544:(insertion or deletion), or chromosomal rearrangement; any such errors may render the gene products coded at that location non-functional. Because this activity can vary depending on the species, cell type, target gene, and nuclease used, it should be monitored when designing new systems. A simple heteroduplex cleavage assay can be run which detects any difference between two alleles amplified by PCR. Cleavage products can be visualized on simple agarose gels or slab gel systems.
64:
20:
462:
cleavage activity. The FokI domain functions as a dimer, requiring two constructs with unique DNA binding domains for sites in the target genome with proper orientation and spacing. Both the number of amino acid residues between the TALE DNA binding domain and the FokI cleavage domain and the number of bases between the two individual TALEN binding sites appear to be important parameters for achieving high levels of activity.
495:
520:
and enter the nucleus to access the genome. Alternatively, TALEN constructs can be delivered to the cells as mRNAs, which removes the possibility of genomic integration of the TALEN-expressing protein. Using an mRNA vector can also dramatically increase the level of homology directed repair (HDR) and
642:
The off-target activity of an active nuclease may lead to unwanted double-strand breaks and may consequently yield chromosomal rearrangements and/or cell death. Studies have been carried out to compare the relative nuclease-associated toxicity of available technologies. Based on these studies and
461:
that are active in a yeast assay. These reagents are also active in plant cells and in animal cells. Initial TALEN studies used the wild-type FokI cleavage domain, but some subsequent TALEN studies also used FokI cleavage domain variants with mutations designed to improve cleavage specificity and
444:
recognition. This straightforward relationship between amino acid sequence and DNA recognition has allowed for the engineering of specific DNA-binding domains by selecting a combination of repeat segments containing the appropriate RVDs. Notably, slight changes in the RVD and the incorporation of
633:
relies on ribonucleotide complex formation instead of protein/DNA recognition. gRNAs have occasionally limitations regarding feasibility due to lack of PAM sites in the target sequence and even though they can be cheaply produced, the current development lead to a remarkable decrease of cost for
486:
oligonucleotide assembly followed by whole gene amplification. A number of modular assembly schemes for generating engineered TALE constructs have also been reported. Both methods offer a systematic approach to engineering DNA binding domains that is conceptually similar to the modular assembly
498:
Workflow of genome editing of Your
Favorite Gene (YFG) using TALEN. The target sequence is identified, a corresponding TALEN sequence is engineered and inserted into a plasmid. The plasmid is inserted into the target cell where it is translated to produce the functional TALEN, which enters the
622:. The DNA binding region of a TAL effector can be combined with the cleavage domain of a meganuclease to create a hybrid architecture combining the ease of engineering and highly specific DNA binding activity of a TAL effector with the low site frequency and specificity of a meganuclease.
588:. Moreover, the method can be used to generate knockin organisms. Wu et al.obtained a Sp110 knockin cattle using Talen nickases to induce increased resistance of tuberculosis. This approach has also been used to generate knockin rats by TALEN mRNA microinjection in one-cell embryos.
380:
which cuts DNA strands). Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in
1146:
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ (February 2011). "A TALE nuclease architecture for efficient genome editing".
629:, TALEN recognizes single nucleotides. It's far more straightforward to engineer interactions between TALEN DNA binding domains and their target nucleotides than it is to create interactions with ZFNs and their target nucleotide triplets. On the other hand,
2561:
Poirot L, Philip B, Schiffer-Mannioui C, Le Clerre D, Chion-Sotinel I, Derniame S, Potrel P, Bas C, Lemaire L, Galetto R, Lebuhotel C, Eyquem J, Cheung GW, Duclert A, Gouble A, Arnould S, Peggs K, Pule M, Scharenberg AM, Smith J (September 2015).
607:. Recently, it was shown that TALEN can be used as tools to harness the immune system to fight cancers; TALEN-mediated targeting can generate T cells that are resistant to chemotherapeutic drugs and show anti-tumor activity.
610:
In theory, the genome-wide specificity of engineered TALEN fusions allows for correction of errors at individual genetic loci via homology-directed repair from a correct exogenous template. In reality, however, the
1394:
Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC (January 2011). "Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures".
