396:. A probe that hybridizes only to a single DNA segment that has not been cut by the restriction enzyme will produce a single band on a Southern blot, whereas multiple bands will likely be observed when the probe hybridizes to several highly similar sequences (e.g., those that may be the result of sequence duplication). To improve specificity and reduce hybridization of the probe to sequences that are less than 100% identical, the hybridization parameters may be changed (for instance, by raising the hybridization temperature or lowering the salt content). Nylon membrane is more durable and has higher binding capacity to DNA fragments than nitrocellulose membrane, so the DNA fragments will be more fixed to the membrane even when the membrane is incubated in high temperatures. In addition, compared to the nitrocellulose membrane which requires a high ionic strength buffer to bind the DNA fragments to the membrane, nylon charged membranes use buffers with very low ionic strength to transfer even small fragments of DNA of about 50 bp to the membrane, usually the DNA to be transferred is separated by polyacrylamide gel. In the blotting step the most efficient method to transfer the DNA from the gel to the membrane is the vacuum transfer since it transfers the DNA more rapidly and quantitatively.
20:
50:
30:
568:
205:
treatment. After the DNA fragments are immobilized on the membrane, prehybridization methods are used to reduce non-specific probe binding. Then the fragments on the membrane are hybridized with either radiolabeled or nonradioactive labeled DNA, RNA, or oligonucleotide probes that are complementary to the target DNA sequence. Then detection methods are used to visualize the target DNA.
196:, when he transferred RNA molecules to DEAE paper. Southern blot was invented in 1973 but it was not published until 1975. Although it was published later the technique was disseminated when Southern introduced the Southern blot technique to a scientist at Cold Spring Harbor Laboratory called Michael Mathews by drawing this technique on a paper.
222:
are used to cut high-molecular-weight DNA strands into smaller fragments. This is done by adding the desired amount of DNA which can be changed according to the probe used and the intricacy of the DNA, with the restriction enzyme, enzyme buffer and purified water. Then everything is incubated at
387:
Hybridization of the probe to a specific DNA fragment on the filter membrane indicates that this fragment contains a DNA sequence that is complementary to the probe. The transfer step of the DNA from the electrophoresis gel to a membrane permits easy binding of the labeled hybridization probe to the
204:
The genomic DNA is digested with either one or more than one restriction enzyme, then the DNA fragments are size-fractionated by gel electrophoresis. Before the DNA fragments are transferred to a solid membrane which is either nylon or nitrocellulose membrane they are first denatured by alkaline
282:
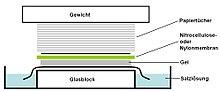
is placed on top of (or below, depending on the direction of the transfer) the gel. Pressure is applied constantly to the gel (either using suction, or by placing a stack of paper towels and a weight on top of the membrane and gel), to ensure good and even contact between gel and membrane. If
352:. In some cases, the hybridization probe may be made from RNA, rather than DNA. To ensure the specificity of the binding of the probe to the sample DNA, most common hybridization methods use salmon or herring sperm DNA for blocking of the membrane surface and target DNA, deionized
313:
Electrophoretic transfer: This method usually uses large electric current which makes it difficult to transfer the DNA efficiently due to the temperature of the buffer used, so these machines can be either equipped with cooling machines or used in a cold
259:) to denature the double-stranded DNA. The denaturation in an alkaline environment may improve binding of the negatively charged thymine residues of DNA to a positively charged amino groups of membrane, separating it into single DNA strands for later
317:
Vacuum transfer: This method uses a buffer from the upper chamber to transfer the DNA from the gel to the nitrocellulose or nylon membrane, the gel is placed directly on the membrane, and the membrane is placed on a porous screen on the vacuum
187:
introduced this technique as
Southern. The second innovation is the gel electrophoresis that is based on separation of mixtures of DNA, RNA, or proteins according to molecular size, which was also developed at Johns Hopkins University, by
714:
Burnette, W. Neal (April 1981). "Western
Blotting: Electrophoretic Transfer of Proteins from Sodium Dodecyl Sulfate-Polyacrylamide Gels to Unmodified Nitrocellulose and Radiographic Detection with Antibody and Radioiodinated Protein A".
