warbrick E, Lane DP, Glover DM, Cox LS (March 1995).
7081:
7036:
5978:
5920:
Gilljam KM, Müller R, Liabakk NB, Otterlei M (2012).
5793:
5639:
4853:
4307:
3768:
3498:
1931:
against proliferating cell nuclear antigen (PCNA) or
637:
6626:
5698:
4348:
3728:
Henneke G, Koundrioukoff S, Hübscher U (July 2003).
2189:
7001:
6662:Matsuoka S, Yamaguchi M, Matsukage A (April 1994).
3252:Matsuoka S, Yamaguchi M, Matsukage A (April 1994).
5580:
4708:
4572:Fujise K, Zhang D, Liu J, Yeh ET (December 2000).
3385:
2046:
2044:
2042:
2025:
2023:
2021:
7002:Szepesi A, Gelfand EW, Lucas JJ (November 1994).
6529:
4571:
3961:
3902:
3063:
1497:(RFC), which is a heteropentameric member of the
877:positive regulation of deoxyribonuclease activity
419:
318:
8807:
6960:
6261:
4042:
3903:Chen U, Chen S, Saha P, Dutta A (October 1996).
3813:Jónsson ZO, Hindges R, Hübscher U (April 1998).
3018:
3016:
3014:
2185:
2183:
872:transcription-coupled nucleotide-excision repair
8027:
6500:
4663:
4661:
4611:
4609:
4482:
4480:
4425:He H, Tan CK, Downey KM, So AG (October 2001).
3997:
3995:
3993:
3295:
3213:"Cdc25C interacts with PCNA at G2/M transition"
3012:
3010:
3008:
3006:
3004:
3002:
3000:
2998:
2996:
2994:
2658:
2039:
2018:
6827:
6391:
5480:
5435:
5433:
5431:
5345:
5343:
4748:
4746:
4001:
3808:
3806:
3590:
3435:
3433:
3291:
3289:
2900:Moldovan GL, Pfander B, Jentsch S (May 2007).
2103:Moldovan GL, Pfander B, Jentsch S (May 2007).
2051:GRCm38: Ensembl release 89: ENSMUSG00000027342
1457:Expression in the nucleus during DNA synthesis
1429:important in the context of genotoxic stress.
8067:
7222:
6792:
4424:
4308:Milutinovic S, Zhuang Q, Szyf M (June 2002).
2793:
2791:
2789:
2180:
7199:(Proliferating cell nuclear antigen) at the
5836:
5787:
5743:
5692:
5633:
5574:
5533:
5384:
5291:
5248:
5207:
5158:
5109:
5068:
5027:
4992:
4986:
4937:
4847:
4795:
4702:
4658:
4606:
4565:
4529:
4486:
4477:
4418:
4383:
4342:
4301:
4260:
4219:
4178:
4110:
3990:
3955:
3762:
3721:
3670:
3619:
3535:
3492:
3330:
3204:
2991:
2893:
2842:
2750:
2744:
2695:
2603:
2554:
2495:
1769:Establishment of Sister Chromatid Cohesion 2
1417:and is essential for replication. PCNA is a
5885:
5474:
5428:
5340:
4888:
4743:
4151:
4083:
4036:
3896:
3855:
3803:
3584:
3430:
3379:
3286:
3245:
2902:"PCNA, the maestro of the replication fork"
2652:
2444:
2395:
2346:
2105:"PCNA, the maestro of the replication fork"
2030:GRCh38: Ensembl release 89: ENSG00000132646
857:nucleotide-excision repair, DNA gap filling
755:DNA polymerase processivity factor activity
203:, ATLD2, proliferating cell nuclear antigen
8074:
8060:
7229:
7215:
6828:Pan ZQ, Chen M, Hurwitz J (January 1993).
5481:Pan ZQ, Chen M, Hurwitz J (January 1993).
2786:
725:dinucleotide insertion or deletion binding
7172:at the U.S. National Library of Medicine
7099:
7064:
7054:
7019:
6863:
6853:
6810:
6775:
6765:
6722:
6679:
6644:
6462:
6427:
6417:
6353:
6343:
6289:
6279:
6238:
6154:
6105:
6095:
6053:
6004:
5955:
5945:
5868:
5819:
5726:
5716:
5675:
5665:
5616:
5606:
5557:
5516:
5506:
5457:
5367:
5323:
5231:
5190:
5141:
5092:
5051:
5010:
4969:
4912:
4871:
4778:
4726:
4685:
4641:
4589:
4512:
4460:
4450:
4366:
4325:
4284:
4243:
4202:
4134:
4019:
3938:
3928:
3879:
3838:
3786:
3745:
3704:
3694:
3559:
3475:
3465:
3362:
3313:
3296:Xiong Y, Zhang H, Beach D (August 1993).
3269:
3228:
3187:
3138:
3089:
3040:
2974:
2917:
2727:
2635:
2586:
2537:
2527:
2427:
2378:
2329:
2215:
2120:
6925:
6793:Morris GF, Mathews MB (September 1990).
6392:Chen M, Pan ZQ, Hurwitz J (April 1992).
2151:
1551:
1382:
922:cellular response to DNA damage stimulus
882:nucleotide-excision repair, DNA incision
6558:
27:Mammalian protein found in Homo sapiens
14:
8808:
2751:Lehmann AR, Fuchs RP (December 2006).
2241:Nature Reviews. Molecular Cell Biology
1904:Werner syndrome ATP-dependent helicase
1774:Flap structure-specific endonuclease 1
967:cellular response to hydrogen peroxide
902:positive regulation of DNA replication
8055:
7210:
6696:
6571:Lawrence Berkeley National Laboratory
6561:"Structure of a clamp–loader complex"
2275:
1511:
1461:PCNA was originally identified as an
424:
385:
380:
323:
282:
277:
6177:
5753:Mechanisms of Ageing and Development
2955:The Journal of Experimental Medicine
1984:City of Hope National Medical Center
1754:Cyclin-dependent kinase inhibitor 1C
7043:The Journal of Biological Chemistry
6799:The Journal of Biological Chemistry
6668:The Journal of Biological Chemistry
6633:The Journal of Biological Chemistry
6592:Somatic Cell and Molecular Genetics
6451:The Journal of Biological Chemistry
5546:The Journal of Biological Chemistry
5446:The Journal of Biological Chemistry
4487:Balajee AS, Geard CR (March 2001).
3258:The Journal of Biological Chemistry
2002:– cellular marker for proliferation
1556:PCNA interacts with many proteins.
24:
6310:
6180:Trends in Pharmacological Sciences
4771:10.1002/j.1460-2075.1996.tb00815.x
2420:10.1002/j.1460-2075.1987.tb04802.x
1392:Proliferating cell nuclear antigen
25:
8847:
8730:Control of chromosome duplication
8296:Autonomously replicating sequence
7236:
7163:
6930:. Vol. 3. pp. 193–210.
952:error-prone translesion synthesis
892:positive regulation of DNA repair
740:histone acetyltransferase binding
7668:Surface epithelial-stromal tumor
7607:Tumor-associated glycoprotein 72
6544:10.1097/00002371-199211000-00001
5316:10.1128/mcb.22.18.6509-6520.2002
5269:10.1046/j.1365-2443.2002.00547.x
5134:10.1128/MCB.21.21.7199-7206.2001
3561:10.1046/j.1365-2443.2002.00584.x
2720:10.1128/MCB.25.21.9350-9359.2005
887:error-free translesion synthesis
791:DNA replication factor C complex
705:receptor tyrosine kinase binding
646:
408:
401:
395:
372:
307:
300:
294:
269:
38:
7847:Early prostate cancer antigen-2
7439:Glial fibrillary acidic protein
6928:Progress in Cell Cycle Research
6255:
6206:
6171:
6122:
6070:
6021:
5972:
5913:
3155:
3131:10.1128/mcb.20.4.1206-1218.2000
3106:
3057:
2942:
2289:
1704:
932:epithelial cell differentiation
7021:10.1182/blood.V84.10.3413.3413
6281:10.1016/j.chembiol.2023.07.001
3168:Molecular and Cellular Biology
3119:Molecular and Cellular Biology
2708:Molecular and Cellular Biology
2232:
2145:
2096:
2078:
2060:
657:More reference expression data
626:More reference expression data
13:
1:
8453:DNA polymerase III holoenzyme
8303:Single-strand binding protein
7837:Glutamate carboxypeptidase II
7101:10.1016/S0960-9822(95)00058-3
7056:10.1016/S0021-9258(18)83257-4
6812:10.1016/S0021-9258(17)46196-5
6703:Journal of Radiation Research
6681:10.1016/S0021-9258(19)78087-9
6231:10.1158/1078-0432.CCR-18-0592
5765:10.1016/s0047-6374(02)00131-8
5183:10.1128/mcb.22.3.784-791.2002
4914:10.1016/s0092-8674(00)81347-1
4551:10.1016/s0167-4781(02)00497-9
3788:10.1016/s1097-2765(01)00272-6
3605:10.1126/science.277.5334.1996
3271:10.1016/S0021-9258(19)78087-9
2628:10.1158/0008-5472.CAN-15-2383
2359:Journal of Clinical Pathology
2011:
1980:post-translationally modified
1538:small ubiquitin-like modifier
1489:) to DNA. PCNA is clamped to
942:cell population proliferation
867:regulation of DNA replication
393:
292:
8816:Genes on human chromosome 20
7466:Melanoma inhibitory activity
6936:10.1007/978-1-4615-5371-7_15
6515:10.1016/0378-1119(94)90433-2
5997:10.1016/j.molcel.2012.05.024
5947:10.1371/journal.pone.0049199
4431:Proc. Natl. Acad. Sci. U.S.A
3683:Proc. Natl. Acad. Sci. U.S.A
2772:10.1016/j.dnarep.2006.07.002
2673:10.1016/0092-8674(92)90416-A
992:response to oxidative stress
7:
2196:The Journal of Cell Biology
1993:
1432:
842:replication fork processing
10:
8852:
8549:Prokaryotic DNA polymerase
8250:Minichromosome maintenance
8197:Origin recognition complex
7832:Prostatic acid phosphatase
7743:Sertoli–Leydig cell tumour
6192:10.1016/j.tips.2014.02.004
2919:10.1016/j.cell.2007.05.003
2322:10.1038/s41467-020-15392-9
2122:10.1016/j.cell.2007.05.003
1678:Poly ADP ribose polymerase
1672:Nucleotide excision repair
546:mucosa of transverse colon
8764:
8738:
8627:Eukaryotic DNA polymerase
8568:
8439:
8430:
8374:
8174:
8107:
8098:
8020:
7993:
7975:
7968:
7921:
7898:
7880:
7827:Prostate-specific antigen
7817:
7780:
7657:
7646:
7620:
7582:
7554:
7547:
7520:
7507:Autocrine motility factor
7487:
7478:
7447:
7429:
7411:
7404:
7382:
7359:
7336:
7303:
7296:
7269:
7251:
7244:
6097:10.1186/s12859-015-0618-9
2086:"Mouse PubMed Reference:"
2068:"Human PubMed Reference:"
1749:Cyclin-dependent kinase 4
1637:Histone acetyltransferase
1465:that is expressed in the
1370:
1365:
1361:
1354:
1338:
1319:
1304:
1300:
1279:
1275:
1268:
1253:
1249:
1228:
1224:
1217:
1204:
1200:
1185:
1181:
1172:
1159:
1155:
1140:
1136:
1127:
1114:
1110:
1095:
1091:
1082:
1067:
1060:
1056:
1040:
1017:leading strand elongation
972:response to dexamethasone
745:estrogen receptor binding
695:identical protein binding
690:MutLalpha complex binding
675:
671:
654:
645:
636:
623:
572:
563:
510:
501:
471:
463:
459:
442:
429:
392:
371:
362:
358:
341:
328:
291:
268:
259:
255:
210:
207:
197:
190:
185:
82:
77:
60:
55:
50:
46:
37:
32:
7622:Hepatocellular carcinoma
7597:Carcinoembryonic antigen
7569:Carcinoembryonic antigen
7497:Carcinoembryonic antigen
7327:Carcinoembryonic antigen
7318:Medullary thyroid cancer
7174:Medical Subject Headings
6969:(Submitted manuscript).