561:
TALEN has been used to efficiently modify plant genomes, creating economically important food crops with favorable nutritional qualities. They have also been harnessed to develop tools for the production of
499:
nucleus and binds and cleaves the target sequence. Depending on the application, this can be used to introduce an error (to knock out a target gene) or to introduce a new DNA sequence into the target gene.
1195:
Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R (July 2011).
2463:
Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von Kalle C, Carlson DF, Maeder ML, Joung JK, Wagner JE, Voytas DF, Blazar BR, Tolar J (June 2013).
540:(NHEJ) directly ligates DNA from either side of a double-strand break where there is very little or no sequence overlap for annealing. This repair mechanism induces errors in the genome via
440:
sequence with divergent 12th and 13th amino acids. These two positions, referred to as the Repeat
Variable Diresidue (RVD), are highly variable and show a strong correlation with specific
767:
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (December 2009). "Breaking the code of DNA binding specificity of TAL-type III effectors".
643:
the maximal theoretical distance between DNA binding and nuclease activity, TALEN constructs are believed to have the greatest precision of the currently available technologies.
2369:
Ramalingam S, Annaluru N, Kandavelou K, Chandrasegaran S (2014). "TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells".
615:
application of TALEN is currently limited by the lack of an efficient delivery mechanism, unknown immunogenic factors, and uncertainty in the specificity of TALEN binding.
1988:
Daboussi F, Leduc S, Maréchal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, Voytas DF, Cavarec L, Duchateau P (May 2014).
470:
The simple relationship between amino acid sequence and DNA recognition of the TALE binding domain allows for the efficient engineering of proteins. In this case,
1247:
Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ (July 2011).
345:
1947:
Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, Mathis L, Voytas DF, Zhang F (September 2014).
478:
of the repetitive sequence found in the TALE binding domain. One solution to this is to use a publicly available software program (DNAWorks) to calculate
1026:"De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks"
2040:
Wienert B, Funnell AP, Norton LJ, Pearson RC, Wilkinson-White LE, Lester K, Vadolas J, Porteus MH, Matthews JM, Quinlan KG, Crossley M (2015).
2603:
Boissel S, Jarjour J, Astrakhan A, Adey A, Gouble A, Duchateau P, Shendure J, Stoddard BL, Certo MT, Baker D, Scharenberg AM (February 2014).
1307:
Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ (August 2011).
2652:
2404:
Dupuy A, Valton J, Leduc S, Armier J, Galetto R, Gouble A, Lebuhotel C, Stary A, Pâques F, Duchateau P, Sarasin A, Daboussi F (2013).
553:
can also introduce foreign DNA at the DSB as the transfected double-stranded sequences are used as templates for the repair enzymes.
338:
652:
396:
294:
869:
Juillerat A, Pessereau C, Dubois G, Guyot V, Maréchal A, Valton J, Daboussi F, Poirot L, Duclert A, Duchateau P (January 2015).
1351:
Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B (August 2011). "Heritable gene targeting in zebrafish using customized TALENs".
591:
TALEN has also been utilized experimentally to correct the genetic errors that underlie disease. For example, it has been used
1658:
547:
Alternatively, DNA can be introduced into a genome through NHEJ in the presence of exogenous double-stranded DNA fragments.
331:
534:
TALEN can be used to edit genomes by inducing double-strand breaks (DSB), which cells respond to with repair mechanisms.
1088:
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (July 2011).
2747:
2732:
166:
2671:
625:
In comparison to other genome editing techniques, TALEN falls in the middle in terms of difficulty and cost. Unlike
1733:"Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes"
2042:"Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin"
2712:
2091:"Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes"
176:
2190:
194:
159:
618:
Another emerging application of TALEN is its ability to combine with other genome engineering tools, such as
571:
85:
76:
2722:
2564:"Multiplex Genome-Edited T-cell Manufacturing Platform for "Off-the-Shelf" Adoptive T-cell Immunotherapies"
136:
45:
2512:
Valton J, Guyot V, Marechal A, Filhol JM, Juillerat A, Duclert A, Duchateau P, Poirot L (September 2015).
732:
Boch J, Bonas U (September 2010). "Xanthomonas AvrBs3 family-type III effectors: discovery and function".
1090:"Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting"
475:
429:
141:
112:
102:
97:
2737:
1490:"Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases"
926:
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (October 2010).
