501:
451:
609:
208:
2755:
248:
270:
27:
2765:
381:
261:
system for exchange of DNA) and Ted (Thermoproteales system for exchange of DNA), appears to be responsible for the transfer of cellular DNA between members of the same species. It has been suggested that in these archaea the conjugation machinery has been fully domesticated for promoting DNA repair through homologous recombination rather than spread of mobile genetic elements.
679:. This is because the presence of pili greatly enhances bacteria's ability to bind to body tissues, which then increases replication rates and ability to interact with the host organism. If a species of bacteria has multiple strains but only some are pathogenic, it is likely that the pathogenic strains will have pili while the nonpathogenic strains do not.
138:. They are also fragile and constantly replaced, sometimes with pili of different composition, resulting in altered antigenicity. Specific host responses to old pili structures are not effective on the new structure. Recombination between genes of some (but not all) pili code for variable (V) and constant (C) regions of the pili (similar to
244:. The connection established by the F-pilus is extremely mechanically and thermochemically resistant thanks to the robust properties of the F-pilus, which ensures successful gene transfer in a variety of environments. Not all bacteria can make conjugative pili, but conjugation can occur between bacteria of different species.
583:
This family was originally identified as "type IV fimbriae" by their appearance under the microscope. This classification survived as it happens to correspond to a clade. It has been shown that some archaeal type IV pilins can exist in 4 different conformations, yielding two pili with dramatically
316:
This appendage ranges from 3–10 nanometers in diameter and can be as much as several micrometers long. Fimbriae are used by bacteria to adhere to one another and to adhere to animal cells and some inanimate objects. A bacterium can have as many as 1,000 fimbriae. Fimbriae are only visible with the
260:
encode pili structurally similar to the bacterial conjugative pili. However, unlike in bacteria, where conjugation apparatus typically mediates the transfer of mobile genetic elements, such as plasmids or transposons, the conjugative machinery of hyperthermophilic archaea, called Ced (Crenarchaeal
613:
adhesions found on the bacteria's fimbriae. This process of bacteria adhering to a host cell can result in the colonization of that host cell as more and more bacteria collect around it, and is integral to the continued survival of the bacteria, enabling them to infect tissues and entire organs.
612:
This figure depicts fimbriae adhesion. In this process the fimbriae of a bacterial cell (right) adhere to specific proteins, called receptors, found on the outer membrane of a host cell (left). They do this by a specific interaction between the receptors of the host cell and the perfectly matched
516:
forces. The external ends of the pili adhere to a solid substrate, either the surface to which the bacterium is attached or to other bacteria. Then, when the pili contract, they pull the bacterium forward like a grappling hook. Movement produced by type IV pili is typically jerky, so it is called
251:
Proposed conjugation mechanisms between donor and recipient cells in archaea (left) and bacteria (right). The schematic shows how ssDNA substrates are generated by the HerA-NurA machinery in the donor archaeal cells and by the plasmid-encoded relaxosome in bacteria. The figure is reproduced from
547:
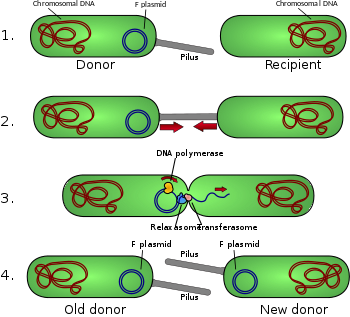
systems. Besides archaella, many archaea produce adhesive type 4 pili, which enable archaeal cells to adhere to different substrates. The N-terminal alpha-helical portions of the archaeal type 4 pilins and archaellins are homologous to the corresponding regions of bacterial T4P; however, the
162:, there has been much interest in the study of pili as an organelle of adhesion and as a vaccine component. The first detailed study of pili was done by Brinton and co-workers who demonstrated the existence of two distinct phases within one bacterial strain: pileated (p+) and non-pileated)
1075:
Beltran, Leticia C.; Cvirkaite-Krupovic, Virginija; Miller, Jessalyn; Wang, Fengbin; Kreutzberger, Mark A. B.; Patkowski, Jonasz B.; Costa, Tiago R. D.; Schouten, Stefan; Levental, Ilya; Conticello, Vincent P.; Egelman, Edward H.; Krupovic, Mart (2023-02-07).
576:. Menningococcal type IV pili bind DNA through the minor pilin ComP via an electropositive stripe that is predicted to be exposed on the filament's surface. ComP displays an exquisite binding preference for selective DUSs. The distribution of DUSs within the
232:, which establishes direct contact and the formation of a controlled pore that allows transfer of DNA from the donor to the recipient. Typically, the DNA transferred consists of the genes required to make and transfer pili (often encoded on a
584:
different structures. Remarkably, the two pili were produced by the same secretion machinery. However, which of the two pili is formed appears to depend on the growth conditions, suggesting that the two pili are functionally distinct.
486:
The secretin protein, PilQ, found on the outer membrane of the cell is necessary for the development/extension of the pilus. PilC is the first proteins to form the pilus and are responsible for overall attachment of the pilus.
491:
Once the Type IV Pilus attaches or interacts with what it needs to, it begins to retract. This occurs with the PilT beginning to degrade the last parts of the PilA in the pilus. The mechanism of PilT is very similar to PilF.
901:
Patkowski, Jonasz B.; Dahlberg, Tobias; Amin, Himani; Gahlot, Dharmender K.; Vijayrajratnam, Sukhithasri; Vogel, Joseph P.; Francis, Matthew S.; Baker, Joseph L.; Andersson, Magnus; Costa, Tiago R. D. (5 April 2023).
1010:
Gschwind, Rémi; Petitjean, Marie; Fournier, Claudine; Lao, Julie; Clermont, Olivier; Nordmann, Patrice; Mellmann, Alexander; Denamur, Erick; Poirel, Laurent; Ruppé, Etienne (2024-04-03). Uhlemann, Anne-Catrin (ed.).