306:
Downward capillary transfer: This method is done by placing the gel on the surface of the membrane (usually nylon charged membrane) and the DNA fragments will be transferred in a downward direction with the flow of the alkaline
104:
to move them through a sieve-like gel or matrix, which allows smaller fragments to move faster than larger fragments. The DNA fragments are transferred out of the gel or matrix onto a solid membrane, which is then exposed to a
340:—a single DNA fragment with a particular sequence whose presence in the target DNA is to be ascertained—is exposed to the membrane. The probe DNA is labelled so that it can be detected, usually by incorporating
263:
to the probe (see below), and destroys any residual RNA that may still be present in the DNA. The choice of alkaline over neutral transfer methods, however, is often empirical and may result in equivalent
299:
interactions bind the DNA to the membrane due to the negative charge of the DNA and positive charge of the membrane. Five methods can be used to transfer DNA fragments to the solid membrane and they are:
109:
labeled with a radioactive, fluorescent, or chemical tag. The tag allows any DNA fragments containing complementary sequences with the DNA probe sequence to be visualized within the
Southern blot.
74:
413:, or other collections of cloned DNA fragments. Sequences that hybridize with the hybridization probe are further analyzed, for example, to obtain the full length sequence of the targeted gene.
212:
The DNA to be studied is isolated from various tissues. The most suitable source of DNA is known as blood tissue. However, it can be isolated from different tissues (hair, semen, saliva, etc.).
62:
463:
cleaves only DNA unmethylated at that site. Therefore, any methylated sites within a sequence analyzed with a particular probe will be cleaved by the former, but not the latter, enzyme.
392:
or other detection methods. Southern blots performed with restriction enzyme-digested genomic DNA may be used to determine the number of sequences (e.g., gene copies) in a
171:
Southern invented
Southern blot after combining three innovations. The first one is the restriction endonucleases, which were developed at Johns Hopkins University by
303:
Upward capillary transfer: This method transfers the DNA fragment upward from the gel to the membrane where the flow of the liquid or the buffer will be upward.
310:
Simultaneous transfer to two membranes: This method is used to transfer DNA fragments of high concentration simultaneously from the gel to two membranes.
476:
Compared to other tests, southern blot is a complex technique that has multiple steps and these steps require equipment and reagents that are expensive.
409:
are designed so that they are complementary to the target sequence. The oligonucleotides are chemically synthesized, radiolabeled, and used to screen a
326:
The membrane is then baked in a vacuum or regular oven at 80 °C for 2 hours (standard conditions; nitrocellulose or nylon membrane) or exposed to
405:
Southern blotting transfer may be used for homology-based cloning based on amino acid sequence of the protein product of the target gene.
295:
to a region of low water potential (usually filter paper and paper tissues) is then used to move the DNA from the gel onto the membrane;
378:
in the case of a radioactive or fluorescent probe, or by development of color on the membrane if a chromogenic detection method is used.
1033:
447:
Southern blotting can also be used to identify methylated sites in particular genes. Particularly useful are the restriction nucleases
762:"Edwin Southern, DNA blotting, and microarray technology: A case study of the shifting role of patents in academic molecular biology"
822:
Glenn, Gary; Andreou, Lefkothea-Vasiliki (2013-01-01), "Chapter Five - Analysis of DNA by
Southern Blotting", in Lorsch, Jon (ed.),
482:
Southern blotting is a time consuming method and can only estimate the size of the DNA since it is a semi-quantitative method.
1137:
1097:
495:
249:
the DNA fragments, breaking the DNA into smaller pieces, thereby allowing more efficient transfer from the gel to membrane.
657:"Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications"
419:
Can be used to find similar sequences in other species or in the genome by decreasing the specificity of hybridization.
841:
717:
349:
977:
1026:
441:
429:
It is useful in identifying changes that occur in genes including insertions, rearrangements, deletions, and
388:
size-fractionated DNA. It also allows for the fixation of the target-probe hybrids, required for analysis by
255:
If alkaline transfer methods are used, the DNA gel is placed into an alkaline solution (typically containing
604:
602:(5 November 1975). "Detection of specific sequences among DNA fragments separated by gel electrophoresis".
1142:
996:
823:
234:
172:
113:
23:
440:
in a restriction mapping. Also it is used to determine which recognition site has been altered due to a
192:
and
Kathleen Danna in 1971. The third innovation is the blotting-through method which was developed by
260:
180:
54:
1019:
357:
327:
96:. Briefly, purified DNA from a biological sample (such as blood or tissue) is digested with
668:
500:
423:
8:
585:
505:
337:
279:
238:
230:
121:
34:
672:
466:
Can be used in personal identification through fingerprinting, and in disease diagnosis.