6646:10.1074/jbc.270.38.22109
6559:Krotz D (28 July 2004).
6532:Journal of Immunotherapy
6464:10.1074/jbc.270.38.22527
6219:Clinical Cancer Research
5459:10.1074/jbc.272.30.18974
4962:10.1093/emboj/20.10.2367
4286:10.1074/jbc.274.35.24766
4136:10.1074/jbc.275.22.16810
4051:(Submitted manuscript).
3930:10.1073/pnas.93.21.11597
3881:10.1074/jbc.272.39.24522
3355:10.1093/emboj/18.13.3834
1332:Chr 2: 132.09 – 132.1 Mb
806:nuclear replication fork
8190:Pre-replication complex
8121:Pre-replication complex
6987:10.1126/science.7973727
6767:10.1073/pnas.0506447102
5667:10.1073/pnas.0800563105
5608:10.1073/pnas.0805685105
4404:10.1242/jcs.114.19.3455
4069:10.1126/science.7973727
3831:10.1093/emboj/17.8.2412
3696:10.1073/pnas.1911936117
3302:Genes & Development
1923:
1914:Y box binding protein 1
1709:PCNA has been shown to
1508:-containing complexes.
1481:clone. PCNA helps hold
1002:response to L-glutamate
957:cellular response to UV
937:response to cadmium ion
538:mucosa of sigmoid colon
7182:—Antibody Patterns.com
6697:Miura M (March 1999).
6419:10.1073/pnas.89.7.2516
6345:10.1073/pnas.84.6.1575
6135:Nucleus (Austin, Tex.)
6034:Nucleic Acids Research
5812:10.1093/nar/28.12.2396
5800:Nucleic Acids Research
5718:10.1186/1471-2199-9-24
5559:10.1074/jbc.M211591200
5369:10.1074/jbc.272.3.1769
5233:10.1074/jbc.M206889200
5094:10.1074/jbc.M208694200
5053:10.1074/jbc.M106990200
5012:10.1074/jbc.M200065200
4873:10.1074/jbc.C300098200
4728:10.1074/jbc.M008463200
4687:10.1074/jbc.C000513200
4591:10.1074/jbc.M006626200
4539:Biochim. Biophys. Acta
4452:10.1073/pnas.221452098
4368:10.1038/sj.onc.1203901
4327:10.1074/jbc.M202504200
4245:10.1074/jbc.M005626200
4204:10.1074/jbc.M005319200
4021:10.1038/sj.onc.1204113
3747:10.1038/sj.onc.1206606
3467:10.1073/pnas.95.4.1392
3230:10.1038/sj.onc.1205229
3070:Nucleic Acids Research
3042:10.1074/jbc.M206194200
2529:10.1073/pnas.96.5.1869
1493:through the action of
1388:
1325:Chr 20: 5.11 – 5.13 Mb
1022:protein ubiquitination
715:DNA polymerase binding
8821:Cell cycle regulators
8613:Replication protein A
8382:Origin of replication
7361:Neuroendocrine tumors
6268:Cell Chemical Biology
5705:BMC Molecular Biology
5406:10.1007/s001090050365
4505:10.1093/nar/29.6.1341
2302:Nature Communications
2208:10.1083/jcb.200903138
1604:DNA methyltransferase
1578:Cell-cycle regulators
1552:PCNA-binding proteins
1529:translesion synthesis
1386:
987:response to estradiol
847:translesion synthesis
786:extracellular exosome
285:Chromosome 20 (human)
8584:Replication factor C
6274:(10): 1235–1247.e6.
3315:10.1101/gad.7.8.1572
3180:10.1128/MCB.01051-09
2967:10.1084/jem.20092241
2371:10.1136/jcp.45.5.416
1583:Chromatin remodeling
1572:Base excision repair
1506:transcription factor
1495:replication factor C
1469:of cells during the
947:telomere maintenance
426:2 F2|2 64.15 cM
387:Chromosome 2 (mouse)
78:List of PDB id codes
51:Available structures
6979:1994Sci...266.1376S
6973:(5189): 1376–1380.
6895:1987Natur.326..471P
6855:10.1073/pnas.90.1.6
6846:1993PNAS...90....6P
6805:(27): 16116–16125.
6758:2005PNAS..10213795M
6752:(39): 13795–13800.
6715:1999JRadR..40....1M
6674:(15): 11030–11036.
6639:(38): 22109–22112.
6457:(38): 22527–22534.
6410:1992PNAS...89.2516C
6336:1987PNAS...84.1575A
5938:2012PLoSO...749199G
5658:2008PNAS..105.3768U
5599:2008PNAS..10512411M
5593:(34): 12411–12416.
5552:(32): 30051–30056.
5508:10.1073/pnas.90.1.6
5499:1993PNAS...90....6P
5452:(30): 18974–18981.
4825:10.1038/nature04209
4817:2005Natur.437.1173R
4443:2001PNAS...9811979H
4061:1994Sci...266.1376S
3970:(42): 12639–12644.
3921:1996PNAS...9311597C
3915:(21): 11597–11602.
3640:2001Natur.410..387H
3599:(5334): 1996–2000.
3458:1998PNAS...95.1392W
3400:1993Natur.366..704S
3264:(15): 11030–11036.
2871:10.1038/nature03665
2863:2005Natur.436..428P
2820:10.1038/nature00991
2812:2002Natur.419..135H
2520:1999PNAS...96.1869Z
2473:10.1038/nature02585
2465:2004Natur.429..724B
2314:2020NatCo..11.1591M
1933:monoclonal antibody
1650:Histone deacetylase
1617:-conjugating enzyme
1471:DNA synthesis phase
917:DNA mismatch repair
862:protein sumoylation
720:damaged DNA binding
522:ganglionic eminence
7139:10.1007/bf00205180
6604:10.1007/BF01534969
6577:on 11 October 2004
6147:10.4161/nucl.36290
6046:10.1093/nar/gkt397
5861:10.1093/nar/gkh556
4634:10.1101/gad.191201
4398:(Pt 19): 3455–62.
3082:10.1093/nar/gkw106
2579:10.1105/tpc.006403
1693:TCP protein domain
1512:Role in DNA repair
1389:
1162:ENSMUSG00000027342
982:liver regeneration
835:Biological process
764:Cellular component
683:Molecular function
8803:
8802:
8760:
8759:
8596:Flap endonuclease
8426:
8425:
8413:Okazaki fragments
8049:
8048:
8045:
8044:
8016:
8015:
7969:General histology
7964:
7963:
7795:Alpha-fetoprotein
7782:Testicular cancer
7707:Alpha-fetoprotein
7642:
7641:
7630:Alpha-fetoprotein
7584:Pancreatic cancer
7556:Colorectal cancer
7543:
7542:
7474:
7473:
7400:
7399:
7392:Homovanillic acid
7292:
7291:
7049:(13): 7466–7472.
7014:(10): 3413–3421.
6945:978-1-4613-7451-0
6889:(6112): 471–475.
6569:. Vol. 243.
6488:(12): 2427–2433.
6379:(10): 1931–1937.
6225:(23): 6053–6065.
6040:(13): 6501–6513.
5849:Nucleic Acids Res
5806:(12): 2396–2405.
5652:(10): 3768–3773.
4680:(47): 36498–501.
4493:Nucleic Acids Res
4055:(5189): 1376–80.
3976:10.1021/bi011117i
3634:(6826): 387–391.
3394:(6456): 704–707.
3349:(13): 3834–3844.
3223:(11): 1717–1726.
3174:(24): 6353–6365.
3076:(11): 5083–5094.
2961:(12): 2631–2645.
2857:(7049): 428–433.
2806:(6903): 135–141.
2766:(12): 1495–1498.
2714:(21): 9350–9359.
2573:(12): 3225–3236.
2459:(6993): 724–730.
1951:. They can be of
1627:Flap endonuclease
1622:ubiquitin ligases
1562:Apoptotic factors
1381:
1380:
1377:
1376:
1350:
1349:
1315:
1314:
1294:
1293:
1264:
1263:
1243:
1242:
1213:
1212:
1194:
1193:
1168:
1167:
1149:
1148:
1123:
1122:
1104:
1103:
1052:
1051:
912:heart development
907:response to lipid
735:chromatin binding
667:
666:
663:
662:
632:
631:
619:
618:
557:
556:
455:
454:
354:
353:
181:
180:
177:
176:
61:Ortholog search:
16:(Redirected from
8843:
8553:DNA polymerase I
8437:
8436:
8397:Replication fork
8289:Licensing factor
8105:
8104:
8076:
8069:
8062:
8053:
8052:
8029:Rhabdomyosarcoma
8025:
8024:
7973:
7972:
7655:
7654:
7552:
7551:
7485:
7484:
7409:
7408:
7338:Pheochromocytoma
7301:
7300:
7249:
7248:
7231:
7224:
7217:
7208:
7207:
7158:
7121:
7103:
7078:
7068:
7058:
7033:
7023:
6998:
6957:
6922:
6903:10.1038/326471a0
6877:
6867:
6857:
6824:
6814:
6789:
6779:
6769:
6736:
6726:
6724:10.1269/jrr.40.1
6693:
6683:
6658:
6648:
6623:
6586:
6584:
6582:
6573:. Archived from
6555:
6526:
6497:
6476:
6466:
6441:
6431:
6421:
6404:(7): 2516–2520.
6388:
6367:
6357:
6347:
6330:(6): 1575–1579.
6304:
6303:
6293:
6283:
6259:
6253:
6252:
6242:
6210:
6204:
6203:
6175:
6169:
6168:
6158:
6126:
6120:
6119:
6109:
6099:
6074:
6068:
6067:
6057:
6025:
6019:
6018:
6008:
5976:
5970:
5969:
5959:
5949:
5917:
5911:
5910:
5889:
5883:
5882:
5872:
5840:
5834:
5833:
5823:
5791:
5785:
5784:
5747:
5741:
5740:
5730:
5720:
5696:
5690:
5689:
5679:
5669:
5637:
5631:
5630:
5620:
5610:
5578:
5572:
5571:
5561:
5537:
5531:
5530:
5520:
5510:
5478:
5472:
5471:
5461:
5437:
5426:
5425:
5388:
5382:
5381:
5371:
5347:
5338:
5337:
5327:
5295:
5289:
5288:
5252:
5246:
5245:
5235:
5226:(50): 48434–40.
5211:
5205:
5204:
5194:
5162:
5156:
5155:
5145:
5128:(21): 7199–206.
5113:
5107:
5106:
5096:
5072:
5066:
5065:
5055:
5046:(52): 49258–66.
5031:
5025:
5024:
5014:
4990:
4984:
4983:
4973:
4941:
4935:
4934:
4916:
4892:
4886:
4885:
4875:
4851:
4845:
4844:
4811:(7062): 1173–8.
4799:
4793:
4792:
4782:
4750:
4741:
4740:
4730:
4706:
4700:
4699:
4689:
4665:
4656:
4655:
4645:
4613:
4604:
4603:
4593:
4584:(50): 39458–65.
4569:
4563:
4562:
4533:
4527:
4526:
4516:
4484:
4475:
4474:
4464:
4454:
4437:(21): 11979–84.
4422:
4416:
4415:
4387:
4381:
4380:
4370:
4346:
4340:
4339:
4329:
4305:
4299:
4298:
4288:
4279:(35): 24766–72.