1979:
Craig, Lisa; Taylor, Ronald (2014). "Chapter 1: The Vibrio cholerae Toxin
Coregulated Pilus: Structure, Assembly, and Function with Implications for Vaccine Design". In Barocchi, Michèle; Telford, John (eds.).
397:
2005:
Rinaudo, Daniela; Moschioni, Monica (2014). "Chapter 13: Pilus-based
Vaccine Development in Streptococci: Variability, Diversity, and Immunological Resposes". In Barocchi, Michèle; Telford, John (eds.).
357:, consists of many aerobic bacteria that adhere to the surface by their fimbriae. Thus, fimbriae allow the aerobic bacteria to remain both on the broth, from which they take nutrients, and near the air.
772:
442:(T4SS). They can be classified into the F-like type (after the F-pilus) and the P-like type. Like their secretion counterparts, the pilus injects material, DNA in this case, into another cell.
724:. The gene for this toxin, once incorporated into the bacterium's genome, is expressed when the gene coding for the pilus is expressed (hence the name "toxin mediated pilus").
364:, as they attach bacteria to host surfaces for colonization during infection. Fimbriae are either located at the poles of a cell or are evenly spread over its entire surface.
1448:
Wang, F; Cvirkaite-Krupovic, V; Kreutzberger, MAB; Su, Z; de
Oliveira, GAP; Osinski, T; Sherman, N; DiMaio, F; Wall, JS; Prangishvili, D; Krupovic, M; Egelman, EH (2019).
228:
in diameter. During conjugation, a pilus emerging from the donor bacterium ensnares the recipient bacterium, draws it in close, and eventually triggers the formation of a
220:. Bacteria produce long extracellular appendages called sex pili, which connect two neighbouring cells and serve as a physical conduit for transfer of DNA. Adapted from
170:
A few names are given to different types of pili by their function. The classification does not always overlap with the structural or evolutionary-based types, as
1754:
Cookson, AL; Cooley, WA; Woodward, MJ (2002), "The role of type 1 and curli fimbriae of Shiga toxin-producing
Escherichia coli in adherence to abiotic surfaces",
305:. The term "fimbria" can refer to many different (structural) types of pilus. Indeed, many different types of pili have been used for adhesion, a case of
240:; however, other pieces of DNA are often co-transferred and this can result in dissemination of genetic traits throughout a bacterial population, such as
2062:
Georgiadou, Michaella; Pelicic, Vladimir (2014). "Chapter 5: Type IV Pili: Functions & Biogenesis". In
Barocchi, Michèle; Telford, John (eds.).
1906:
682:
The development of attachment pili may then result in the development of further virulence traits. Fimbriae are one of the primary mechanisms of
834:"Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective"
142:
diversity). As the primary antigenic determinants, virulence factors and impunity factors on the cell surface of a number of species of
1925:
Epstein, EA; Reizian, MA; Chapman, MR (2009), "Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly.",
790:
Brinton, Charles (1954). "Electrophoresis and phage susceptibility studies on a filament-producing variant of the E. coli bacterium".
194:, because they allow for the exchange of genes via the formation of "mating pairs". Perhaps the most well-studied is the F-pilus of
706:
bacteria. Their presence greatly enhances the bacteria's ability to attach to the host and cause disease. Nonpathogenic strains of
554:
is the process by which a recipient bacterial cell takes up DNA from a neighboring cell and integrates this DNA into its genome by
1497:
Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V (2013).
904:"The F-pilus biomechanical adaptability accelerates conjugative dissemination of antimicrobial resistance and biofilm formation"
2071:
2015:
1989:
1432:
2694:
777:
367:
This term was also used in a lax sense to refer to all pili, by those who use "pilus" to specifically refer to sex pili.
46:
Both cells recircularize their plasmids, synthesize second strands, and reproduce pili; both cells are now viable donors.
883:
2461:
2201:
474:, removes a leader sequence, thus making the Pre-PilA shorter and into PilA, the main building-block protein of Pili.
1702:
Liu, J; Eastep, GN; Cvirkaite-Krupovic, V; Rich-New, ST; Kreutzberger, MAB; Egelman, EH; Krupovic, M; Wang, F (2024).
2799:
1364:
580:
genome favors certain genes, suggesting that there is a bias for genes involved in genomic maintenance and repair.
313:
system does not treat fimbriae as a distinct type of appendage, using the generic pilus (GO:0009289) type instead.
953:"Phylum barrier and Escherichia coli intra-species phylogeny drive the acquisition of antibiotic-resistance genes"
2451:
2378:
2575:
2306:
625:." Curli are a type of fimbriae. Curli are composed of proteins called curlins. Some of the genes involved are
201:
2416:
2768:
618:
551:
1842:"Pyelonephritic Escherichia coli expressing P fimbriae decrease immune response of the mouse kidney"
1359:. Foster, John Watkins (Fourth ed.). New York: W. W. Norton & Company. pp. 1000–1002.
596:
is responsible for moving many types of fimbriae out of the cell, including type 1 fimbriae and the
2570:
2493:
2456:
2252:
2173:
2163:
2153:
555:
540:
439:
393:
42:
The mobile plasmid is nicked and a single strand of DNA is then transferred to the recipient cell.
1214:"Pili in Gram-negative and Gram-positive bacteria — structure, assembly and their role in disease"
301:
that is used to attach the bacterium to a surface, sometimes also called an "attachment pilus" or
593:
479:
124:
951:
Petitjean, Marie; Condamine, Bénédicte; Burdet, Charles; Denamur, Erick; Ruppé, Etienne (2021).
884:"Gut bacteria use super-polymers to dodge antibiotics | Imperial News | Imperial College London"
2646:
2565:
2544:
2194:
1791:"Everything You Always Wanted to Know About Salmonella Type 1 Fimbriae, but Were Afraid to Ask"
560:
147:
84:
67:
2686:
2328:
2169:
548:
C-terminal beta-strand-rich domains appear to be unrelated in bacterial and archaeal pilins.
500:
496:
Degradation of the pilus into the components to be utilized and synthesized into PilA again.