537:
261:
126:
121:
90:
1542:"A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity"
1440:"Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases"
1439:
818:
Moscou MJ, Bogdanove AJ (December 2009). "A simple cipher governs DNA recognition by TAL effectors".
483:
471:
266:
216:
131:
107:
550:
1594:"Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription"
2742:
2727:
1949:"Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family"
299:
226:
171:
977:"TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain"
576:
1780:
Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J (2011). Shiu SH (ed.).
1641:
Hoover D (2012). "Using DNAWorks in
Designing Oligonucleotides for PCR-Based Gene Synthesis".
2199:"TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis"
1898:
Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF (January 2013).
634:
TALENs, so that they are in a similar price and time range like CRISPR based genome editing.
604:
600:
400:
1731:
Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B (August 2011).
2417:
2269:
2210:
2102:
2053:
2001:
1900:"Transcription activator-like effector nucleases enable efficient plant genome engineering"
1852:
1793:
1260:
1037:
882:
827:
776:
657:
626:
314:
8:
596:
567:
54:
2421:
2273:
2214:
2106:
2057:
2005:
1856:
1797:
1264:
1041:
886:
831:
780:
745:
2629:
2604:
2538:
2513:
2489:
2464:
2440:
2405:
2346:
2321:
2292:
2257:
2233:
2198:
2174:
2149:
2125:
2090:
1924:
1899:
1875:
1840:
1816:
1781:
1757:
1732:
1708:
1683:
1618:
1593:
1566:
1541:
1514:
1489:
1470:
1420:
1376:
1281:
1248:
1221:
1196:
1172:
1114:
1089:
1060:
1025:
1001:
976:
952:
927:
903:
870:
851:
800:
714:
373:
366:
2514:"A Multidrug-resistant Engineered CAR T Cell for Allogeneic Combination Immunotherapy"
2382:
1540:
Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T (November 2011).
309:
23:
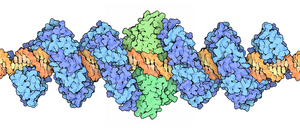
Spacefill drawing of dimeric TALE-FokI fusion (blue: TALE; green: FokI) bound to DNA (
2634:
2585:
2543:
2494:
2445:
2386:
2351:
2297:
2238:
2179:
2130:
2071:
2019:
1970:
1929:
1880:
1821:
1762:
1713:
1664:
1654:
1623:
1571:
1519:
1462:
1412:
1368:
1330:
1286:
1226:
1164:
1119:
1065:
1006:
957:
908:
843:
804:
792:
749:
706:
319:
199:
25:
2605:"megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering"
2406:"Targeted gene therapy of xeroderma pigmentosum cells using meganuclease and TALEN™"
2148:
Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR (August 2011).
1990:"Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology"
1839:
Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S (2011). Bendahmane M (ed.).
1474:
1424:
1380:
1176:
2624:
2616:
2575:
2533:
2525:
2484:
2476:
2435:
2425:
2378:
2341:
2333:
2287:
2277:
2228:
2218:
2169:
2161:
2120:
2110:
2061:
2009:
1960:
1919:
1911:
1870:
1860:
1811:
1801:
1752:
1744:
1703:
1695:
1646:
1613:
1605:
1561:
1553:
1509:
1501:
1454:
1404:
1360:
1320:
1276:
1268:
1216:
1208:
1156:
1109:
1101:
1055:
1045:
996:
988:
947:
939:
898:
890:
855:
835:
784:
741:
698:
479:
369:
that can be engineered to cut specific sequences of DNA. They are made by fusing a
2580:
2563:
718:
2430:
2282:
2115:
1865:
1806:
1650:
975:
Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B (January 2011).
517:
433:
204:
2696:
943:
2203:
Proceedings of the
National Academy of Sciences of the United States of America
1438:
Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T (July 2007).
1030:
Proceedings of the
National Academy of Sciences of the United States of America
408:
382:
304:
233:
2258:"Generation of TALEN-mediated GRdim knock-in rats by homologous recombination"
1505:
2706:
2150:"Targeted gene disruption in somatic zebrafish cells using engineered TALENs"
1782:"Transcriptional activators of human genes with programmable DNA-specificity"
1024:
Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK (February 2011).
278:
211:
2223:
2089:
Davies B, Davies G, Preece C, Puliyadi R, Szumska D, Bhattacharya S (2013).