4264:
4258:
4257:
4247:
4223:
4217:
4216:
4206:
4182:
4176:
4175:
4155:
4149:
4148:
4138:
4114:
4108:
4107:
4087:
4081:
4080:
4040:
4034:
4033:
4023:
3999:
3988:
3987:
3959:
3953:
3952:
3942:
3932:
3900:
3894:
3893:
3883:
3859:
3853:
3852:
3842:
3810:
3801:
3800:
3790:
3766:
3760:
3759:
3749:
3725:
3719:
3718:
3708:
3698:
3689:(2): 1081–1089.
3674:
3668:
3667:
3648:10.1038/35066610
3623:
3617:
3616:
3588:
3582:
3581:
3563:
3554:(10): 997–1007.
3539:
3533:
3532:
3496:
3490:
3489:
3479:
3469:
3452:(4): 1392–1397.
3437:
3428:
3427:
3408:10.1038/366704a0
3383:
3377:
3376:
3366:
3343:The EMBO Journal
3334:
3328:
3327:
3317:
3308:(8): 1572–1583.
3293:
3284:
3283:
3273:
3249:
3243:
3242:
3232:
3208:
3202:
3201:
3191:
3159:
3153:
3152:
3142:
3125:(4): 1206–1218.
3110:
3104:
3103:
3093:
3061:
3055:
3054:
3044:
3020:
2989:
2988:
2978:
2946:
2940:
2939:
2921:
2897:
2891:
2890:
2846:
2840:
2839:
2795:
2784:
2783:
2757:
2748:
2742:
2741:
2731:
2699:
2693:
2692:
2656:
2650:
2649:
2639:
2622:(6): 1463–1475.
2607:
2601:
2600:
2590:
2558:
2552:
2551:
2541:
2531:
2514:(5): 1869–1874.
2499:
2493:
2492:
2448:
2442:
2441:
2431:
2408:The EMBO Journal
2399:
2393:
2392:
2382:
2350:
2344:
2343:
2333:
2293:
2287:
2286:
2279:
2273:
2272:
2236:
2230:
2229:
2219:
2187:
2178:
2177:
2149:
2143:
2142:
2124:
2100:
2094:
2093:
2082:
2076:
2075:
2064:
2058:
2048:
2037:
2027:
1939:can be used for
1661:Licensing factor
1408:DNA polymerase δ
1363:
1362:
1334:
1327:
1310:
1298:
1297:
1289:
1273:
1272:
1269:RefSeq (protein)
1259:
1247:
1246:
1238:
1222:
1221:
1198:
1197:
1179:
1178:
1153:
1152:
1134:
1133:
1108:
1107:
1089:
1088:
1058:
1057:
816:replication fork
811:PCNA-p21 complex
673:
672:
659:
650:
643:
642:
628:
588:genital tubercle
584:ventricular zone
568:
566:Top expressed in
561:
560:
526:ventricular zone
518:secondary oocyte
506:
504:Top expressed in
499:
498:
478:
477:
461:
460:
451:
438:
427:
412:
405:
399:
388:
376:
360:
359:
350:
337:
326:
311:
304:
298:
287:
273:
257:
256:
251:
249:PCNA - orthologs
202:
195:
172:
75:
74:
69:
48:
47:
42:
30:
29:
21:
8851:
8850:
8846:
8845:
8844:
8842:
8841:
8840:
8826:DNA replication
8806:
8805:
8804:
8799:
8756:
8734:
8574:
8570:
8564:
8558:Klenow fragment
8441:
8422:
8406:leading strands
8370:
8180:
8176:
8170:
8109:
8094:
8083:DNA replication
8080:
8050:
8041:
8021:Musculoskeletal
8012:
7989:
7960:
7941:erbB-3 receptor
7936:erbB-2 receptor
7917:
7908:erbB-3 receptor
7894:
7882:Germ cell tumor
7876:
7842:erbB-3 receptor
7819:Prostate cancer
7813:
7776:
7715:Choriocarcinoma
7650:
7648:
7638:
7616:
7578:
7539:
7522:Hemangiosarcoma
7516:
7480:
7479:Cardiovascular/
7470:
7443:
7425:
7396:
7378:
7355:
7346:Normetanephrine
7332:
7288:
7265:
7240:
7235:
7166:
7161:
7088:Current Biology
6946:
6580:
6578:
6313:
6311:Further reading
6308:
6307:
6260:
6256:
6211:
6207:
6176:
6172:
6127:
6123:
6075:
6071:
6026:
6022:
5977:
5973:
5918:
5914:
5895:Cancer Research
5890:
5886:
5855:(7): 2193–201.
5841:
5837:
5792:
5788:
5748:
5744:
5697:
5693:
5638:
5634:
5579:
5575:
5538:
5534:
5479:
5475:
5438:
5429:
5389:
5385:
5348:
5341:
5310:(18): 6509–20.
5304:Mol. Cell. Biol
5296:
5292:
5253:
5249:
5212:
5208:
5171:Mol. Cell. Biol
5163:
5159:
5122:Mol. Cell. Biol
5114:
5110:
5087:(12): 10041–7.
5073:
5069:
5032:
5028:
5005:(27): 24340–5.
4991:
4987:
4956:(10): 2367–75.
4942:
4938:
4893:
4889:
4866:(41): 39265–8.
4852:
4848:
4800:
4796:
4765:(16): 4423–33.
4751:
4744:
4707:
4703:
4666:
4659:
4614:
4607:
4570:
4566:
4534:
4530:
4485:
4478:
4423:
4419:
4388:
4384:
4347:
4343:
4320:(23): 20974–8.
4306:
4302:
4265:
4261:
4224:
4220:
4197:(47): 36892–8.
4183:
4179:
4166:(12): 2427–33.
4156:
4152:
4129:(22): 16810–9.
4115:
4111:
4088:
4084:
4041:
4037:
4000:
3991:
3960:
3956:
3901:
3897:
3874:(39): 24522–9.
3860:
3856:
3811:
3804:
3767:
3763:
3740:(28): 4301–13.
3726:
3722:
3675:
3671:
3624:
3620:
3589:
3585:
3540:
3536:
3501:Nature Genetics
3497:
3493:
3438:
3431:
3384:
3380:
3335:
3331:
3294:
3287:
3250:
3246:
3209:
3205:
3160:
3156:
3111:
3107:
3062:
3058:
3035:(43): 40362–7.
3021:
2992:
2947:
2943:
2898:
2894:
2847:
2843:
2796:
2787:
2755:
2749:
2745:
2700:
2696:
2657:
2653:
2616:Cancer Research
2608:
2604:
2559:
2555:
2500:
2496:
2449:
2445:
2400:
2396:
2351:
2347:
2294:
2290:
2281:
2280:
2276:
2253:10.1038/nrm3562
2237:
2233:
2188:
2181:
2150:
2146:
2101:
2097:
2084:
2083:
2079:
2066:
2065:
2061:
2049:
2040:
2028:
2019:
2014:
1996:
1926:
1918:
1707:
1702:
1688:Protein kinases
1655:Mismatch repair
1609:DNA polymerases
1554:
1514:
1459:
1435:
1402:that acts as a
1372:View/Edit Mouse
1367:View/Edit Human
1330:
1323:
1320:Location (UCSC)
1306:
1285:
1281:
1255:
1234:
1230:
1143:ENSG00000132646
1036:
852:DNA replication
830:
759:
730:protein binding
655:
624:
615:
610:
606:
602:
598:
594:
590:
586:
582:
578:
564:
553:
548:
544:
540:
536:
532:
530:trabecular bone
528:
524:
520:
516:
502:
446:
433:
425:
415:
414:
413:
406:
386:
363:Gene location (
345:
332:
324:
314:
313:
312:
305:
283:
260:Gene location (
211:
198:
191:
84:
62:
28:
23:
22:
15:
12:
11:
5:
8849:
8839:
8838:
8833:
8828:
8823:
8818:
8801:
8800:
8798:
8797:
8796:
8795:
8790:
8785:
8770:
8768:
8762:
8761:
8758:
8757:
8755:
8754:
8749:
8742:
8740:
8736:
8735:
8733:
8732:
8726:
8725:
8724:
8723:
8712:
8711:
8710:
8709:
8704:
8699:
8694:
8684:
8683:
8682:
8677:
8672:
8667:
8657:
8656:
8655:
8650:
8645:
8640:
8630:
8623:
8622:
8621:
8620:
8610:
8605:
8604:
8603:
8593:
8592:
8591:
8580:
8578:
8566:
8565:
8563:
8562:
8561:
8560:
8545:
8544:
8543:
8542:
8532:
8527:
8522:
8517:
8516:
8515:
8510:
8505:
8500:
8495:
8490:
8485:
8480:
8475:
8470:
8465:
8460:
8449:
8447:
8434:
8428:
8427:
8424:
8423:
8421:
8420:
8415:
8410:
8409:
8408:
8393:
8392:
8378:
8376:
8372:
8371:
8369:
8368:
8363:
8353:
8352:
8342:
8341:
8340:
8339:
8334:
8323:
8322:
8321:
8320:
8315:
8310:
8299:
8298:
8292:
8291:
8285:
8284:
8283:
8282:
8277:
8272:
8267:
8262:
8257:
8246:
8245:
8239:
8238:
8232:
8231:
8230:
8229:
8224:
8219:
8214:
8209:
8204:
8193:
8192:
8186:
8184:
8179:preparation in
8172:
8171:
8169:
8168:
8167:
8166:
8155:
8154:
8153:
8152:
8147:
8142:
8131:
8130:
8124:
8123:
8117:
8115:
8102:
8096:
8095:
8079:
8078:
8071:
8064:
8056:
8047:
8046:
8043:
8042:
8040:
8039:
8033:
8031:
8022:
8018:
8017:
8014:
8013:
8011:
8010:
8004:
8002:
7991:
7990:
7988:
7987:
7981:
7979:
7970:
7966:
7965:
7962:
7961:
7959:
7958:
7953:
7948:
7943:
7938:
7933:
7927:
7925:
7919:
7918:
7916:
7915:
7910:
7904:
7902:
7900:Bladder cancer
7896:
7895:
7893:
7892:
7886:
7884:
7878:
7877:
7875:
7874:
7869:
7864:
7859:
7854:
7849:
7844:
7839:
7834:
7829:
7823:
7821:
7815:
7814:
7812:
7811:
7806:
7801:
7792:
7786:
7784:
7778:
7777:
7775:
7774:
7767:
7766:
7765:
7753:
7752:
7751:
7739:
7738:
7737:
7725:
7724:
7723:
7711:
7710:
7709:
7697:
7696:
7695:
7690:
7678:
7677:
7676:
7663:
7661:
7652:
7644:
7643:
7640:
7639:
7637:
7636:
7626:
7624:
7618:
7617:
7615:
7614:
7609:
7604:
7599:
7594:
7588:
7586:
7580:
7579:
7577:
7576:
7571:
7566:
7560:
7558:
7549:
7545:
7544:
7541:
7540:
7538:
7537:
7531:
7529:
7518:
7517:
7515:
7514:
7509:
7504:
7499:
7493:
7491:
7482:
7476:
7475:
7472:
7471:
7469:
7468:
7463:
7457:
7455:
7445:
7444:
7442:
7441:
7435:
7433:
7427:
7426:
7424:
7423:
7417:
7415:
7406:
7405:Nervous system
7402:
7401:
7398:
7397:
7395:
7394:
7388:
7386:
7380:
7379:
7377:
7376:
7374:Chromogranin A
7371:
7365:
7363:
7357:
7356:
7354:
7353:
7348:
7342:
7340:
7334:
7333:
7331:
7330:
7324:
7315:
7309:
7307:
7305:Thyroid cancer
7298:
7294:
7293:
7290:
7289:
7287:
7286:
7281:
7275:
7273:
7267:
7266:
7264:
7263:
7257:
7255:
7246:
7242:
7241:
7234:
7233:
7226:
7219:
7211:
7205:
7204:
7183:
7177:
7165:
7164:External links
7162:
7160:
7159:
7127:Human Genetics
7122:
7094:(3): 275–282.