241:
217:
187:
97:
717:
438:
The Tra (transfer) family includes all known sex pili (as of 2010). They are related to the
2526:
2401:
2384:
2222:
2159:
2100:
1510:
1401:
1158:
1089:
694:
306:
171:
115:
Dozens of these structures can exist on the bacterial and archaeal surface. Some bacteria,
1882:
1840:
Rice JC, Peng T, Spence JS, Wang HQ, Goldblum RM, Corthésy B, Nowicki BJ (December 2005).
1704:"Two distinct archaeal type IV pili structures formed by proteins with identical sequence"
298:
8:
2789:
2651:
2536:
2531:
2424:
2394:
2247:
2232:
1142:
1013:"Inter-phylum circulation of a beta-lactamase-encoding gene: a rare but observable event"
733:
661:
Pili are responsible for virulence in the pathogenic strains of many bacteria, including
565:
450:
318:
191:
2104:
1728:
1703:
1514:
1284:
1240:
1213:
1162:
1093:
1045:
1012:
928:
903:
406:
Please expand the section to include this information. Further details may exist on the
2681:
2580:
2389:
2301:
2294:
1947:
1817:
1790:
1676:
1652:"Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek"
1651:
1627:
1606:
1533:
1498:
1474:
1449:
1332:
1307:
1253:
1189:
1146:
1118:
1077:
987:
952:
860:
833:
592:
Another type are called type 1 fimbriae. They contain FimH adhesins at the "tips". The
527:
518:
2089:"Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract"
1582:
1557:
2794:
2758:
2621:
2587:
2554:
2411:
2406:
2364:
2311:
2187:
2149:
2128:
2123:
2088:
2067:
2011:
1985:
1952:
1900:
1863:
1822:
1771:
1733:
1681:
1632:
1587:
1538:
1479:
1428:
1405:
1370:
1360:
1337:
1288:
1245:
1194:
1176:
1123:
1105:
1050:
1032:
992:
974:
933:
865:
807:
803:
532:
325:
20:
2473:
2349:
2284:
2118:
2108:
1942:
1934:
1853:
1812:
1802:
1763:
1723:
1715:
1671:
1663:
1622:
1614:
1577:
1569:
1528:
1518:
1469:
1461:
1397:
1327:
1319:
1280:
1235:
1225:
1184:
1166:
1113:
1097:
1040:
1024:
982:
964:
923:
915:
855:
845:
799:
688:
662:
608:
544:
350:
346:
339:
274:
212:
196:
1618:
1257:
354:
2699:
2598:
2483:
2359:
2289:
2242:
707:
669:
105:
2087:
Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C (September 1996).
2033:"Textbook of Bacteriology: Bacterial Structure in Relationship to Pathogenicity"
1078:"Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery"
564:(also called meningococcus), DNA transformation requires the presence of short
328:
which attach them to some sort of substratum so that the bacteria can withstand
2354:
2318:
1719:
1151:
Proceedings of the
National Academy of Sciences of the United States of America
1101:
919:
535:. Bacterial type IV pili are similar in structure to the component proteins of
139:
2032:
1558:"Biased distribution of DNA uptake sequences towards genome maintenance genes"
1465:
1230:
2783:
2708:
2593:
2549:
2521:
2512:
2502:
2478:
2374:
1807:
1374:
1180:
1109:
1036:
978:
850:
721:
675:
569:
433:
310:
229:
143:
120:
2113:
1767:
1523:
1271:
Ottow, JC (1975). "Ecology, physiology, and genetics of fimbriae and pili".
1171:
2738:
2723:
2603:
2344:
2262:
2210:
1956:
1867:
1858:
1841:
1826:
1775:
1737:
1685:
1636:
1591:
1542:
1483:
1409:
1341:
1249:
1198:
1127:
1054:
996:
937:
869:
811:
521:, as opposed to other forms of bacterial motility such as that produced by
128:
2132:
1667:
1292:
969:
92:) can be used interchangeably, although some researchers reserve the term
2626:
2179:
1573:
1556:
Davidsen T, Rødland EA, Lagesen K, Seeberg E, Rognes T, Tønjum T (2004).
1028:
753:
713:
572:
of the donor DNA. Specific recognition of DUSs is mediated by a type IV
329:
237:
1938:
1450:"An extensively glycosylated archaeal pilus survives extreme conditions"
1323:
207:
2733:
2718:
2636:
2369:
748:
597:
536:
407:
1607:"Meningococcal carriage and disease--population biology and evolution"
2728:
2559:
2517:
2429:
2323:
1789:
Kolenda, Rafal; Ugorski, Maciej; Grzymajlo, Krzysztof (14 May 2019).
738:
683:
522:
462:
Pre-PilA is made in the cytoplasm and moves into the inner membrane.
247:
225:
1447:
1140:
712:
first evolved pili, allowing them to bind to human tissues and form
2237:
2214:
1701:
1074:
513:
471:
269:
109:
71:
1308:"A variety of bacterial pili involved in horizontal gene transfer"
832:
Chagnot, C; Zorgani, MA; Astruc, T; Desvaux, M (14 October 2013).
482:-Binding protein that provides energy for Type IV Pili Assembly.
2667:
2631:
2267:
743:
361:
334:
257:
233:
135:
75:
38:
Pilus attaches to recipient cell, brings the two cells together.
380:
2713:
2468:
1141:
van
Wolferen, Marleen; Wagner, Alexander; van der Does, Chris;
950:
26:
16:
A proteinaceous hair-like appendage on the surface of bacteria
1555:
622:
573:
401:
286:
116:
101:
55:
1009:
1496:
1423:
Jarrell; et al. (2009). "Archaeal
Flagella and Pili".
900:
831:
1388:
Mattick JS (2002). "Type IV pili and twitching motility".