914:
833:
796:
761:
637:
437:
97:
931:
Biochemistry 3rd
Edition, Matthews, Van Holde et al, Addison Wesley Publishing, pg 977
691:
656:
617:
151:) that employ similar principles, but using RNA or protein, have later been named for
1115:
1091:
918:
906:
898:
847:
837:
783:
742:
734:
730:
696:
629:
621:
530:
525:
242:
176:
148:
93:
19:
991:
641:
549:
1109:
1103:
888:
829:
791:
773:
726:
686:
676:
613:
288:
256:
193:
144:
287:
buffer is used to ensure a seal and prevent drying of the gel. Buffer transfer by
179:. Those restriction endonucleases are used to cut the DNA at a specific sequence.
1042:
406:
389:
375:
292:
101:
78:
422:
In a mixture having different sizes of digested DNA, it is used to identify the
599:
430:
366:
After hybridization, excess probe is washed from the membrane (typically using
271:
189:
163:. The names for other blotting methods may follow this convention, by analogy.
132:
128:
117:
73:
38:
1131:
1062:
902:
787:
778:
738:
625:
572:
510:
416:
Normal chromosomal or gene rearrangement can be studied using this technique.
341:
184:
140:
241:, then before blotting, the gel may be treated with an acid, such as dilute
1074:
1068:
910:
851:
681:
520:
515:
436:
Moreover it is used to identify a specific region that uses many different
345:
330:(nylon membrane) to permanently attach the transferred DNA to the membrane.
296:
246:
219:
136:
89:
61:
951:
893:
876:
746:
633:
237:
to separate them by size. If some of the DNA fragments are larger than 15
700:
410:
160:
66:
49:
455:, both of which recognize and cleave within the same sequence. However,
1006:
367:
284:
29:
760:
Tofano, Daidree; Wiechers, Ilse R.; Cook-Deegan, Robert (2006-08-15).
353:
106:
952:"Southern Blot- Definition, Principle, Steps, Results, Applications"
1011:
155:
directions as a sort of pun from
Southern's name. As the label is
116:-separated DNA fragments to a filter membrane in a process called
571:
This article incorporates text from this source, which is in the
152:
33:
Tray with a stack consisting top down of a weight, paper towels,
88:
is a method used for detection and quantification of a specific
393:
156:
371:
275:
42:
459:
requires that a C within that site be methylated, whereas
759:
485:
It cannot be used to detect mutations at base-pair level.
655:
Towbin; Staehelin, T; Gordon, J; et al. (1979).
654:
370:), and the pattern of hybridization is visualized on
875:Green, Michael R.; Sambrook, Joseph (July 2021).
828:, vol. 529, Academic Press, pp. 47–63,
479:High quality and large amounts of DNA are needed.
159:, Southern is capitalized, as is conventional of
1129:
127:The method is named after the British biologist
112:The Southern blotting combines the transfer of
1027:
874:
821:
382:
360:to reduce non-specific binding of the probe.
550:"Talking Glossary of Genetic Terms | NHGRI"
444:that changes a specific restriction enzyme.
120:, and the subsequent fragment detection by
1034:
1020:
892:
795:
777:
690:
680:
131:, who first published it in 1975. Other
45:, gel, salt solution and a slab of glass.
713:
598:
72:
60:
48:
28:
18:
1112:(lipid:post translational modification)
949:
92:in DNA samples. This method is used in
1130:
877:"Analysis of DNA by Southern Blotting"
102:separated by using an electric current
100:, and the resulting DNA fragments are
1015:
945:
943:
941:
939:
937:
1098:Electrophoretic mobility shift assay
1041:
870:
868:
866:
817:
815:
813:
811:
809:
807:
496:Gel electrophoresis of nucleic acids
13:
934:
834:10.1016/b978-0-12-418687-3.00005-7
433:that affect the restriction sites.
14:
1154:
1077:(post translational modification)
971:
863:
804:
566:
65:Southern blot agarose gel under
950:Sapkota, Anupama (2021-06-03).
556:. National Institutes of Health
399:
344:or tagging the molecule with a
925:
753:
707:
648:
592:
578:
542:
470:
442:single nucleotide polymorphism
1:
618:10.1016/S0022-2836(75)80083-0
536:
53:Southern blot membrane after
1138:Molecular biology techniques
997:Resources in other libraries
881:Cold Spring Harbor Protocols
766:Genomics, Society and Policy
731:10.1016/0003-2697(81)90281-5
605:Journal of Molecular Biology
7:
489:
229:The DNA fragments are then
10:
1159:
166:
1084:
1049:
992:Resources in your library
383:Interpretation of results
356:, and detergents such as
283:transferring by suction,
199:
779:10.1186/1746-5354-2-2-50
718:Analytical Biochemistry
682:10.1073/pnas.76.9.4350
600:Southern, Edwin Mellor
291:from a region of high
82:
70:
58:
46:
26:
16:DNA analysis technique
894:10.1101/pdb.top100396
328:ultraviolet radiation
223:37 °C overnight.