7079:
7034:
6999:
6958:
6944:
6923:
6878:
6825:
6790:
6737:
6694:
6659:
6624:
6598:(4): 297–307.
6587:
6556:
6538:(4): 219–223.
6527:
6509:(2): 243–250.
6498:
6477:
6442:
6389:
6368:
6314:
6312:
6309:
6306:
6305:
6254:
6205:
6186:(4): 178–186.
6170:
6141:(6): 590–600.
6121:
6084:BMC Bioinform.
6069:
6020:
5991:(3): 396–409.
5985:Molecular Cell
5971:
5932:(11): e49199.
5912:
5901:(2): 342–346.
5884:
5835:
5786:
5759:(2): 167–174.
5742:
5691:
5632:
5573:
5532:
5473:
5427:
5383:
5362:(3): 1769–76.
5339:
5290:
5247:
5206:
5157:
5108:
5067:
5026:
4985:
4936:
4907:(2): 297–306.
4887:
4846:
4794:
4742:
4721:(8): 5547–55.
4701:
4657:
4605:
4564:
4545:(1–3): 59–72.
4528:
4499:(6): 1341–51.
4476:
4417:
4382:
4361:(46): 5291–7.
4341:
4300:
4259:
4238:(4): 2766–74.
4218:
4177:
4150:
4109:
4098:(10): 1931–7.
4082:
4035:
4014:(4): 484–489.
3989:
3954:
3895:
3854:
3825:(8): 2412–25.
3802:
3781:(6): 1221–31.
3761:
3720:
3669:
3618:
3583:
3548:Genes to Cells
3534:
3507:(3): 269–277.
3491:
3429:
3378:
3329:
3285:
3244:
3203:
3154:
3105:
3056:
2990:
2941:
2912:(4): 665–679.
2892:
2841:
2785:
2743:
2694:
2667:(2): 367–374.
2651:
2602:
2567:The Plant Cell
2553:
2494:
2443:
2414:(3): 637–642.
2394:
2365:(5): 416–419.
2345:
2288:
2274:
2247:(5): 269–282.
2231:
2202:(5): 645–654.
2179:
2160:(3): 195–199.
2144:
2115:(4): 665–679.
2095:
2077:
2059:
2038:
2016:
2015:
2013:
2010:
2009:
2008:
2003:
1995:
1992:
1925:
1922:
1917:
1916:
1911:
1906:
1901:
1896:
1891:
1886:
1881:
1876:
1871:
1866:
1861:
1856:
1851:
1846:
1841:
1836:
1831:
1826:
1821:
1816:
1811:
1806:
1801:
1796:
1791:
1786:
1781:
1776:
1771:
1766:
1761:
1756:
1751:
1746:
1741:
1736:
1731:
1726:
1721:
1715:
1706:
1703:
1701:
1700:
1695:
1690:
1685:
1680:
1675:
1669:
1663:
1658:
1652:
1647:
1639:
1634:
1629:
1624:
1618:
1611:
1606:
1601:
1596:
1591:
1586:
1580:
1575:
1569:
1564:
1558:
1553:
1550:
1518:DNA polymerase
1513:
1510:
1483:DNA polymerase
1458:
1455:
1434:
1431:
1379:
1378:
1375:
1374:
1369:
1359:
1358:
1352:
1351:
1348:
1347:
1345:
1343:
1336:
1335:
1328:
1321:
1317:
1316:
1313:
1312:
1302:
1301:
1295:
1292:
1291:
1277:
1276:
1270:
1266:
1265:
1262:
1261:
1251:
1250:
1244:
1241:
1240:
1226:
1225:
1219:
1215:
1214:
1211:
1210:
1202:
1201:
1195:
1192:
1191:
1183:
1182:
1176:
1170:
1169:
1166:
1165:
1157:
1156:
1150:
1147:
1146:
1138:
1137:
1131:
1125:
1124:
1121:
1120:
1112:
1111:
1105:
1102:
1101:
1093:
1092:
1086:
1080:
1079:
1074:
1069:
1065:
1064:
1054:
1053:
1050:
1049:
1038:
1037:
1035:
1034:
1029:
1024:
1019:
1014:
1009:
1004:
999:
994:
989:
984:
979:
974:
969:
964:
959:
954:
949:
944:
939:
934:
929:
924:
919:
914:
909:
904:
899:
894:
889:
884:
879:
874:
869:
864:
859:
854:
849:
844:
838:
836:
832:
831:
829:
828:
823:
818:
813:
808:
803:
798:
793:
788:
783:
778:
773:
767:
765:
761:
760:
758:
757:
752:
747:
742:
737:
732:
727:
722:
717:
712:
707:
702:
700:enzyme binding
697:
692:
686:
684:
680:
679:
669:
668:
665:
664:
661:
660:
652:
651:
640:
634:
633:
630:
629:
621:
620:
617:
616:
614:
613:
609:
605:
601:
597:
593:
589:
585:
581:
580:tail of embryo
577:
573:
570:
569:
558:
555:
554:
552:
551:
547:
543:
539:
535:
531:
527:
523:
519:
515:
511:
508:
507:
495:
494:
486:
475:
469:
468:
465:RNA expression
457:
456:
453:
452:
444:
440:
439:
431:
428:
423:
417:
416:
407:
400:
394:
390:
389:
384:
378:
377:
369:
368:
356:
355:
352:
351:
343:
339:
338:
330:
327:
322:
316:
315:
306:
299:
293:
289:
288:
281:
275:
274:
266:
265:
253:
252:
209:
205:
204:
196:
188:
187:
183:
182:
179:
178:
175:
174:
80:
79:
71:
70:
59:
53:
52:
44:
43:
35:
34:
26:
9:
6:
4:
3:
2:
8848:
8837:
8834:
8832:
8829:
8827:
8824:
8822:
8819:
8817:
8814:
8813:
8811:
8794:
8791:
8789:
8786:
8784:
8781:
8780:
8779:
8775:
8772:
8771:
8769:
8767:
8763:
8753:
8750:
8748:
8744:
8743:
8741:
8737:
8731:
8728:
8727:
8722:
8719:
8718:
8717:
8714:
8713:
8708:
8705:
8703:
8700:
8698:
8695:
8693:
8690:
8689:
8688:
8685:
8681:
8678:
8676:
8673:
8671:
8668:
8666:
8663:
8662:
8661:
8658:
8654:
8651:
8649:
8646:
8644:
8641:
8639:
8636:
8635:
8634:
8631:
8628:
8625:
8624:
8619:
8616:
8615:
8614:
8611:
8609:
8608:Topoisomerase
8606:
8602:
8599:
8598:
8597:
8594:
8590:
8587:
8586:
8585:
8582:
8581:
8579:
8576:
8567:
8559:
8556:
8555:
8554:
8550:
8547:
8546:
8541:
8538:
8537:
8536:
8535:Topoisomerase
8533:
8531:
8528:
8526:
8523:
8521:
8518:
8514:
8511:
8509:
8506:
8504:
8501:
8499:
8496:
8494:
8491:
8489:
8486:
8484:
8481:
8479:
8476:
8474:
8471:
8469:
8466:
8464:
8461:
8459:
8456:
8455:
8454:
8451:
8450:
8448:
8445:
8438:
8435:
8433:
8429:
8419:
8416:
8414:
8411:
8407:
8403:
8400:
8399:
8398:
8395:
8394:
8391:
8387:
8383:
8380:
8379:
8377:
8373:
8367:
8364:
8362:
8358:
8355:
8354:
8351:
8347:
8344:
8343:
8338:
8335:
8333:
8330:
8329:
8328:
8325:
8324:
8319:
8316:
8314:
8311:
8309:
8306:
8305:
8304:
8301:
8300:
8297:
8294:
8293:
8290:
8287:
8286:
8281:
8278:
8276:
8273:
8271:
8268:
8266:
8263:
8261:
8258:
8256:
8253:
8252:
8251:
8248:
8247:
8244:
8241:
8240:
8237:
8234:
8233:
8228:
8225:
8223:
8220:
8218:
8215:
8213:
8210:
8208:
8205:
8203:
8200:
8199:
8198:
8195:
8194:
8191:
8188:
8187:
8185:
8182:
8173:
8165:
8162:
8161:
8160:
8157:
8156:
8151:
8148:
8146:
8143:
8141:
8138:
8137:
8136:
8133:
8132:
8129:
8126:
8125:
8122:
8119:
8118:
8116:
8113:
8106:
8103:
8101:
8097:
8092:
8088:
8084:
8077:
8072:
8070:
8065:
8063:
8058:
8057:
8054:
8038:
8035:
8034:
8032:
8030:
8026:
8023:
8019:
8009:
8006:
8005:
8003:
8000:
7996:
7992:
7986:
7983:
7982:
7980:
7978:
7974:
7971:
7967:
7957:
7954:
7952:
7949:
7947:
7944:
7942:
7939:
7937:
7934:
7932:
7929:
7928:
7926:
7924:
7923:Breast cancer
7920:
7914:
7911:
7909:
7906:
7905:
7903:
7901:
7897:
7891:
7888:
7887:
7885:
7883:
7879:
7873:
7870:
7868:
7865:
7863:
7860:
7858:
7855:
7853:
7850:
7848:
7845:
7843:
7840:
7838:
7835:
7833:
7830:
7828:
7825:
7824:
7822:
7820:
7816:
7810:
7807:
7805:
7802:
7800:
7796:
7793:
7791:
7788:
7787:
7785:
7783:
7779:
7773:
7772:
7768:
7764:
7761:
7760:
7759:
7758:
7754:
7750:
7747:
7746:
7745:
7744:
7740:
7736:
7733:
7732:
7731:
7730:
7726:
7722:
7719:
7718:
7717:
7716:
7712:
7708:
7705:
7704:
7703:
7702:
7698:
7694:
7691:
7689:
7686:
7685:
7684:
7683:
7679:
7675:
7672:
7671:
7670:
7669:
7665:
7664:
7662:
7660:
7659:Ovarian tumor
7656:
7653:
7647:Reproductive/
7645:
7635:
7631:
7628:
7627:
7625:
7623:
7619:
7613:
7610:
7608:
7605:
7603:
7600:
7598:
7595:
7593:
7590:
7589:
7587:
7585:
7581:
7575:
7572:
7570:
7567:
7565:
7562:
7561:
7559:
7557:
7553:
7550:
7546:
7536:
7533:
7532:
7530:
7527:
7523:
7519:
7513:
7510:
7508:
7505:
7503:
7500:
7498:
7495:
7494:
7492:
7490:
7486:
7483:
7477:
7467:
7464:
7462:
7459:
7458:
7456:
7454:
7450:
7446:
7440:
7437:
7436:
7434:
7432:
7428:
7422:
7419:
7418:
7416:
7414:
7410:
7407:
7403:
7393:
7390:
7389:
7387:
7385:
7384:Neuroblastoma
7381:
7375:
7372:
7370:
7369:Synaptophysin
7367:
7366:
7364:
7362:
7358:
7352:
7349:
7347:
7344:
7343:
7341:
7339:
7335:
7328:
7325:
7323:
7319:
7316:
7314:
7313:Thyroglobulin
7311:
7310:
7308:
7306:
7302:
7299:
7295:
7285:
7282:
7280:
7277:
7276:
7274:
7272:
7271:Lymphosarcoma
7268:
7262:
7259:
7258:
7256:
7254:
7250:
7247:
7243:
7239:
7238:Tumor markers
7232:
7227:
7225:
7220:
7218:
7213:
7212:
7209:
7202:
7198:
7197:
7192:
7188:
7184:
7181:
7178:
7175:
7171:
7168:
7167:
7156:
7152:
7148:
7144:
7140:
7136:
7132:
7128:
7123:
7119:
7115:
7111:
7107:
7102:
7097:
7093:
7089:
7085:
7080:
7076:
7072:
7067:
7062:
7057:
7052:
7048:
7044:
7040:
7035:
7031:
7027:
7022:
7017:
7013:
7009:
7005:
7000:
6996:
6992:
6988:
6984:
6980:
6976:
6972:
6968:
6964:
6959:
6955:
6951:
6947:
6941:
6937:
6933:
6929:
6924:
6920:
6916:
6912:
6908:
6904:
6900:
6896:
6892:
6888:
6884:
6879:
6875:
6871:
6866:
6861:
6856:
6851:
6847:
6843:
6839:
6835:
6831:
6826:
6822:
6818:
6813:
6808:
6804:
6800:
6796:
6791:
6787:
6783:
6778:
6773:
6768:
6763:
6759:
6755:
6751:
6747:
6743:
6738:
6734:
6730:
6725:
6720:
6716:
6712:
6708:
6704:
6700:
6695:
6691:
6687:
6682:
6677:
6673:
6669:
6665:
6660:
6656:
6652:
6647:
6642:
6638:
6634:
6630:
6625:
6621:
6617:
6613:
6609:
6605:
6601:
6597:
6593:
6588:
6576:
6572:
6568:
6567:
6562:
6557:
6553:
6549:
6545:
6541:
6537:
6533:
6528:
6524:
6520:
6516:
6512:
6508:
6504:
6499:
6495:
6491:
6487:
6483:
6478:
6474:
6470:
6465:
6460:
6456:
6452:
6448:
6443:
6439:
6435:
6430:
6425:
6420:
6415:
6411:
6407:
6403:
6399:
6395:
6390:
6386:
6382:
6378:
6374:
6369:
6365:
6361:
6356:
6351:
6346:
6341:
6337:
6333:
6329:
6325:
6321:
6316:
6315:
6301:
6297:
6292:
6287:
6282:
6277:
6273:
6269:
6265:
6258:
6250:
6246:
6241:
6236:
6232:
6228:
6224:
6220:
6216:
6209:
6201:
6197:
6193:
6189:
6185:
6181:
6174:
6166:
6162:
6157:
6152:
6148:
6144:
6140:
6136:
6132:
6125:
6117:
6113:
6108:
6103:
6098:
6093:
6089:
6086:
6085:
6080:
6073:
6065:
6061:
6056:
6051:
6047:
6043:
6039:
6035:
6031:
6024:
6016:
6012:
6007:
6002:
5998:
5994:
5990:
5986:
5982:
5975:
5967:
5963:
5958:
5953:
5948:
5943:
5939:
5935:
5931:
5927:
5923:
5916:
5908:
5904:
5900:
5896:
5888:
5880:
5876:
5871:
5866:
5862:
5858:
5854:
5850:
5846:
5839:
5831:
5827:
5822:
5817:
5813:
5809:
5805:
5801:
5797:
5790:
5782:
5778:
5774:
5770:
5766:
5762:
5758:
5754:
5746:
5738:
5734:
5729:
5724:
5719:
5714:
5710:
5706:
5702:
5695:
5687:
5683:
5678:
5673:
5668:
5663:
5659:
5655:
5651:
5647:
5643:
5636:
5628:
5624:
5619:
5614:
5609:
5604:
5600:
5596:
5592:
5588:
5584:
5577:
5569:
5565:
5560:
5555:
5551:
5547:
5543:
5536:
5528:
5524:
5519:
5514:
5509:
5504:
5500:
5496:
5492:
5488:
5484:
5477:
5469:
5465:
5460:
5455:
5451:
5447:
5443:
5436:
5434:
5432:
5423:
5419:
5415:
5411:
5407:
5403:
5400:(4): 386–92.