370:
2086:
2064:
Bacterial Pili: Structure, Synthesis, and Role in
Disease
2008:
Bacterial Pili: Structure, Synthesis, and Role in
Disease
1982:
Bacterial Pili: Structure, Synthesis, and Role in Disease
183:
1788:
1499:"Specific DNA recognition mediated by a type IV pilin"
190:. They are sometimes called "sex pili", in analogy to
1924:
1753:
1425:
Pili and Flagella: Current Research and Future Trends
1697:
1695:
1643:
1883:"Colonization and Invasion by Bacterial Pathogens"
1839:
716:. These pili then served as binding sites for the
100:. All conjugative pili are primarily composed of
2061:
539:(archaeal flagella), and both are related to the
2781:
2004:
1920:
1918:
1916:
1749:
1747:
1692:
466:Pre-PilA is inserted into the inner membrane.
827:
825:
823:
821:
404:), LPXTG including type 3 pilus (T3P; spaHIG).
2195:
1913:
1846:Journal of the American Society of Nephrology
1744:
1604:
1905:: CS1 maint: multiple names: authors list (
1598:
1549:
1490:
1381:
1299:
30:Schematic drawing of bacterial conjugation.
2080:
1978:
818:
568:(DUSs) which are 9-10 monomers residing in
398:extracellular nucleation-precipitation pili
360:Fimbriae are required for the formation of
349:form a very thin layer at the surface of a
182:Conjugative pili allow for the transfer of
2209:
2202:
2188:
2010:. C.A.B. International. pp. 182–202.
1656:Microbiology and Molecular Biology Reviews
1416:
1387:
1211:
504:Type IVa pilus machine architectural model
2172:at the U.S. National Library of Medicine
2162:at the U.S. National Library of Medicine
2152:at the U.S. National Library of Medicine
2122:
2112:
1946:
1857:
1816:
1806:
1727:
1675:
1626:
1581:
1532:
1522:
1473:
1331:
1239:
1229:
1212:Proft, T.; Baker, E. N. (February 2009).
1188:
1170:
1117:
1044:
986:
968:
927:
859:
849:
543:(T2SS); they are unified by the group of
2066:. C.A.B. International. pp. 71–84.
607:
499:
449:
268:
246:
206:
25:
1984:. C.A.B. International. pp. 1–16.
1422:
1357:Microbiology : an evolving science
1305:
789:
371:Types by assembling system or structure
297:) is a term used for a short pilus, an
2782:
1649:
1402:10.1146/annurev.micro.56.012302.160938
525:. However, some bacteria, for example
392:about mention of other types: various
2183:
2057:
2055:
2053:
1974:
1972:
1970:
1968:
1966:
1270:
1147:"The archaeal Ced system imports DNA"
1017:Antimicrobial Agents and Chemotherapy
2764:
1354:
1218:Cellular and Molecular Life Sciences
1070:
1068:
1066:
1064:
374:
332:and obtain nutrients. For example,
321:. They may be straight or flexible.
186:between bacteria, in the process of
165:
1285:10.1146/annurev.mi.29.100175.000455
290:
177:
59:
13:
2050:
1963:
587:
14:
2811:
2143:
2030:
1061:
720:that carries the disease-causing
617:"Gram-negative bacteria assemble
2763:
2754:
2753:
456:Type IV Pilus Twitching Motility
379:
224:A sex pilus is typically 6 to 7
2452:Bacterial cellular morphologies
2024:
1998:
1874:
1833:
1782:
1650:Nuccio SP, et al. (2007).
1441:
1348:
1264:
1205:
445:
96:for the appendage required for
1887:www.textbookofbacteriology.net
1880:
1605:Caugant DA, Maiden MC (2009).
1134:
1003:
944:
894:
876:
783:
766:
127:on pili at the start of their
1:
1619:10.1016/j.vaccine.2009.04.061
1273:Annual Review of Microbiology
792:Biochimica et Biophysica Acta
759:
88:(Latin for 'fringe'; plural:
19:For the sea snail genus, see
2093:Proc. Natl. Acad. Sci. U.S.A
1881:WI, Kenneth Todar, Madison.
1503:Proc. Natl. Acad. Sci. U.S.A
804:10.1016/0006-3002(54)90011-6
778:Dorland's Medical Dictionary
656:
7:
727:
427:
264:
34:Donor cell produces pilus.
10:
2816:
2695:Bacteria (classifications)
2417:Primary nutritional groups
1720:10.1038/s41467-024-45062-z
1613:. 27 Suppl 2 (4): B64–70.
1427:. Caister Academic Press.
1355:Joan, Slonczewski (2017).
1102:10.1038/s41467-023-36349-8
920:10.1038/s41467-023-37600-y
431:
18:
2749:
2680:
2660:
2612:
2501:
2492:
2444:
2337:
2275:
2261:
2221:
1795:Frontiers in Microbiology
1466:10.1038/s41564-019-0458-x
1231:10.1007/s00018-008-8477-4
838:Frontiers in Microbiology
400:built by T8SS (including
2800:Prokaryotic cell anatomy
2571:Bacterial outer membrane
2174:Medical Subject Headings
2164:Medical Subject Headings
2154:Medical Subject Headings
2037:Textbook of Bacteriology
1808:10.3389/fmicb.2019.01017
1306:Filloux, A (July 2010).