1272:
1050:
839:
788:
2717:
2638:
2589:
2547:
2498:
2449:
2390:
2355:
2301:
2242:
2183:
2134:
2075:
2023:
1974:
1933:
1884:
1825:
1766:
1717:
1668:
1627:
1592:
Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P (February 2011).
1575:
1523:
1466:
1416:
1372:
1334:
1290:
1230:
1168:
1123:
1069:
1010:
961:
912:
847:
796:
753:
710:
662:
619:
581:
513:
419:
370:
273:
221:
63:
2620:
2368:
1915:
1748:
1699:
1557:
1105:
992:
488:
424:
19:
2529:
2337:
2480:
2066:
2041:
2014:
1989:
1408:
441:
437:
2560:
1965:
1948:
894:
508:
Once the TALEN constructs have been assembled, they are inserted into
33:
29:
2165:
1684:"Assembly of custom TALE-type DNA binding domains by modular cloning"
1609:
1364:
1325:
1308:
1212:
1197:"Genetic engineering of human pluripotent cells using TALE nucleases"
1160:
702:
585:
2256:
Ponce de León V, Mérillat AM, Tesson L, Anegón I, Hummler E (2014).
1458:
574:(IPSCs) clones and human erythroid cell lines, to generate knockout
436:. The DNA binding domain contains a repeated highly conserved 33–34
871:"Optimized tuning of TALEN specificity using non-conventional RVDs"
566:. In addition, it has been used to engineer stably modified human
458:
445:"nonconventional" RVD sequences can improve targeting specificity.
377:
2197:
Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y (March 2015).
2255:
563:
509:
391:
1645:. Methods in Molecular Biology. Vol. 852. pp. 215–23.
1539:
928:"Targeting DNA double-strand breaks with TAL effector nucleases"
2699:
An entry in the
Protein Database's monthly structural highlight
1681:
667:
630:
404:
386:
243:
1249:"Targeted genome editing across species using ZFNs and TALENs"
1437:
868:
541:
494:
238:
2672:"Boston Consulting Group - Report on Gene Editing Precision"
1309:"Knockout rats generated by embryo microinjection of TALENs"
1023:
766:
595:
to correct the genetic defects that cause disorders such as
2602:
2039:
1987:
1841:"Assembly of designer TAL effectors by Golden Gate cloning"
1779:
1682:
Morbitzer R, Elsaesser J, Hausner J, Lahaye T (July 2011).
925:
454:
2462:
2088:
1838:
2690:
2511:
1946:
453:
The non-specific DNA cleavage domain from the end of the
2319:
1087:
2465:"TALEN-based gene correction for epidermolysis bullosa"
2403:
2320:
Carlson DF, Fahrenkrug SC, Hackett PB (January 2012).
2147:
1393:
1306:
1194:
1591:
689:
Boch J (February 2011). "TALEs of genome targeting".
725:
1246:
1145:
1897:
974:
637:
521:the success of introgression during gene editing.
2141:
1730:
1190:
1188:
1186:
2704:
1535:
1533:
1350:
2653:"Pros and Cons Of ZFNS, TALENS, AND CRISPR/CAS"
2196:
2035:
2033:
817:
465:
359:Transcription activator-like effector nucleases
1183:
1675:
1530:
1302:
1300:
1141:
1139:
1137:
1135:
1133:
516:with the plasmids, and the gene products are
457:endonuclease can be used to construct hybrid
339:
2030:
1773:
1587:
1585:
1487:
1431:
1346:
1344:
1083:
1081:
1079:
919:
407:, TALEN is a prominent tool in the field of
2669:
1242:
1240:
2362:
2315:
2313:
2311:
1634:
1297:
1130:
684:
682:
414:
346:
332:
2628:
2579:
2537:
2488:
2439:
2429:
2345:
2291:
2281:
2232:
2222:
2173:
2124:
2114:
2065:
2013:
1964:
1923:
1874:
1864:
1832:
1815:
1805:
1756:
1707:
1617:
1582:
1565:
1513:
1387:
1341:
1324:
1280:
1220:
1113:
1076:
1059:
1049:
1000:
951:
902:
731:
1237:
653:Genome editing with engineered nucleases
493:
397:genome editing with engineered nucleases
18:
16:Enzymes that cleave DNA in specific ways
2322:"Targeting DNA With Fingers and TALENs"
2308:
1481:
679:
295:Genetically modified food controversies
2705:
2456:
1724:
1640:
448:
2693:A comprehensive tool for TALEN design
2397:
1488:Guo J, Gaj T, Barbas CF (July 2010).