5399:
5395:
5387:
5379:
5375:
5370:
5365:
5361:
5357:
5356:J. Biol. Chem
5353:
5346:
5344:
5335:
5331:
5326:
5321:
5317:
5313:
5309:
5305:
5301:
5294:
5286:
5282:
5278:
5274:
5270:
5266:
5263:(7): 639–51.
5262:
5258:
5251:
5243:
5239:
5234:
5229:
5225:
5221:
5220:J. Biol. Chem
5217:
5210:
5202:
5198:
5193:
5188:
5184:
5180:
5177:(3): 784–91.
5176:
5172:
5168:
5161:
5153:
5149:
5144:
5139:
5135:
5131:
5127:
5123:
5119:
5112:
5104:
5100:
5095:
5090:
5086:
5082:
5081:J. Biol. Chem
5078:
5071:
5063:
5059:
5054:
5049:
5045:
5041:
5040:J. Biol. Chem
5037:
5030:
5022:
5018:
5013:
5008:
5004:
5000:
4999:J. Biol. Chem
4996:
4989:
4981:
4977:
4972:
4967:
4963:
4959:
4955:
4951:
4947:
4940:
4932:
4928:
4924:
4920:
4915:
4910:
4906:
4902:
4898:
4891:
4883:
4879:
4874:
4869:
4865:
4861:
4860:J. Biol. Chem
4857:
4850:
4842:
4838:
4834:
4830:
4826:
4822:
4818:
4814:
4810:
4806:
4798:
4790:
4786:
4781:
4776:
4772:
4768:
4764:
4760:
4756:
4749:
4747:
4738:
4734:
4729:
4724:
4720:
4716:
4715:J. Biol. Chem
4712:
4705:
4697:
4693:
4688:
4683:
4679:
4675:
4674:J. Biol. Chem
4671:
4664:
4662:
4653:
4649:
4644:
4639:
4635:
4631:
4628:(6): 724–36.
4627:
4623:
4619:
4612:
4610:
4601:
4597:
4592:
4587:
4583:
4579:
4578:J. Biol. Chem
4575:
4568:
4560:
4556:
4552:
4548:
4544:
4540:
4532:
4524:
4520:
4515:
4510:
4506:
4502:
4498:
4494:
4490:
4483:
4481:
4472:
4468:
4463:
4458:
4453:
4448:
4444:
4440:
4436:
4432:
4428:
4421:
4413:
4409:
4405:
4401:
4397:
4393:
4386:
4378:
4374:
4369:
4364:
4360:
4356:
4352:
4345:
4337:
4333:
4328:
4323:
4319:
4315:
4314:J. Biol. Chem
4311:
4304:
4296:
4292:
4287:
4282:
4278:
4274:
4273:J. Biol. Chem
4270:
4263:
4255:
4251:
4246:
4241:
4237:
4233:
4232:J. Biol. Chem
4229:
4222:
4214:
4210:
4205:
4200:
4196:
4192:
4191:J. Biol. Chem
4188:
4181:
4173:
4169:
4165:
4161:
4154:
4146:
4142:
4137:
4132:
4128:
4124:
4123:J. Biol. Chem
4120:
4113:
4105:
4101:
4097:
4093:
4086:
4078:
4074:
4070:
4066:
4062:
4058:
4054:
4050:
4046:
4039:
4031:
4027:
4022:
4017:
4013:
4009:
4005:
3998:
3996:
3994:
3985:
3981:
3977:
3973:
3969:
3965:
3958:
3950:
3946:
3941:
3936:
3931:
3926:
3922:
3918:
3914:
3910:
3906:
3899:
3891:
3887:
3882:
3877:
3873:
3869:
3868:J. Biol. Chem
3865:
3858:
3850:
3846:
3841:
3836:
3832:
3828:
3824:
3820:
3816:
3809:
3807:
3798:
3794:
3789:
3784:
3780:
3776:
3772:
3765:
3757:
3753:
3748:
3743:
3739:
3735:
3731:
3724:
3716:
3712:
3707:
3702:
3697:
3692:
3688:
3684:
3680:
3673:
3665:
3661:
3657:
3653:
3649:
3645:
3641:
3637:
3633:
3629:
3622:
3614:
3610:
3606:
3602:
3598:
3594:
3587:
3579:
3575:
3571:
3567:
3562:
3557:
3553:
3549:
3545:
3538:
3530:
3526:
3522:
3518:
3514:
3513:10.1038/77023
3510:
3506:
3502:
3495:
3487:
3483:
3478:
3473:
3468:
3463:
3459:
3455:
3451:
3447:
3443:
3436:
3434:
3425:
3421:
3417:
3413:
3409:
3405:
3401:
3397:
3393:
3389:
3382:
3374:
3370:
3365:
3360:
3356:
3352:
3348:
3344:
3340:
3333:
3325:
3321:
3316:
3311:
3307:
3303:
3299:
3292:
3290:
3281:
3277:
3272:
3267:
3263:
3259:
3255:
3248:
3240:
3236:
3231:
3226:
3222:
3218:
3214:
3207:
3199:
3195:
3190:
3185:
3181:
3177:
3173:
3169:
3165:
3158:
3150:
3146:
3141:
3136:
3132:
3128:
3124:
3120:
3116:
3109:
3101:
3097:
3092:
3087:
3083:
3079:
3075:
3071:
3067:
3060:
3052:
3048:
3043:
3038:
3034:
3030:
3029:J. Biol. Chem
3026:
3019:
3017:
3015:
3013:
3011:
3009:
3007:
3005:
3003:
3001:
2999:
2997:
2995:
2986:
2982:
2977:
2972:
2968:
2964:
2960:
2956:
2952:
2945:
2937:
2933:
2929:
2925:
2920:
2915:
2911:
2907:
2903:
2896:
2888:
2884:
2880:
2876:
2872:
2868:
2864:
2860:
2856:
2852:
2845:
2837:
2833:
2829:
2825:
2821:
2817:
2813:
2809:
2805:
2801:
2794:
2792:
2790:
2781:
2777:
2773:
2769:
2765:
2761:
2754:
2747:
2739:
2735:
2730:
2725:
2721:
2717:
2713:
2709:
2705:
2698:
2690:
2686:
2682:
2678:
2674:
2670:
2666:
2662:
2655:
2647:
2643:
2638:
2633:
2629:
2625:
2621:
2617:
2613:
2606:
2598:
2594:
2589:
2584:
2580:
2576:
2572:
2568:
2564:
2557:
2549:
2545:
2540:
2535:
2530:
2525:
2521:
2517:
2513:
2509:
2505:
2498:
2490:
2486:
2482:
2478:
2474:
2470:
2466:
2462:
2458:
2454:
2447:
2439:
2435:
2430:
2425:
2421:
2417:
2413:
2409:
2405:
2398:
2390:
2386:
2381:
2376:
2372:
2368:
2364:
2360:
2356:
2349:
2341:
2337:
2332:
2327:
2323:
2319:
2315:
2311:
2307:
2303:
2299:
2292:
2284:
2278:
2270:
2266:
2262:
2258:
2254:
2250:
2246:
2242:
2235:
2227:
2223:
2218:
2213:
2209:
2205:
2201:
2197:
2193:
2186:
2184:
2175:
2171:
2167:
2163:
2159:
2155:
2148:
2140:
2136:
2132:
2128:
2123:
2118:
2114:
2110:
2106:
2099:
2091:
2087:
2081:
2073:
2069:
2063:
2056:
2052:
2047:
2045:
2043:
2035:
2031:
2026:
2024:
2022:
2017:
2007:
2006:Transcription
2004:
2001:
1998:
1997:
1991:
1989:
1985:
1981:
1976:
1974:
1968:
1966:
1962:
1958:
1954:
1950:
1946:
1943:of different
1942:
1938:
1934:
1930:
1921:
1915:
1912:
1910:
1907:
1905:
1902:
1900:
1897:
1895:
1892:
1890:
1887:
1885:
1882:
1880:
1877:
1875:
1872:
1870:
1867:
1865:
1862:
1860:
1857:
1855:
1852:
1850:
1847:
1845:
1842:
1840:
1837:
1835:
1832:
1830:
1827:
1825:
1822:
1820:
1817:
1815:
1812:
1810:
1807:
1805:
1802:
1800:
1797:
1795:
1792:
1790:
1787:
1785:
1782:
1780:
1777:
1775:
1772:
1770:
1767:
1765:
1762:
1760:
1757:
1755:
1752:
1750:
1747:
1745:
1742:
1740:
1737:
1735:
1732:
1730:
1727:
1725:
1722:
1720:
1717:
1716:
1714:
1712:
1699:
1698:Topoisomerase
1696:
1694:
1691:
1689:
1686:
1684:
1681:
1679:
1676:
1673:
1670:
1667:
1664:
1662:
1659:
1656:
1653:
1651:
1648:
1646:
1643:
1640:
1638:
1635:
1633:
1630:
1628:
1625:
1623:
1619:
1616:
1612:
1610:
1607:
1605:
1602:
1600:
1597:
1595:
1592:
1590:
1587:
1584:
1581:
1579:
1576:
1573:
1570:
1568:
1565:
1563:
1560:
1559:
1557:
1549:
1547:
1543:
1539:
1535:
1530:
1525:
1523:
1519:
1509:
1507:
1504:
1500:
1496:
1492:
1488:
1484:
1480:
1476:
1472:
1468:
1464:
1454:
1452:
1447:
1445:
1441:
1440:ubiquitinated
1430:
1426:
1424:
1420:
1416:
1413:
1409:
1405:
1401:
1397:
1393:
1385:
1373:
1368:
1364:
1360:
1357:
1353:
1346:
1344:
1341:
1337:
1333:
1329:
1326:
1322:
1318:
1311:
1309:
1303:
1299:
1296:
1290:
1288:
1284:
1278:
1274:
1271:
1267:
1260:
1258:
1252:
1248:
1245:
1239:
1237:
1233:
1227:
1223:
1220:
1218:RefSeq (mRNA)
1216:
1209:
1208:
1203:
1199:
1196:
1190:
1189:
1184:
1180:
1177:
1175:
1171:
1164:
1163:
1158:
1154:
1151:
1145:
1144:
1139:
1135:
1132:
1130:
1126:
1119:
1118:
1113:
1109:
1106:
1100:
1099:
1094:
1090:
1087:
1085:
1081:
1078:
1075:
1073:
1070:
1066:
1063:
1059:
1055:
1048:
1044:
1039:
1033:
1032:viral process
1030:
1028:
1025:
1023:
1020:
1018:
1015:
1013:
1010:
1008:
1005:
1003:
1000:
998:
995:
993:
990:
988:
985:
983:
980:
978:
975:
973:
970:
968:
965:
963:
962:estrous cycle
960:
958:
955:
953:
950:
948:
945:
943:
940:
938:
935:
933:
930:
928:
925:
923:
920:
918:
915:
913:
910:
908:
905:
903:
900:
898:
895:
893:
890:
888:
885:
883:
880:
878:
875:
873:
870:
868:
865:
863:
860:
858:
855:
853:
850:
848:
845:
843:
840:
839:
837:
834:
833:
827:
824:
822:
819:
817:
814:
812:
809:
807:
804:
802:
799:
797:
794:
792:
789:
787:
784:
782:
779:
777:
774:
772:
769:
768:
766:
763:
762:
756:
753:
751:
748:
746:
743:
741:
738:
736:
733:
731:
728:
726:
723:
721:
718:
716:
713:
711:
708:
706:
703:
701:
698:
696:
693:
691:
688:
687:
685:
682:
681:
678:
677:Gene ontology
674:
670:
658:
653:
649:
644:
641:
639:
635:
627:
622:
611:
607:
603:
599:
595:
591:
587:
583:
579:
575:
574:
571:
567:
562:
559:
549:
545:
541:
537:
533:
529:
525:
521:
517:
513:
512:
509:
505:
500:
497:
496:
493:
491:
487:
485:
484:
480:
479:
476:
474:
470:
466:
462:
458:
450:
445:
441:
437:
432:
422:
418:
411:
404:
398:
391:
383:
379:
375:
370:
366:
361:
357:
349:
344:
340:
336:
331:
321:
317:
310:
303:
297:
290:
286:
280:
276:
272:
267:
263:
258:
254:
250:
246:
242:
238:
234:
230:
226:
222:
218:
214:
206:
201:
194:
189:
184:
173:
171:
167:
163:
159:
155:
151:
147:
143:
139:
135:
131:
127:
123:
119:
115:
111:
107:
103:
99:
95:
91:
87:
81:
76:
73:
72:
68:
65:
58:
54:
49:
45:
41:
36:
31:
19:
8747:Processivity
8720:
8573:synthesis in
7769:
7755:
7749:Testosterone
7741:
7729:Dysgerminoma
7727:
7713:
7699:
7680:
7666:
7461:S100 protein
7420:
7194:
7133:(1): 84–86.
7130:
7126:
7091:
7087:
7066:11380/811513
7046:
7042:
7011:
7007:
6970:
6966:
6927:
6886:
6882:
6837:
6833:
6802:
6798:
6749:
6745:
6706:
6702:
6671:
6667:
6636:
6632:
6595:
6591:
6579:. Retrieved
6575:the original
6564:
6535:
6531:
6506:
6502:
6485:
6481:
6454:
6450:
6401:
6397:
6376:
6372:
6327:
6323:
6271:
6267:
6257:
6222:
6218:
6208:
6183:
6179:
6173:
6138:
6134:
6124:
6090:(180): 180.
6087:
6082:
6072:
6037:
6033:
6023:
5988:
5984:
5974:
5929:
5925:
5915:
5898:
5894:
5887:
5852:
5848:
5838:
5803:
5799:
5789:
5756:
5752:
5745:
5708:
5704:
5694:
5649:
5645:
5635:
5590:
5586:
5576:
5549:
5545:
5535:
5490:
5486:
5476:
5449:
5445:
5397:
5393:
5386:
5359:
5355:
5307:
5303:
5293:
5260:
5256:
5250:
5223:
5219:
5209:
5174:
5170:
5160:
5125:
5121:
5111:
5084:
5080:
5070:
5043:
5039:
5029:
5002:
4998:
4988:
4953:
4949:
4939:
4904:
4900:
4890:
4863:
4859:
4849:
4808:
4804:
4797:
4762:
4758:
4718:
4714:
4704:
4677:
4673:
4625:
4621:
4581:
4577:
4567:
4542:
4538:
4531:
4496:
4492:
4434:
4430:
4420:
4395:
4391:
4385:
4358:
4354:
4344:
4317:
4313:
4303:
4276:
4272:
4262:
4235:
4231:
4221:
4194:
4190:
4180:
4163:
4159:
4153:
4126:
4122:
4112:
4095:
4091:
4085:
4052:
4048:
4038:
4011:
4007:
3967:
3964:Biochemistry
3963:
3957:
3912:
3908:
3898:
3871:
3867:
3857:
3822:
3818:
3778:
3774:
3764:
3737:
3733:
3723:
3686:
3682:
3672:
3631:
3627:
3621:
3596:
3592:
3586:
3551:
3547:
3537:
3504:
3500:
3494:
3449:
3445:
3391:
3387:
3381:
3346:
3342:
3332:
3305:
3301:
3261:
3257:
3247:
3220:
3216:
3206:
3171:
3167:
3157:
3122:
3118:
3108:
3073:
3069:
3059:
3032:
3028:
2958:
2954:
2944:
2909:
2905:
2895:
2854:
2850:
2844:
2803:
2799:
2763:
2759:
2746:
2711:
2707:
2697:
2664:
2660:
2654:
2619:
2615:
2605:
2570:
2566:
2556:
2511:
2507:
2497:
2456:
2452:
2446:
2411:
2407:
2397:
2362:
2358:
2348:
2305:
2301:
2291:
2277:
2244:
2240:
2234:
2199:
2195:
2157:
2153:
2147:
2112:
2108:
2098:
2089:
2080:
2071:
2062:
1977:
1972:
1969:
1927:
1919:
1708:
1705:Interactions
1589:Clamp loader
1555:
1541:
1526:
1515:
1486:
1460:
1448:
1436:
1427:
1404:processivity
1395:
1391:
1390:
1305:
1280:
1254:
1229:
1205:
1186:
1160:
1141:
1115:
1096:
1076:
1071:
1012:DNA ligation
821:nuclear body
801:PCNA complex
488:
481:
447:132,095,234
434:132,091,082
208:External IDs
83:
8766:Termination
8440:Prokaryotic
8432:Replication
8108:Prokaryotic
8087:prokaryotic
8085:(comparing
8008:Cytokeratin
7946:Cathepsin D
7535:Factor VIII
7526:endothelium
7489:Lung cancer
7481:respiratory
7431:Astrocytoma
7413:Brain tumor
6840:(1): 6–10.
6709:(1): 1–12.
5493:(1): 6–10.
5394:J. Mol. Med
5257:Genes Cells
4392:J. Cell Sci
2308:(1): 1591.
1949:astrocytoma
1899:Ubiquitin C
1683:Procaspases
1444:Pseudogenes
1423:epigenetics
1406:factor for
781:nucleoplasm
710:DNA binding
600:bone marrow
186:Identifiers
8831:DNA repair
8810:Categories
8778:Telomerase
8752:DNA ligase
8745:Movement:
8569:Eukaryotic
8540:DNA gyrase
8525:DNA ligase
8444:elongation
8175:Eukaryotic
8112:initiation
8100:Initiation
8091:eukaryotic
7999:epithelium
7322:Calcitonin
7261:Neprilysin
2760:DNA Repair
2057:, May 2017
2036:, May 2017
2012:References
1978:caPCNA, a
1965:antibodies
1957:prognostic
1953:diagnostic
1929:Antibodies
1719:Annexin A2
1599:DNA ligase
1522:DNA repair
1475:cell cycle
1419:homotrimer
1412:eukaryotic
997:DNA repair
796:centrosome
492:(ortholog)
346:5,126,626
333:5,114,953
229:HomoloGene
8716:DNA clamp
8530:DNA clamp
8520:Replisome
7995:Carcinoma
7548:Digestive
7502:Enolase 2
7351:Enolase 2
7297:Endocrine
4622:Genes Dev
3775:Mol. Cell
2836:205209495
2154:BioEssays
1945:neoplasms
1739:Cyclin D1
1645:chaperone
1632:Helicases
1534:ubiquitin
1400:DNA clamp
1308:NP_035175
1287:NP_872590
1283:NP_002583
1257:NM_011045
1236:NM_002592
1232:NM_182649
1062:Orthologs
826:chromatin
776:replisome
237:GeneCards
8836:Proteins
8774:Telomere
8390:Replicon
8346:Helicase
8337:RNASEH2A
8181:G1 phase
8135:Helicase
7985:Vimentin
7951:CA 27-29
7649:urinary/
7453:Melanoma
7253:Lymphoma
7155:27107553
6786:16169902
6733:10408173
6620:27217843
6581:3 August
6482:Oncogene
6373:Oncogene
6300:37531956
6291:10592352
6249:29967249
6200:24655521
6165:25484186
6116:26022740
6064:23677613
6015:22704558
5966:23152873
5926:PLOS ONE
5879:15107487
5830:10871373
5781:37287691
5773:12633936
5737:18284681
5686:18316726
5627:18719106
5568:12766176
5422:22183443
5414:10353443
5334:12192049
5285:29714829
5277:12081642
5242:12368291
5201:11784855
5152:11585903
5103:12522211
5062:11595739
5021:11986310
4980:11350925
4931:17461501
4882:12930846
4833:16189514
4737:11092888
4696:11005803
4652:11274057
4600:10978339
4559:12393188
4523:11239001
4471:11593007
4412:11682605
4377:11077446
4355:Oncogene
4336:11929879
4295:10455148
4254:11022036
4213:10973963
4160:Oncogene
4145:10828065
4092:Oncogene
4030:11313979
4008:Oncogene
3984:11601988
3797:11430825
3756:12853968
3734:Oncogene
3715:31879348
3656:11268218
3578:25310911
3570:12354094
3529:26149386
3521:10888872
3373:10393198
3239:11896603
3217:Oncogene
3198:19822659
3149:10648606
3100:26908650
3051:12171929
2985:20975039
2928:17512402
2879:15931174
2828:12226657
2780:16956796
2738:16227586
2689:12260457
2646:26833126
2597:12468739
2548:10051561
2481:15201901
2340:32221299
2269:25952152
2261:23594953
2226:19736315
2131:17512402
2053:–
2032:–
1994:See also
1809:KIAA0101
1744:Cyclin O
1711:interact
1668:receptor
1542:inhibits
1433:Function
1356:Wikidata
1041:Sources:
596:yolk sac
592:epiblast
550:appendix
8687:epsilon
8575:S phase
8402:Lagging
8357:Primase
8332:RNASEH1
8327:RNase H
8159:Primase
7977:Sarcoma
7931:CA 15-3
7867:TMPRSS2
7763:Inhibin
7201:PDBe-KB
7191:UniProt
7147:1979311
7118:1559243
7110:7780738
7075:2565339
7030:7949095
6995:7973727
6975:Bibcode
6967:Science
6954:9552415
6919:4336365
6911:2882422
6891:Bibcode
6874:8093561
6842:Bibcode
6821:1975809
6777:1236569
6754:Bibcode
6711:Bibcode
6690:7908906
6655:7673186
6612:2569765
6566:ALSNews
6552:1477073
6523:7821789
6494:7784094
6473:7673244
6438:1313560
6406:Bibcode
6385:7478510
6364:2882507
6332:Bibcode
6240:6279569
6156:4615156
6107:4448323
6055:3711418
6006:3613862
5957:3496702
5934:Bibcode
5907:9927044
5728:2263069
5677:2268824
5654:Bibcode
5618:2518831
5595:Bibcode
5527:8093561
5495:Bibcode
5468:9228079
5378:8999859
4923:8861913
4841:4427026
4813:Bibcode
4789:8861969
4439:Bibcode
4172:7784094
4104:7478510
4077:7973727
4057:Bibcode
4049:Science
3949:8876181
3917:Bibcode
3890:9305916
3849:9545252
3840:1170584
3706:6969535
3664:2129847
3636:Bibcode
3613:9302295
3593:Science
3486:9465025
3454:Bibcode
3424:4368128
3416:8259215
3396:Bibcode
3364:1171460
3324:8101826
3280:7908906
3189:2786881
3091:4914081
2976:2989777
2936:3547069
2887:4316517
2859:Bibcode
2808:Bibcode
2729:1265825
2681:1348971
2637:4794399
2516:Bibcode
2489:4346799
2461:Bibcode
2438:2884104
2389:1350788
2331:7101311
2310:Bibcode
2217:2742182
2174:9631646
2139:3547069
2055:Ensembl
2034:Ensembl
1988:AOH1996
1973:in situ
1961:S phase
1947:, e.g.