851:10.3389/fmicb.2013.00303
603:
556:homologous recombination
541:Type II secretion system
440:type IV secretion system
394:chaperone-usher fimbriae
2114:10.1073/pnas.93.18.9827
1768:10.1078/1438-4221-00203
1524:10.1073/pnas.1218832110
1312:Journal of Bacteriology
1172:10.1073/pnas.1513740113
718:lysogenic bacteriophage
594:chaperone-usher pathway
353:. This layer, called a
338:uses them to attach to
236:), and so is a kind of
2566:Gram-negative bacteria
2545:Gram-positive bacteria
1859:10.1681/ASN.2005030243
673:, and many strains of
621:surface fibers called
614:
561:Neisseria meningitidis
552:Genetic transformation
505:
497:
390:is missing information
279:
253:
221:
148:gram-positive bacteria
68:cell-surface appendage
47:
2421:Substrate preference
1708:Nature Communications
1668:10.1128/MMBR.00014-07
1082:Nature Communications
970:10.1099/mgen.0.000489
908:Nature Communications
611:
503:
453:
272:
250:
242:antibiotic resistance
210:
188:bacterial conjugation
98:bacterial conjugation
29:
2402:Microbial metabolism
1390:Annu. Rev. Microbiol
1143:Albers, Sonja-Verena
1029:10.1128/aac.01459-23
695:Bordetella pertussis
566:DNA uptake sequences
307:convergent evolution
172:convergent evolution
2652:Non-motile bacteria
2248:Pathogenic bacteria
2105:1996PNAS...93.9827C
1939:10.1128/JB.01244-08
1756:Int J Med Microbiol
1515:2013PNAS..110.3065C
1454:Nature Microbiology
1324:10.1128/JB.00424-10
1163:2016PNAS..113.2496V
1094:2023NatCo..14..666B
734:Bacterial nanowires
319:electron microscope
192:sexual reproduction
2581:Lipopolysaccharide
1574:10.1093/nar/gkh255
957:Microbial Genomics
619:functional amyloid
615:
528:Myxococcus xanthus
519:twitching motility
508:Some pili, called
506:
498:
280:
256:Hyperthermophilic
254:
222:
152:Enterobacteriaceae
48:
2777:
2776:
2676:
2675:
2622:Bacterial capsule
2588:Periplasmic space
2555:Lipoteichoic acid
2440:
2439:
2412:Microbial ecology
2407:Nitrogen fixation
2170:Fimbriae+Proteins
2073:978-1-78064-255-0
2017:978-1-78064-255-0
1991:978-1-78064-255-0
1562:Nucleic Acids Res
1434:978-1-904455-48-6
425:
424:
340:mannose receptors
324:Fimbriae possess
200:, encoded by the
166:Types by function
66:) is a hair-like
21:Pilus (gastropod)
2807:
2767:
2766:
2757:
2756:
2705:Former groupings
2499:
2498:
2350:Human microbiome
2273:
2272:
2204:
2197:
2190:
2181:
2180:
2137:
2136:
2126:
2116:
2084:
2078:
2077:
2059:
2048:
2047:
2045:
2043:
2031:Todar, Kenneth.
2028:
2022:
2021:
2002:
1996:
1995:
1976:
1961:
1960:
1950:
1922:
1911:
1910:
1904:
1896:
1894:
1893:
1878:
1872:
1871:
1861:
1837:
1831:
1830:
1820:
1810:
1786:
1780:
1779:
1762:(3–4): 195–205,
1751:
1742:
1741:
1731:
1699:
1690:
1689:
1679:
1647:
1641:
1640:
1630:
1602:
1596:
1595:
1585:
1553:
1547:
1546:
1536:
1526:
1494:
1488:
1487:
1477:
1460:(8): 1401–1410.
1445:
1439:
1438:
1420:
1414:
1413:
1385:
1379:
1378:
1352:
1346:
1345:
1335:
1303:
1297:
1296:
1268:
1262:
1261:
1243:
1233:
1209:
1203:
1202:
1192:
1174:
1157:(9): 2496–2501.
1138:
1132:
1131:
1121:
1072:
1059:
1058:
1048:
1007:
1001:
1000:
990:
972:
948:
942:
941:
931:
898:
892:
891:
880:
874:
873:
863:
853:
829:
816:
815:
787:
781:
770:
545:Type IV filament
533:gliding motility
512:(T4P), generate
420:
417:
411:
383:
375:
347:aerobic bacteria
292:
275:Escherichia coli
213:Escherichia coli
197:Escherichia coli
178:Conjugative pili
156:Pseudomonadaceae
106:fibrous proteins
61:
2815:
2814:
2810:
2809:
2808:
2806:
2805:
2804:
2780:
2779:
2778:
2773:
2745:
2700:Bacterial phyla
2684:
2672:
2656:
2614:
2608:
2599:Arabinogalactan
2504:
2488:
2436:
2333:
2277:
2265:
2257:
2243:Lysogenic cycle
2224:
2217:
2208:
2160:Bacterial+Pilus
2146:
2141:
2140:
2099:(18): 9827–32.
2085:
2081:
2074:
2060:
2051:
2041:
2039:
2029:
2025:
2018:
2003:
1999:
1992:
1977:
1964:
1923:
1914:
1898:
1897:
1891:
1889:
1879:
1875:
1852:(12): 3583–91.
1838:
1834:
1787:
1783:
1752:
1745:
1700:
1693:
1648:
1644:
1603:
1599:
1554:
1550:
1495:
1491:
1446:
1442:
1435:
1421:
1417:
1386:
1382:
1367:
1353:
1349:
1304:
1300:
1269:
1265:
1210:
1206:
1139:
1135:
1073:
1062:
1023:(4): e0145923.
1008:
1004:
949:
945:
899:
895:
882:
881:
877:
830:
819:
788:
784:
771:
767:
762:
730:
670:Vibrio cholerae
659:
606:
590:
588:Type 1 fimbriae
578:N. meningitides
448:
436:
430:
421:
415:
412:
405:
396:built by T7SS,
384:
373:
267:
180:
168:
24:
17:
12:
11:
5:
2813:
2803:
2802:
2797:
2792:
2775:
2774:
2772:
2771:
2761:
2750:
2747:
2746:
2744:
2743:
2742:
2741:
2736:
2731:
2726:
2716:
2711:
2702:
2697:
2691:
2689:
2678:
2677:
2674:
2673:
2671:
2670:
2664:
2662:
2658:
2657:
2655:
2654:
2649:
2644:
2639:
2634:
2629:
2624:
2618:
2616:
2610:
2609:
2607:
2606:
2601:
2590:
2585:
2584:
2583:
2578:
2562:
2557:
2552:
2541:
2540:
2539:
2534:
2529:
2515:
2509:
2507:
2496:
2490:
2489:
2487:
2486:
2481:
2476:
2471:
2466:
2465:
2464:
2459:
2457:cell structure
2448:
2446:
2442:
2441:
2438:
2437:
2435:
2434:
2433:
2432:
2430:Saccharophilic
2427:
2419:
2414:
2409:
2404:
2399:
2398:
2397:
2392:
2387:
2382:
2372:
2367:
2362:
2357:
2347:
2341:
2339:
2335:
2334:
2332:
2331:
2326:
2321:
2319:Microaerophile
2316:
2315:
2314:
2309:
2299:
2298:
2297:
2292:
2281:
2279:
2270:
2259:
2258:
2256:
2255:
2250:
2245:
2240:
2235:
2229:
2227:
2219:
2218:
2207:
2206:
2199:
2192:
2184:
2178:
2177:
2167:
2157:
2145:
2144:External links
2142:
2139:
2138:
2079:
2072:
2049:
2023:
2016:
1997:
1990:
1962:
1933:(2): 608–615,
1912:
1873:
1832:
1781:
1743:
1691:
1662:(4): 551–575.