688:
482:suitable for assembly in a two step
746:10.1146/annurev-phyto-080508-081936
474:is problematic because of improper
13:
422:are proteins that are secreted by
14:
2759:
2684:
2670:Boglioli, Elsy; Richard, Magali.
2383:10.2174/1566523214666140918101725
524:
2326:Molecular Therapy: Nucleic Acids
62:
2663:
2645:
2596:
2554:
2505:
2249:
2082:
1981:
1940:
1891:
734:Annual Review of Phytopathology
638:TAL effector nuclease precision
556:
503:
177:Cartagena Protocol on Biosafety
1017:
968:
862:
811:
760:
584:, knockout mice, and knockout
77:Genetically modified organisms
1:
2581:10.1158/0008-5472.CAN-14-3321
673:
572:induced pluripotent stem cell
529:
2431:10.1371/journal.pone.0078678
2283:10.1371/journal.pone.0088146
2116:10.1371/journal.pone.0060216
1866:10.1371/journal.pone.0019722
1807:10.1371/journal.pone.0019509
1651:10.1007/978-1-61779-564-0_16
1494:Journal of Molecular Biology
512:; the target cells are then
466:Engineering TALEN constructs
376:to a DNA cleavage domain (a
7:
1953:Plant Biotechnology Journal
944:10.1534/genetics.110.120717
646:
37:), by David Goodsell
10:
2764:
538:Non-homologous end joining
262:Genetically modified crops
2697:PDB Molecule of the Month
1506:10.1016/j.jmb.2010.04.060
491:DNA recognition domains.
472:artificial gene synthesis
430:type III secretion system
2748:Repetitive DNA sequences
2733:History of biotechnology
551:Homology directed repair
2224:10.1073/pnas.1421587112
1273:10.1126/science.1207773
1051:10.1073/pnas.1019533108
840:10.1126/science.1178817
789:10.1126/science.1178811
415:TALE DNA-binding domain
395:, a technique known as
300:GMO conspiracy theories
172:Substantial equivalence
2713:Biological engineering
2657:The Jackson Laboratory
2609:Nucleic Acids Research
1737:Nucleic Acids Research
1688:Nucleic Acids Research
1546:Nucleic Acids Research
1094:Nucleic Acids Research
981:Nucleic Acids Research
500:
487:method for generating
152:History and regulation
38:
2046:Nature Communications
1994:Nature Communications
1916:10.1104/pp.112.205179
605:epidermolysis bullosa
601:xeroderma pigmentosum
497:
401:zinc finger nucleases
22:
2371:Current Gene Therapy
2154:Nature Biotechnology
1598:Nature Biotechnology
1447:Nature Biotechnology
1353:Nature Biotechnology
1313:Nature Biotechnology
1201:Nature Biotechnology
1149:Nature Biotechnology
691:Nature Biotechnology
658:Zinc finger nuclease
315:StarLink corn recall
2723:Genetic engineering
2621:10.1093/nar/gkt1224
2530:10.1038/mt.2015.104
2422:2013PLoSO...878678D
2338:10.1038/mtna.2011.5
2274:2014PLoSO...988146P
2215:2015PNAS..112E1530W
2107:2013PLoSO...860216D
2058:2015NatCo...6.7085W
2006:2014NatCo...5.3831D
1857:2011PLoSO...619722W
1798:2011PLoSO...619509G
1265:2011Sci...333..307W
1042:2011PNAS..108.2623M
887:2015NatSR...5E8150J
832:2009Sci...326.1501M
781:2009Sci...326.1509B
597:sickle cell disease
568:embryonic stem cell
449:DNA cleavage domain
428:bacteria via their
367:restriction enzymes
55:Genetic engineering
2481:10.1038/mt.2013.56
2067:10.1038/ncomms8085
2015:10.1038/ncomms4831
1749:10.1093/nar/gkr188
1700:10.1093/nar/gkr151
1558:10.1093/nar/gkr597
1409:10.1038/nmeth.1539
1106:10.1093/nar/gkr218
993:10.1093/nar/gkq704
875:Scientific Reports
501:
374:DNA-binding domain
39:
2738:Molecular biology
2518:Molecular Therapy
2469:Molecular Therapy
1966:10.1111/pbi.12201
1660:978-1-61779-563-3
895:10.1038/srep08150
775:(5959): 1509–12.