1941:grading
1935:termed
1859:POLDIP2
1784:GADD45G
1779:GADD45A
1657:enzymes
1642:Histone
1594:Cohesin
1574:enzymes
1567:ATPases
1485:delta (
1473:of the
1463:antigen
1398:) is a
1174:UniProt
1129:Ensembl
1068:Species
1047:QuickGO
771:nucleus
467:pattern
325:20p12.3
193:Aliases
8418:Primer
8037:Desmin
7852:SPINK1
7799:AFP-L3
7674:CA-125
7651:breast
7634:AFP-L3
7602:CA 242
7592:CA19-9
7564:CA19-9
7196:P12004
7176:(MeSH)
7153:
7145:
7116:
7108:
7073:
7028:
6993:
6952:
6942:
6917:
6909:
6883:Nature
6872:
6862:
6819:
6784:
6774:
6731:
6688:
6653:
6618:
6610:
6550:
6521:
6492:
6471:
6436:
6426:
6383:
6362:
6355:304478
6352:
6298:
6288:
6247:
6237:
6198:
6163:
6153:
6114:
6104:
6062:
6052:
6013:
6003:
5964:
5954:
5905:
5877:
5870:407833
5867:
5828:
5821:102739
5818:
5779:
5771:
5735:
5725:
5711:: 24.
5684:
5674:
5625:
5615:
5566:
5525:
5515:
5466:
5420:
5412:
5376:
5332:
5325:135621
5322:
5283:
5275:
5240:
5199:
5192:133560
5189:
5150:
5140:
5101:
5060:
5019:
4978:
4971:125454
4968:
4950:EMBO J
4929:
4921:
4880:
4839:
4831:
4805:Nature
4787:
4780:452166
4777:
4759:EMBO J
4735:
4694:
4650:
4643:312660
4640:
4598:
4557:
4521:
4511:
4469:
4459:
4410:
4375:
4334:
4293:
4252:
4211:
4170:
4143:
4102:
4075:
4028:
3982:
3947:
3937:
3888:
3847:
3837:
3819:EMBO J
3795:
3754:
3713:
3703:
3662:
3654:
3628:Nature
3611:
3576:
3568:
3527:
3519:
3484:
3474:
3422:
3414:
3388:Nature
3371:
3361:
3322:
3278:
3237:
3196:
3186:
3147:
3137:
3098:
3088:
3049:
2983:
2973:
2934:
2926:
2885:
2877:
2851:Nature
2834:
2826:
2800:Nature
2778:
2736:
2726:
2687:
2679:
2644:
2634:
2595:
2588:151214
2585:
2546:
2536:
2487:
2479:
2453:Nature
2436:
2429:553445
2426:
2387:
2380:495304
2377:
2338:
2328:
2267:
2259:
2224:
2214:
2172:
2137:
2129:
1804:KCTD13
1734:CHTF18
1729:CDC25C
1713:with:
1674:enzyme
1585:factor
1516:Since
1467:nuclei
1342:search
1340:PubMed
1207:P17918
1188:P12004
1084:Entrez
638:BioGPS
612:zygote
608:embryo
604:embryo
576:thymus
542:rectum
514:oocyte
217:176740
8707:POLE4
8702:POLE3
8697:POLE2
8680:POLD4
8675:POLD3
8670:POLD2
8665:POLD1
8660:delta
8653:PRIM2
8648:PRIM1
8643:POLA2
8638:POLA1
8633:alpha
8366:PRIM2
8361:PRIM1
8318:SSBP4
8313:SSBP3
8308:SSBP2
7956:hPG80
7913:NMP22
7890:NANOG
7872:hPG80
7857:GOLM1
7809:hPG80
7771:hPG80
7612:hPG80
7574:hPG80
7512:hPG80
7284:CD79a
7245:Blood
7151:S2CID
7114:S2CID
7008:Blood
6915:S2CID
6865:45588
6616:S2CID
6429:48692
5777:S2CID
5518:45588
5418:S2CID
5281:S2CID
5143:99895
4927:S2CID
4837:S2CID
4514:29758
4462:59753
3940:38103
3660:S2CID
3574:S2CID
3525:S2CID
3477:19016
3420:S2CID
3140:85246
2932:S2CID
2883:S2CID
2832:S2CID
2756:(PDF)
2685:S2CID
2539:26703
2485:S2CID
2265:S2CID
2135:S2CID
2000:Ki-67
1937:Ki-67
1909:XRCC1
1854:POLD3
1849:POLD2
1839:MUTYH
1789:HDAC1
1764:EP300
1759:DNMT1
1724:CAF-1
1666:NKp44
1546:Rad51
1487:Pol δ
1415:cells
1117:18538
1077:Mouse
1072:Human
1043:Amigo
534:gonad
490:Mouse
483:Human
430:Start
365:Mouse
329:Start
262:Human
225:97503
8793:DKC1
8788:TERC
8783:TERT
8739:Both
8721:PCNA
8692:POLE
8618:RPA1
8601:FEN1
8589:RFC1
8513:holE
8508:holD
8503:holC
8498:holB
8493:holA
8488:dnaX
8483:dnaT
8478:dnaQ
8473:dnaN
8468:dnaH
8463:dnaE
8458:dnaC
8404:and
8375:Both
8350:HFM1
8280:MCM7
8275:MCM6
8270:MCM5
8265:MCM4
8260:MCM3
8255:MCM2
8243:Cdt1
8236:Cdc6
8227:ORC6
8222:ORC5
8217:ORC4
8212:ORC3
8207:ORC2
8202:ORC1
8164:dnaG
8145:dnaB
8140:dnaA
8128:dnaC
7862:PCA3
7804:CD30
7790:βhCG
7688:CD30
7421:PCNA
7189:for
7170:PCNA
7143:PMID
7106:PMID
7071:PMID
7026:PMID
6991:PMID
6950:PMID
6940:ISBN
6907:PMID
6870:PMID
6817:PMID
6782:PMID
6729:PMID
6686:PMID
6651:PMID
6608:PMID
6583:2023
6548:PMID
6519:PMID
6503:Gene
6490:PMID
6469:PMID
6434:PMID
6381:PMID
6360:PMID
6296:PMID
6245:PMID
6196:PMID
6161:PMID
6112:PMID
6060:PMID
6011:PMID
5962:PMID
5903:PMID
5875:PMID
5826:PMID
5769:PMID
5733:PMID
5682:PMID
5623:PMID
5564:PMID
5523:PMID
5464:PMID
5410:PMID
5374:PMID
5330:PMID
5273:PMID
5238:PMID
5197:PMID
5148:PMID
5099:PMID
5058:PMID
5017:PMID
4976:PMID
4919:PMID
4901:Cell
4878:PMID
4829:PMID
4785:PMID
4733:PMID
4692:PMID
4648:PMID
4596:PMID
4555:PMID
4543:1578
4519:PMID
4467:PMID
4408:PMID
4373:PMID
4332:PMID
4291:PMID
4250:PMID
4209:PMID
4168:PMID
4141:PMID
4100:PMID
4073:PMID
4026:PMID
3980:PMID
3945:PMID
3886:PMID
3845:PMID
3793:PMID
3752:PMID
3711:PMID
3652:PMID
3609:PMID
3566:PMID
3517:PMID
3482:PMID
3412:PMID
3369:PMID
3320:PMID
3276:PMID
3235:PMID
3194:PMID
3145:PMID
3096:PMID
3047:PMID
2981:PMID
2924:PMID
2906:Cell
2875:PMID
2824:PMID
2776:PMID
2734:PMID
2677:PMID
2661:Cell
2642:PMID
2593:PMID
2544:PMID
2477:PMID
2434:PMID
2385:PMID
2336:PMID
2257:PMID
2222:PMID
2170:PMID
2127:PMID
2109:Cell
1955:and
1924:Uses
1894:RFC5
1889:RFC4
1884:RFC3
1879:RFC2
1874:RFC1
1869:POLL
1864:POLH
1834:MSH6
1829:MSH3
1824:MCL1
1819:Ku80
1814:Ku70
1799:ING1
1794:HUS1
1615:SUMO
1499:AAA+
1479:cDNA
1451:polD
1396:PCNA
1098:5111
473:Bgee
421:Band
382:Chr.
320:Band
279:Chr.