1642:
1597:
1548:
1509:(8): 3065–70.
1489:
1440:
1433:
1415:
1396:(1): 289–314.
1380:
1365:
1347:
1318:(13): 3243–5.
1298:
1263:
1224:(4): 613–635.
1204:
1145:(2016-03-01).
1133:
1060:
1002:
943:
893:
875:
817:
798:(4): 533–542.
782:
764:
763:
761:
758:
757:
756:
751:
746:
741:
736:
729:
726:
700:Staphylococcus
658:
655:
605:
602:
589:
586:
570:coding regions
447:
444:
432:Main article:
429:
426:
423:
422:
387:
385:
378:
372:
369:
303:adhesive pilus
289:for 'fringe',
266:
263:
179:
176:
167:
164:
140:immunoglobulin
121:bacteriophages
70:found on many
15:
9:
6:
4:
3:
2:
2812:
2801:
2798:
2796:
2793:
2791:
2788:
2787:
2785:
2770:
2762:
2760:
2752:
2751:
2748:
2740:
2737:
2735:
2732:
2730:
2727:
2725:
2722:
2721:
2720:
2717:
2715:
2712:
2710:
2709:Schizomycetes
2706:
2703:
2701:
2698:
2696:
2693:
2692:
2690:
2688:
2683:
2679:
2669:
2666:
2665:
2663:
2659:
2653:
2650:
2648:
2645:
2643:
2640:
2638:
2635:
2633:
2630:
2628:
2625:
2623:
2620:
2619:
2617:
2611:
2605:
2602:
2600:
2597:
2595:
2591:
2589:
2586:
2582:
2579:
2577:
2574:
2573:
2572:
2569:
2567:
2563:
2561:
2558:
2556:
2553:
2551:
2550:Teichoic acid
2548:
2546:
2542:
2538:
2535:
2533:
2530:
2528:
2525:
2524:
2523:
2522:Peptidoglycan
2519:
2516:
2514:
2513:Cell membrane
2511:
2510:
2508:
2506:
2500:
2497:
2495:
2491:
2485:
2482:
2480:
2477:
2475:
2472:
2470:
2467:
2463:
2460:
2458:
2455:
2454:
2453:
2450:
2449:
2447:
2443:
2431:
2428:
2426:
2423:
2422:
2420:
2418:
2415:
2413:
2410:
2408:
2405:
2403:
2400:
2396:
2393:
2391:
2388:
2386:
2383:
2380:
2376:
2373:
2371:
2368:
2366:
2363:
2361:
2358:
2356:
2353:
2352:
2351:
2348:
2346:
2343:
2342:
2340:
2336:
2330:
2327:
2325:
2322:
2320:
2317:
2313:
2310:
2308:
2305:
2304:
2303:
2300:
2296:
2293:
2291:
2288:
2287:
2286:
2283:
2282:
2280:
2274:
2271:
2269:
2264:
2260:
2254:
2251:
2249:
2246:
2244:
2241:
2239:
2236:
2234:
2231:
2230:
2228:
2226:
2220:
2216:
2212:
2205:
2200:
2198:
2193:
2191:
2186:
2185:
2182:
2175:
2171:
2168:
2165:
2161:
2158:
2155:
2151:
2148:
2147:
2134:
2130:
2125:
2120:
2115:
2110:
2106:
2102:
2098:
2094:
2090:
2083:
2075:
2069:
2065:
2058:
2056:
2054:
2038:
2034:
2027:
2019:
2013:
2009:
2001:
1993:
1987:
1983:
1975:
1973:
1971:
1969:
1967:
1958:
1954:
1949:
1944:
1940:
1936:
1932:
1928:
1921:
1919:
1917:
1908:
1902:
1888:
1884:
1877:
1869:
1865:
1860:
1855:
1851:
1847:
1843:
1836:
1828:
1824:
1819:
1814:
1809:
1804:
1800:
1796:
1792:
1785:
1777:
1773:
1769:
1765:
1761:
1757:
1750:
1748:
1739:
1735:
1730:
1725:
1721:
1717:
1713:
1709:
1705:
1698:
1696:
1687:
1683:
1678:
1673:
1669:
1665:
1661:
1657:
1653:
1646:
1638:
1634:
1629:
1624:
1620:
1616:
1612:
1608:
1601:
1593:
1589:
1584:
1579:
1575:
1571:
1568:(3): 1050–8.
1567:
1563:
1559:
1552:
1544:
1540:
1535:
1530:
1525:
1520:
1516:
1512:
1508:
1504:
1500:
1493:
1485:
1481:
1476:
1471:
1467:
1463:
1459:
1455:
1451:
1444:
1436:
1430:
1426:
1419:
1411:
1407:
1403:
1399:
1395:
1391:
1384:
1376:
1372:
1368:
1366:9780393614039
1362:
1358:
1351:
1343:
1339:
1334:
1329:
1325:
1321:
1317:
1313:
1309:
1302:
1294:
1290:
1286:
1282:
1278:
1274:
1267:
1259:
1255:
1251:
1247:
1242:
1237:
1232:
1227:
1223:
1219:
1215:
1208:
1200:
1196:
1191:
1186:
1182:
1178:
1173:
1168:
1164:
1160:
1156:
1152:
1148:
1144:
1137:
1129:
1125:
1120:
1115:
1111:
1107:
1103:
1099:
1095:
1091:
1087:
1083:
1079:
1071:
1069:
1067:
1065:
1056:
1052:
1047:
1042:
1038:
1034:
1030:
1026:
1022:
1018:
1014:
1006:
998:
994:
989:
984:
980:
976:
971:
966:
963:(8): 000489.