356:
355:
320:He Jiankui affair
200:Molecular cloning
2755:
2679:
2678:
2676:
2667:
2661:
2660:
2649:
2643:
2642:
2632:
2600:
2594:
2593:
2583:
2558:
2552:
2551:
2541:
2509:
2503:
2502:
2492:
2460:
2454:
2453:
2443:
2433:
2401:
2395:
2394:
2366:
2360:
2359:
2349:
2317:
2306:
2305:
2295:
2285:
2253:
2247:
2246:
2236:
2226:
2194:
2188:
2187:
2177:
2166:10.1038/nbt.1934
2145:
2139:
2138:
2128:
2118:
2086:
2080:
2079:
2069:
2037:
2028:
2027:
2017:
1985:
1979:
1978:
1968:
1944:
1938:
1937:
1927:
1904:Plant Physiology
1895:
1889:
1888:
1878:
1868:
1836:
1830:
1829:
1819:
1809:
1777:
1771:
1770:
1760:
1728:
1722:
1721:
1711:
1679:
1673:
1672:
1638:
1632:
1631:
1621:
1610:10.1038/nbt.1775
1589:
1580:
1579:
1569:
1537:
1528:
1527:
1517:
1485:
1479:
1478:
1444:
1435:
1429:
1428:
1391:
1385:
1384:
1365:10.1038/nbt.1939
1348:
1339:
1338:
1328:
1326:10.1038/nbt.1940
1304:
1295:
1294:
1284:
1244:
1235:
1234:
1224:
1213:10.1038/nbt.1927
1192:
1181:
1180:
1161:10.1038/nbt.1755
1143:
1128:
1127:
1117:
1085:
1074:
1073:
1063:
1053:
1021:
1015:
1014:
1004:
972:
966:
965:
955:
923:
917:
916:
906:
866:
860:
859:
815:
809:
808:
764:
758:
757:
729:
723:
722:
703:10.1038/nbt.1767
686:
480:oligonucleotides
348:
341:
334:
66:
41:
40:
36:
2763:
2762:
2758:
2757:
2756:
2754:
2753:
2752:
2703:
2702:
2687:
2682:
2674:
2668:
2664:
2651:
2650:
2646:
2615:(4): 2591–601.
2601:
2597:
2574:(18): 3853–64.
2568:Cancer Research
2559:
2555:
2510:
2506:
2461:
2457:
2402:
2398:
2367:
2363:
2318:
2309:
2254:
2250:
2209:(13): E1530-9.
2195:
2191:
2146:
2142:
2087:
2083:
2038:
2031:
1986:
1982:
1945:
1941:
1896:
1892:
1837:
1833:
1778:
1774:
1743:(14): 6315–25.
1729:
1725:
1680:
1676:
1661:
1639:
1635:
1590:
1583:
1552:(21): 9283–93.
1538:
1531:
1486:
1482:
1459:10.1038/nbt1317
1442:
1436:
1432:
1392:
1388:
1349:
1342:
1305:
1298:
1245:
1238:
1193:
1184:
1144:
1131:
1086:
1077:
1022:
1018:
973:
969:
924:
920:
867:
863:
816:
812:
765:
761:
730:
726:
687:
680:
676:
649:
640:
559:
532:
527:
506:
468:
451:
417:
352:
310:Séralini affair
205:Recombinant DNA
24:
17:
12:
11:
5:
2761:
2751:
2750:
2745:
2743:Non-coding RNA
2740:
2735:
2730:
2728:Genome editing
2725:
2720:
2715:
2701:
2700:
2694:
2686:
2685:External links
2683:
2681:
2680:
2662:
2644:
2595:
2553:
2524:(9): 1507–18.
2504:
2455:
2416:(11): e78678.