241:PCNA
233:1945
213:OMIM
200:PCNA
170:5E0V
166:5E0T
162:5E0U
158:5IY4
154:4ZTD
150:3JA9
146:4RJF
142:3WGW
138:3VKX
134:3TBL
130:3P87
126:2ZVM
122:2ZVL
118:2ZVK
114:1W60
110:1VYM
106:1VYJ
102:1UL1
98:1U7B
94:1U76
90:1AXC
86:4D2G
67:RCSB
64:PDBe
33:PCNA
18:PCNA
8386:Ori
8089:to
7757:GCT
7735:LDH
7721:hCG
7701:EST
7693:hCG
7279:CD3
7187:PDB
7135:doi
7096:doi
7061:hdl
7051:doi
7047:264
7016:doi
6983:doi
6971:266
6932:doi
6899:doi
6887:326
6860:PMC
6850:doi
6807:doi
6803:265
6772:PMC
6762:doi
6750:102
6719:doi
6676:doi
6672:269
6641:doi
6637:270
6600:doi
6540:doi
6511:doi
6507:150
6459:doi
6455:270
6424:PMC
6414:doi
6350:PMC
6340:doi
6286:PMC
6276:doi
6235:PMC
6227:doi
6188:doi
6151:PMC
6143:doi
6102:PMC
6092:doi
6050:PMC
6042:doi
6001:PMC
5993:doi
5952:PMC
5942:doi
5865:PMC
5857:doi
5816:PMC
5808:doi
5761:doi
5757:124
5723:PMC
5713:doi
5672:PMC
5662:doi
5650:105
5613:PMC
5603:doi
5591:105
5554:doi
5550:278
5513:PMC
5503:doi
5454:doi
5450:272
5402:doi
5364:doi
5360:272
5320:PMC
5312:doi
5265:doi
5228:doi
5224:277
5187:PMC
5179:doi
5138:PMC
5130:doi
5089:doi
5085:278
5048:doi
5044:276
5007:doi
5003:277
4966:PMC
4958:doi
4909:doi
4868:doi
4864:278
4821:doi
4809:437
4775:PMC
4767:doi
4723:doi
4719:276
4682:doi
4678:275
4638:PMC
4630:doi
4586:doi
4582:275
4547:doi
4509:PMC
4501:doi
4457:PMC
4447:doi
4400:doi
4396:114
4363:doi
4322:doi
4318:277
4281:doi
4277:274
4240:doi
4236:276
4199:doi
4195:275
4131:doi
4127:275
4065:doi
4053:266
4016:doi
3972:doi
3935:PMC
3925:doi
3876:doi
3872:272
3835:PMC
3827:doi
3783:doi
3742:doi
3701:PMC
3691:doi
3687:117
3644:doi
3632:410
3601:doi
3597:277
3556:doi
3509:doi
3472:PMC
3462:doi
3404:doi
3392:366
3359:PMC
3351:doi
3310:doi
3266:doi
3262:269
3225:doi
3184:PMC
3176:doi
3135:PMC
3127:doi
3086:PMC
3078:doi
3037:doi
3033:277
2971:PMC
2963:doi
2959:207
2914:doi
2910:129
2867:doi
2855:436
2816:doi
2804:419
2768:doi
2724:PMC
2716:doi
2669:doi
2632:PMC
2624:doi
2583:PMC
2575:doi
2534:PMC
2524:doi
2469:doi
2457:429
2424:PMC
2416:doi
2375:PMC
2367:doi
2326:PMC
2318:doi
2249:doi
2212:PMC
2204:doi
2200:186
2162:doi
2117:doi
2113:129
1844:P21
1620:E3
1613:E2
1503:E2F
1491:DNA
1410:in
443:End
342:End
245:OMA
221:MGI
57:PDB
8812::
8776::
8551::
8359::
8348::
8150:T7
7682:EC
7449:NC
7193::
7149:.
7141:.
7131:86
7129:.
7112:.
7104:.
7090:.
7086:.
7069:.
7059:.
7045:.
7041:.
7024:.
7012:84
7010:.
7006:.
6989:.
6981:.
6965:.
6948:.
6938:.
6913:.
6905:.
6897:.
6885:.
6868:.
6858:.
6848:.
6838:90
6836:.
6832:.
6815:.
6801:.
6797:.
6780:.
6770:.
6760:.
6748:.
6744:.
6727:.
6717:.
6707:40
6705:.
6701:.
6684:.
6670:.
6666:.
6649:.
6635:.
6631:.
6614:.
6606:.
6596:15
6594:.
6563:.
6546:.
6536:12
6534:.
6517:.
6505:.
6486:10
6484:.
6467:.
6453:.
6449:.
6432:.
6422:.
6412:.
6402:89
6400:.
6396:.
6377:11
6375:.
6358:.
6348:.
6338:.
6328:84
6326:.
6322:.
6294:.
6284:.
6272:30
6270:.
6266:.
6243:.
6233:.
6223:24
6221:.
6217:.
6194:.
6184:35
6182:.
6159:.
6149:.
6137:.
6133:.
6110:.
6100:.
6088:16
6081:.
6058:.
6048:.
6038:41
6036:.
6032:.
6009:.
5999:.
5989:47
5987:.
5983:.
5960:.
5950:.
5940:.
5928:.
5924:.
5899:59
5897:.
5873:.
5863:.
5853:32
5851:.
5847:.
5824:.
5814:.
5804:28
5802:.
5798:.
5775:.
5767:.
5755:.
5731:.
5721:.
5707:.
5703:.
5680:.
5670:.
5660:.
5648:.
5644:.
5621:.
5611:.
5601:.
5589:.
5585:.
5562:.
5548:.
5544:.
5521:.
5511:.
5501:.
5491:90
5489:.
5485:.
5462:.
5448:.
5444:.
5430:^
5416:.
5408:.
5398:77
5396:.
5372:.
5358:.
5354:.
5342:^
5328:.
5318:.
5308:22
5306:.
5302:.
5279:.
5271:.
5259:.
5236:.
5222:.
5218:.
5195:.
5185:.
5175:22
5173:.
5169:.
5146:.
5136:.
5126:21
5124:.
5120:.
5097:.
5083:.
5079:.
5056:.
5042:.
5038:.
5015:.
5001:.
4997:.
4974:.
4964:.
4954:20
4952:.
4948:.
4925:.
4917:.
4905:87
4903:.
4899:.
4876:.
4862:.
4858:.
4835:.
4827:.
4819:.
4807:.
4783:.
4773:.
4763:15
4761:.
4757:.
4745:^
4731:.
4717:.
4713:.
4690:.
4676:.
4672:.
4660:^
4646:.
4636:.
4626:15
4624:.
4620:.
4608:^
4594:.
4580:.
4576:.
4553:.
4541:.
4517:.
4507:.
4497:29
4495:.
4491:.
4479:^
4465:.
4455:.
4445:.
4435:98
4433:.
4429:.
4406:.
4394:.
4371:.
4359:19
4357:.
4353:.
4330:.
4316:.
4312:.
4289:.
4275:.
4271:.
4248:.
4234:.
4230:.
4207:.
4193:.
4189:.
4164:10
4162:.
4139:.
4125:.
4121:.
4096:11
4094:.
4071:.
4063:.
4047:.
4024:.
4012:20
4010:.
4006:.
3992:^
3978:.
3968:40
3966:.
3943:.
3933:.
3923:.
3913:93
3911:.
3907:.
3884:.
3870:.
3866:.
3843:.
3833:.
3823:17
3821:.
3817:.
3805:^
3791:.
3777:.
3773:.
3750:.
3738:22
3736:.
3732:.
3709:.
3699:.
3685:.
3681:.
3658:.
3650:.
3642:.
3630:.
3607:.
3595:.
3572:.
3564:.
3550:.
3546:.
3523:.
3515:.
3505:25
3503:.
3480:.
3470:.
3460:.
3450:95
3448:.
3444:.
3432:^
3418:.
3410:.
3402:.
3390:.
3367:.
3357:.
3347:18
3345:.
3341:.
3318:.
3304:.
3300:.
3288:^
3274:.
3260:.
3256:.
3233:.
3221:21
3219:.
3215:.
3192:.
3182:.
3172:29
3170:.
3166:.
3143:.
3133:.
3123:20
3121:.
3117:.
3094:.
3084:.
3074:44
3072:.
3068:.
3045:.
3031:.
3027:.
2993:^
2979:.
2969:.
2957:.
2953:.
2930:.
2922:.
2908:.
2904:.
2881:.
2873:.
2865:.
2853:.
2830:.
2822:.
2814:.
2802:.
2788:^
2774:.
2762:.
2758:.
2732:.
2722:.
2712:25
2710:.
2706:.
2683:.
2675:.
2665:69
2663:.
2640:.
2630:.
2620:76
2618:.
2614:.
2591:.
2581:.
2571:14
2569:.
2565:.
2542:.
2532:.
2522:.
2512:96
2510:.
2506:.
2483:.
2475:.
2467:.
2455:.
2432:.
2422:.
2410:.
2406:.
2383:.
2373:.
2363:45
2361:.
2357:.
2334:.
2324:.
2316:.
2306:11
2304:.
2300:.
2263:.
2255:.
2245:14
2243:.
2220:.
2210:.
2198:.
2194:.
2182:^
2168:.
2158:20
2156:.
2133:.
2125:.
2111:.
2107:.
2088:.
2070:.
2041:^
2020:^
1425:.
1045:/
449:bp
436:bp
348:bp
335:bp
243:;
239::
235:;
231::
227:;
223::
219:;
215::
168:,
164:,
160:,
156:,
152:,
148:,
144:,
140:,
136:,
132:,
128:,
124:,
120:,
116:,
112:,
108:,
104:,
100:,
96:,
92:,
88:,
8629::
8577:)
8571:(
8446:)
8442:(
8388:/
8384:/
8183:)
8177:(
8114:)
8110:(
8093:)
8075:e
8068:t
8061:v
8001:)
7997:(
7797:/
7632:/
7528:)
7524:(
7451:/
7329:)
7320:(
7230:e
7223:t
7216:v
7203:.
7157:.
7137::
7120:.
7098::
7092:5
7077:.
7063::
7053::
7032:.
7018::
6997:.
6985::
6977::
6956:.
6934::
6921:.
6901::
6893::
6876:.
6852::
6844::
6823:.
6809::
6788:.
6764::
6756::
6735:.
6721::
6713::
6692:.
6678::
6657:.
6643::
6622:.
6602::
6585:.
6554:.
6542::
6525:.
6513::
6496:.
6475:.
6461::
6440:.
6416::
6408::
6387:.
6366:.
6342::
6334::
6302:.
6278::
6251:.
6229::
6202:.
6190::
6167:.
6145::
6139:5
6118:.
6094::
6066:.
6044::
6017:.
5995::
5968:.
5944::
5936::
5930:7
5909:.
5881:.
5859::
5832:.
5810::
5783:.
5763::
5739:.
5715::
5709:9
5688:.
5664::
5656::
5629:.
5605::
5597::
5570:.
5556::
5529:.
5505::
5497::
5470:.
5456::
5424:.
5404::
5380:.
5366::
5336:.
5314::
5287:.
5267::
5261:7
5244:.
5230::
5203:.
5181::
5154:.
5132::
5105:.
5091::
5064:.
5050::
5023:.
5009::
4982:.
4960::
4933:.
4911::
4884:.
4870::
4843:.
4823::
4815::
4791:.
4769::
4739:.
4725::
4698:.
4684::
4654:.
4632::
4602:.
4588::
4561:.
4549::
4525:.
4503::
4473:.
4449::
4441::
4414:.
4402::
4379:.
4365::
4338:.
4324::
4297:.
4283::
4256:.
4242::
4215:.
4201::
4174:.
4147:.
4133::
4106:.
4079:.
4067::
4059::
4032:.
4018::
3986:.
3974::
3951:.
3927::
3919::
3892:.
3878::
3851:.
3829::
3799:.
3785::
3779:7
3758:.
3744::
3717:.
3693::
3666:.
3646::
3638::
3615:.
3603::
3580:.
3558::
3552:7
3531:.
3511::
3488:.
3464::
3456::
3426:.
3406::
3398::
3375:.
3353::
3326:.
3312::
3306:7
3282:.
3268::
3241:.
3227::
3200:.
3178::
3151:.
3129::
3102:.
3080::
3053:.
3039::
2987:.
2965::
2938:.
2916::
2889:.
2869::
2861::
2838:.
2818::
2810::
2782:.
2770::
2764:5
2740:.
2718::
2691:.
2671::
2648:.
2626::
2599:.
2577::
2550:.
2526::
2518::
2491:.
2471::
2463::
2440:.
2418::
2412:6
2391:.
2369::
2342:.
2320::
2312::
2285:.
2271:.
2251::
2228:.
2206::
2176:.
2164::
2141:.
2119::
2092:.
2074:.
1394:(
367:)
264:)
247::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.