962:
958:
954:
947:
939:
935:
930:
925:
921:
917:
913:
909:
905:
897:
889:
888:Imperial News
885:
879:
871:
867:
862:
857:
852:
847:
843:
839:
835:
828:
826:
824:
822:
813:
809:
805:
801:
797:
793:
786:
780:
779:
774:
769:
765:
755:
752:
750:
747:
745:
742:
740:
737:
735:
732:
731:
725:
723:
719:
715:
714:microcolonies
711:
710:
705:
704:Streptococcus
701:
697:
696:
691:
690:
685:
680:
678:
677:
676:Streptococcus
672:
671:
666:
665:
654:
652:
648:
644:
640:
636:
632:
628:
624:
620:
610:
601:
599:
595:
585:
581:
579:
575:
571:
567:
563:
562:
557:
553:
549:
546:
542:
538:
534:
530:
529:
524:
520:
515:
511:
502:
495:
490:
485:
481:
477:
473:
469:
465:
461:
458:
457:
452:
443:
441:
435:
434:Transfer gene
419:
416:December 2020
409:
403:
399:
395:
391:
388:This section
386:
382:
377:
376:
368:
365:
363:
358:
356:
352:
351:broth culture
348:
343:
341:
337:
336:
331:
327:
322:
320:
314:
312:
311:Gene Ontology
308:
304:
300:
296:
288:
284:
277:
276:
271:
262:
259:
249:
245:
243:
239:
235:
231:
230:mating bridge
227:
219:
215:
214:
209:
205:
203:
199:
198:
193:
189:
185:
175:
173:
163:
161:
160:Neisseriaceae
157:
153:
149:
145:
144:gram-negative
141:
137:
132:
130:
126:
122:
118:
113:
111:
107:
103:
99:
95:
91:
87:
86:
81:
78:. The terms
77:
73:
69:
65:
57:
53:
45:
41:
37:
33:
28:
22:
2739:Mendosicutes
2724:Gracilicutes
2704:
2641:
2604:Mycolic acid
2594:Mycobacteria
2592:
2564:
2543:
2479:Coccobacilli
2379:in pregnancy
2345:Extremophile
2329:Aerotolerant
2263:Biochemistry
2225:microbiology
2211:Microbiology
2096:
2092:
2082:
2063:
2040:. Retrieved
2036:
2026:
2007:
2000:
1981:
1930:
1926:
1890:. Retrieved
1886:
1876:
1849:
1845:
1835:
1798:
1794:
1784:
1759:
1755:
1711:
1707:
1659:
1655:
1645:
1610:
1600:
1565:
1561:
1551:
1506:
1502:
1492:
1457:
1453:
1443:
1424:
1418:
1393:
1389:
1383:
1356:
1350:
1315:
1311:
1301:
1276:
1272:
1266:
1221:
1217:
1207:
1154:
1150:
1136:
1085:
1081:
1020:
1016:
1005:
960:
956:
946:
911:
907:
896:
887:
878:
841:
837:
795:
791:
785:
776:
768:
708:
703:
699:
693:
687:
681:
674:
668:
663:
660:
650:
646:
642:
638:
634:
630:
626:
616:
591:
582:
577:
559:
550:
526:
510:type IV pili
509:
507:
493:
488:
483:
475:
467:
463:
459:
455:
454:
446:Type IV pili
437:
413:
389:
366:
359:
344:
333:
330:shear forces
323:
315:
302:
294:
282:
281:
273:
255:
223:
211:
202:F sex factor
195:
181:
169:
159:
155:
151:
150:, including
133:
129:reproductive
114:
108:, which are
93:
89:
83:
79:
63:
58:for 'hair';
51:
49:
43:
39:
35:
31:
2627:Slime layer
2307:Facultative
2295:Facultative
2042:24 November
1927:J Bacteriol
1714:(1): 5049.
914:(1): 1879.
754:PilZ domain
709:V. cholerae
238:selfish DNA
218:conjugation
216:undergoing
2790:Organelles
2784:Categories
2734:Mollicutes
2729:Firmicutes
2719:Prokaryota
2637:Glycocalyx
2462:plasticity
2425:Lipophilic
2278:preference
2253:Resistance
1892:2016-12-03
1279:: 79–108.
1088:(1): 666.
760:References
749:P fimbriae
598:P fimbriae
531:, exhibit
317:use of an
123:attach to
110:oligomeric
2687:evolution
2661:Composite
2560:Endospore
2518:Cell wall
2494:Structure
2385:Placental
2324:Nanaerobe
2302:Anaerobic
2233:Infection
2150:Sex+Pilus
1375:951925510
1181:1091-6490
1110:2041-1723
1037:0066-4804
979:2057-5858
739:Flagellum
684:virulence
657:Virulence
537:archaella
472:peptidase
408:talk page
299:appendage
174:occurs.
146:and some
136:antigenic
134:Pili are
125:receptors
2795:Bacteria
2759:Category
2682:Taxonomy
2615:envelope
2505:envelope
2395:Salivary
2312:Obligate
2290:Obligate
2238:Exotoxin
2215:Bacteria
1957:19011034
1901:cite web
1868:16236807
1827:31139165
1801:: 1017.
1776:12398210
1738:38877064
1729:11178852
1686:18063717
1637:19464092
1592:14960717
1543:23386723
1484:31110358
1410:12142488
1342:20418394
1250:18953686
1241:11131518
1199:26884154
1128:36750723
1055:38441061
1046:10989005
997:34435947
938:37019921
929:10076315
870:24133488
812:13230101
728:See also
523:flagella
478:PilF, a
470:PilD, a
428:Transfer
355:pellicle
326:adhesins
295:fimbriae
265:Fimbriae
90:fimbriae
72:bacteria
2769:Commons
2668:Biofilm
2647:Fimbria
2632:S-layer
2613:Outside
2474:Bacilli
2390:Uterine
2375:Vaginal
2285:Aerobic
2268:ecology
2223:Medical
2133:8790416
2101:Bibcode
1948:2620823
1818:6527747
1677:2168650
1628:2719693
1611:Vaccine
1534:3581936
1511:Bibcode
1475:6656605
1333:2897649
1293:1180526
1190:4780597
1159:Bibcode
1119:9905601
1090:Bibcode
988:8549366
861:3796261
844:: 303.