76:
64:
52:
32:
22:
887:(7): pdb.top100396.
501:Restriction fragment
424:restriction fragment
274:(or, alternatively,
227:Gel electrophoresis:
673:1979PNAS...76.4350T
506:Genetic fingerprint
438:restriction enzymes
426:of a specific size.
338:hybridization probe
122:probe hybridization
98:restriction enzymes
1143:Eponyms in biology
83:
71:
59:
47:
27:
1125:
1124:
1116:Northwestern blot
1106:(protein:protein)
1092:Southwestern blot
978:Library resources
531:Northwestern blot
526:Southwestern blot
149:southwestern blot
94:molecular biology
1150:
1110:Far-eastern blot
1104:Far-western blot
1043:Molecular probes
1036:
1029:
1022:
1013:
1012:
966:
965:
963:
962:
947:
932:
929:
923:
922:
896:
872:
861:
860:
859:
858:
819:
802:
801:
799:
781:
757:
751:
750:
711:
705:
704:
694:
684:
652:
646:
645:
596:
590:
589:
582:
576:
570:
569:
565:
563:
561:
546:
407:Oligonucleotides
289:capillary action
257:sodium hydroxide
194:Frederick Sanger
1158:
1157:
1153:
1152:
1151:
1149:
1148:
1147:
1128:
1127:
1126:
1121:
1080:
1045:
1040:
1003:
1002:
1001:
986:
985:
981:
974:
969:
960:
958:
948:
935:
930:
926:
873:
864:
856:
854:
844:
820:
805:
758:
754:
712:
708:
653:
649:
597:
593:
586:"Southern Blot"
584:
583:
579:
567:
559:
557:
548:
547:
543:
539:
492:
473:
431:point mutations
402:
390:autoradiography
385:
376:autoradiography
350:chromogenic dye
324:Immobilization:
293:water potential
231:electrophoresed
202:
169:
135:methods (i.e.,
114:electrophoresis
17:
12:
11:
5:
1156:
1146:
1145:
1140:
1123:
1122:
1120:
1119:
1113:
1107:
1101:
1095:
1088:
1086:
1082:
1081:
1079:
1078:
1072:
1066:
1060:
1053:
1051:
1047:
1046:
1039:
1038:
1031:
1024:
1016:
1010:
1009:
1000:
999:
994:
988:
987:
976:
975:
973:
972:External links
970:
968:
967:
933:
924:
862:
842:
803:
752:
725:(2): 195–203.
706:
647:
612:(3): 503–517.
591:
577:
554:www.genome.gov
540:
538:
535:
534:
533:
528:
523:
518:
513:
508:
503:
498:
491:
488:
487:
486:
483:
480:
477:
472:
469:
468:
467:
464:
445:
434:
427:
420:
417:
414:
401:
398:
384:
381:
380:
379:
361:
336:After that, a
334:Hybridization:
331:
321:
320:
319:
315:
311:
308:
304:
272:nitrocellulose
265:
250:
224:
216:DNA digestion:
213:
210:DNA Isolation:
201:
198:
190:Daniel Nathans
177:Hamilton Smith
168:
165:
129:Edwin Southern
77:Southern blot
39:nitrocellulose
15:
9:
6:
4:
3:
2:
1155:
1144:
1141:
1139:
1136:
1135:
1133:
1118:(RNA:protein)
1117:
1114:
1111:
1108:
1105:
1102:
1100:(DNA:protein)
1099:
1096:
1094:(protein:DNA)
1093:
1090:
1089:
1087:
1083:
1076:
1073:
1070:
1067:
1064:
1063:Northern blot
1061:
1058:
1057:Southern blot
1055:
1054:
1052:
1048:
1044:
1037:
1032:
1030:
1025:
1023:
1018:
1017:
1014:
1008:
1005:
1004:
998:
995:
993:
990:
989:
984:
983:Southern blot
979:
957:
956:Microbe Notes
953:
946:
944:
942:
940:
938:
928:
920:
916:
912:
908:
904:
900:
895:
890:
886:
882:
878:
871:
869:
867:
853:
849:
845:
843:9780124186873
839:
835:
831:
827:
826:
818:
816:
814:
812:
810:
808:
798:
793:
789:
785:
780:
775:
771:
767:
763:
756:
748:
744:
740:
736:
732:
728:
724:
720:
719:
710:
702:
698:
693:
688:
683:
678:
674:
670:
667:(9): 4350–4.
666:
662:
658:
651:
643:
639:
635:
631:
627:
623:
619:
615:
611:
607:
606:
601:
595:
587:
581:
574:
573:public domain
555:
551:
545:
541:
532:
529:
527:
524:
522:
519:
517:
514:
512:
511:Northern blot
509:
507:
504:
502:
499:
497:
494:
493:
484:
481:
478:
475:
474:
465:
462:
458:
454:
450:
446:
443:
439:
435:
432:
428:
425:
421:
418:
415:
412:
408:
404:
403:
397:
395:
391:
377:
373:
369:
365:
362:
359:
355:
351:
347:
343:
342:radioactivity
339:
335:
332:
329:
325:
322:
316:
312:
309:
305:
302:
301:
298:
294:
290:
286:
281:
277:
273:
269:
266:
262:
261:hybridization
258:
254:
253:Denaturation:
251:
248:
244:
240:
236:
232:
228:
225:
221:
220:endonucleases
217:
214:
211:
208:
207:
206:
197:
195:
191:
186:
185:Noreen Murray
182:
178:
174:
164:
162:
158:
154:
150:
146:
142:
141:northern blot
138:
134:
130:
125:
123:
119:
115:
110:
108:
103:
99:
95:
91:
87:
86:Southern blot
80:
79:autoradiogram
75:
69:illumination.
68:
63:
56:
55:hybridization
51:
44:
40:
36:
31:
25:
21:
1085:Interactions
1075:Eastern blot
1069:Western blot
1056:
982:
959:. Retrieved
955:
927:
884:
880:
855:, retrieved
824:
769:
765:
755:
722:
716:
709:
664:
660:
650:
609:
603:
594:
580:
558:. Retrieved
553:
544:
521:Eastern blot
516:Western blot
460:
456:
452:
448:
400:Applications
386:
363:
333:
323:
297:ion exchange
267:
252:
226:
218:Restriction
215:
209:
203:
170:
161:proper nouns
145:eastern blot
137:western blot
126:
111:
90:DNA sequence
85:
84:
57:and rinsing.
1007:OpenWetWare
471:Limitations
411:DNA library
346:fluorescent
270:A sheet of
247:depurinates
235:agarose gel
67:ultraviolet
24:Agarose gel
1132:Categories
961:2023-01-04
857:2023-01-04
560:24 January
537:References
368:SSC buffer
364:Detection:
1071:(protein)
919:235710916
903:1940-3402
788:1746-5354
739:0003-2697
626:0022-2836
354:formamide
268:Blotting:
173:Tom Kelly
157:eponymous
107:DNA probe
911:34210774
852:24011036
642:20126741
490:See also
374:film by
318:chamber.
280:membrane
264:results.
133:blotting
118:blotting
35:membrane
1050:General
797:5424904
747:6266278
669:Bibcode
634:1195397
307:buffer.
285:20X SSC
245:. This
181:Kenneth
167:History
153:compass
980:about
917:
909:
901:
850:
840:
794:
786:
745:
737:
701:388439
699:
692:411572
689:
640:
632:
624:
394:genome
233:on an
200:Method
1065:(RNA)
1059:(DNA)
915:S2CID
772:(2).
638:S2CID
457:HpaII
453:HpaII
372:X-ray
314:area.
276:nylon
43:nylon
907:PMID
899:ISSN
885:2021
848:PMID
838:ISBN
784:ISSN
743:PMID
735:ISSN
697:PMID
661:PNAS
630:PMID
622:ISSN
562:2023
461:MspI
451:and
449:MspI
183:and
175:and
889:doi
830:doi
825:DNA
792:PMC
774:doi
727:doi
723:112
687:PMC
677:doi
614:doi
358:SDS
348:or
243:HCl
41:or
37:of
1134::
954:.
936:^
913:.
905:.
897:.
883:.
879:.
865:^
846:,
836:,
806:^
790:.
782:.
768:.
764:.
741:.
733:.
721:.
695:.
685:.
675:.
665:76
663:.
659:.
636:.
628:.
620:.
610:98
608:.
552:.
278:)
239:kb
147:,
143:,
139:,
124:.
1035:e
1028:t
1021:v
964:.
921:.
891::
832::
800:.
776::
770:2
749:.
729::
703:.
679::
671::
644:.
616::
588:.
575:.
564:.
81:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.