773:"pilus"
744:Sortase
689:E. coli
664:E. coli
362:biofilm
335:E. coli
283:Fimbria
258:archaea
234:plasmid
131:cycle.
117:viruses
85:fimbria
76:archaea
2714:Monera
2484:Spiral
2276:Oxygen
2176:(MeSH)
2166:(MeSH)
2156:(MeSH)
2131:
2121:
2070:
2014:
1988:
1955:
1945:
1866:
1825:
1815:
1774:
1736:
1726:
1684:
1674:
1635:
1625:
1590:
1583:373393
1580:
1541:
1531:
1482:
1472:
1431:
1408:
1373:
1363:
1340:
1330:
1291:
1258:860681
1256:
1248:
1238:
1197:
1187:
1179:
1126:
1116:
1108:
1053:
1043:
1035:
995:
985:
977:
936:
926:
868:
858:
810:
649:, and
558:. In
514:motile
309:. The
158:, and
2642:Pilus
2596:only:
2576:Porin
2568:only:
2547:only:
2469:Cocci
2445:Shape
2365:Mouth
2338:Other
2124:38514
1254:S2CID
722:toxin
623:curli
604:Curli
574:pilin
402:curli
345:Some
287:Latin
102:pilin
94:pilus
80:pilus
56:Latin
52:pilus
2685:and
2503:Cell
2370:Skin
2360:Lung
2266:and
2129:PMID
2068:ISBN
2044:2017
2012:ISBN
1986:ISBN
1953:PMID
1907:link
1864:PMID
1823:PMID
1772:PMID
1734:PMID
1682:PMID
1633:PMID
1588:PMID
1539:PMID
1480:PMID
1429:ISBN
1406:PMID
1371:OCLC
1361:ISBN
1338:PMID
1289:PMID
1246:PMID
1195:PMID
1177:ISSN
1124:PMID
1106:ISSN
1051:PMID
1033:ISSN
993:PMID
975:ISSN
934:PMID
866:PMID
808:PMID
702:and
686:for
651:CsgG
647:CsgF
643:CsgE
639:CsgD
635:CsgC
631:CsgB
627:CsgA
82:and
74:and
64:pili
2537:DAP
2532:NAG
2527:NAM
2355:Gut
2119:PMC
2109:doi
1943:PMC
1935:doi
1931:191
1854:doi
1813:PMC
1803:doi
1764:doi
1760:292
1724:PMC
1716:doi
1672:PMC
1664:doi
1623:PMC
1615:doi
1578:PMC
1570:doi
1529:PMC
1519:doi
1507:110
1470:PMC
1462:doi
1398:doi
1328:PMC
1320:doi
1316:192
1281:doi
1236:PMC
1226:doi
1185:PMC
1167:doi
1155:113
1114:PMC
1098:doi
1041:PMC
1025:doi
983:PMC
965:doi
924:PMC
916:doi
856:PMC
846:doi
800:doi
775:at
480:NTP
342:.
291:pl.
184:DNA
119:or
60:pl.
2786::
2707::
2520::
2213::
2127:.
2117:.
2107:.
2097:93
2095:.
2091:.
2052:^
2035:.
1965:^
1951:,
1941:,
1929:,
1915:^
1903:}}
1899:{{
1885:.
1862:.
1850:16
1848:.
1844:.
1821:.
1811:.
1799:10
1797:.
1793:.
1770:,
1758:,
1746:^
1732:.
1722:.
1712:15
1710:.
1706:.
1694:^
1680:.
1670:.
1660:71
1658:.
1654:.
1631:.
1621:.
1609:.
1586:.
1576:.
1566:32
1564:.
1560:.
1537:.
1527:.
1517:.
1505:.
1501:.
1478:.
1468:.
1456:.
1452:.
1404:.
1394:56
1392:.
1369:.
1336:.
1326:.
1314:.
1310:.
1287:.
1277:29
1275:.
1252:.
1244:.
1234:.
1222:66
1220:.
1216:.
1193:.
1183:.
1175:.
1165:.
1153:.
1149:.
1122:.
1112:.
1104:.
1096:.
1086:14
1084:.
1080:.
1063:^
1049:.
1039:.
1031:.
1021:68
1019:.
1015:.
991:.
981:.
973:.
959:.
955:.
932:.
922:.
912:14
910:.
906:.
886:.
864:.
854:.
840:.
836:.
820:^
806:.
796:15
794:.
698:,
692:,
667:,
653:.
645:,
641:,
637:,
633:,
629:,
600:.
494:7.
489:6.
484:5.
476:4.
468:3.
464:2.
460:1.
293::
226:nm
204:.
154:,
112:.
104:–
62::
50:A
44:4-
40:3-
36:2-
32:1-
2381:)
2377:(
2203:e
2196:t
2189:v
2135:.
2111::
2103::
2076:.
2046:.
2020:.
1994:.
1959:.
1937::
1909:)
1895:.
1870:.
1856::
1829:.
1805::
1778:.
1766::
1740:.
1718::
1688:.
1666::
1639:.
1617::
1594:.
1572::
1545:.
1521::
1513::
1486:.
1464::
1458:4
1437:.
1412:.
1400::
1377:.
1344:.
1322::
1295:.
1283::
1260:.
1228::
1201:.
1169::
1161::
1130:.
1100::
1092::
1057:.
1027::
999:.
967::
961:7
940:.
918::
890:.
872:.
848::
842:4
814:.
802::
418:)
414:(
410:.
285:(
278:.
54:(
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.