2396:
2361:
2307:
2248:
2189:
2140:
2081:
2029:
1980:
1939:
1890:
1831:
1772:
1723:
1694:(13): 5790–9.
1674:
1659:
1643:Gene Synthesis
1633:
1581:
1529:
1480:
1430:
1397:Nature Methods
1386:
1359:(8): 699–700.
1340:
1296:
1236:
1182:
1129:
1075:
1016:
967:
918:
861:
826:(5959): 1501.
810:
759:
724:
677:
675:
672:
671:
670:
665:
660:
655:
648:
645:
639:
636:
558:
555:
531:
528:
526:
525:Genome editing
523:
505:
502:
467:
464:
450:
447:
416:
413:
409:genome editing
354:
353:
351:
350:
343:
336:
328:
325:
324:
323:
322:
317:
312:
307:
305:Pusztai affair
302:
297:
289:
288:
284:
283:
282:
281:
276:
271:
270:
269:
256:
255:
251:
250:
249:
248:
247:
246:
241:
234:Genome editing
231:
230:
229:
224:
219:
217:Transformation
209:
208:
207:
197:
189:
188:
184:
183:
182:
181:
180:
179:
174:
163:
162:
154:
153:
149:
148:
147:
146:
145:
144:
139:
134:
129:
118:
117:
116:
115:
110:
105:
94:
93:
88:
80:
79:
73:
72:
68:
67:
59:
58:
50:
49:
15:
9:
6:
4:
3:
2:
2760:
2749:
2746:
2744:
2741:
2739:
2736:
2734:
2731:
2729:
2726:
2724:
2721:
2719:
2716:
2714:
2711:
2710:
2708:
2698:
2695:
2692:
2689:
2688:
2673:
2666:
2659:. March 2014.
2658:
2654:
2648:
2640:
2636:
2631:
2626:
2622:
2618:
2614:
2610:
2606:
2599:
2591:
2587:
2582:
2577:
2573:
2569:
2565:
2557:
2549:
2545:
2540:
2535:
2531:
2527:
2523:
2519:
2515:
2508:
2500:
2496:
2491:
2486:
2482:
2478:
2475:(6): 1151–9.
2474:
2470:
2466:
2459:
2451:
2447:
2442:
2437:
2432:
2427:
2423:
2419:
2415:
2411:
2407:
2400:
2392:
2388:
2384:
2380:
2377:(6): 461–72.
2376:
2372:
2365:
2357:
2353:
2348:
2343:
2339:
2335:
2331:
2327:
2323:
2316:
2314:
2312:
2303:
2299:
2294:
2289:
2284:
2279:
2275:
2271:
2268:(2): e88146.
2267:
2263:
2259:
2252:
2244:
2240:
2235:
2230:
2225:
2220:
2216:
2212:
2208:
2204:
2200:
2193:
2185:
2181:
2176:
2171:
2167:
2163:
2159:
2155:
2151:
2144:
2136:
2132:
2127:
2122:
2117:
2112:
2108:
2104:
2101:(3): e60216.
2100:
2096:
2092:
2085:
2077:
2073:
2068:
2063:
2059:
2055:
2051:
2047:
2043:
2036:
2034:
2025:
2021:
2016:
2011:
2007:
2003:
1999:
1995:
1991:
1984:
1976:
1972:
1967:
1962:
1959:(7): 934–40.
1958:
1954:
1950:
1943:
1935:
1931:
1926:
1921:
1917:
1913:
1909:
1905:
1901:
1894:
1886:
1882:
1877:
1872:
1867:
1862:
1858:
1854:
1851:(5): e19722.
1850:
1846:
1842:
1835:
1827:
1823:
1818:
1813:
1808:
1803:
1799:
1795:
1792:(5): e19509.
1791:
1787:
1783:
1776:
1768:
1764:
1759:
1754:
1750:
1746:
1742:
1738:
1734:
1727:
1719:
1715:
1710:
1705:
1701:
1697:
1693:
1689:
1685:
1678:
1670:
1666:
1662:
1656:
1652:
1648:
1644:
1637:
1629:
1625:
1620:
1615:
1611:
1607:
1604:(2): 149–53.
1603:
1599:
1595:
1588:
1586:
1577:
1573:
1568:
1563:
1559:
1555:
1551:
1547:
1543:
1536:
1534:
1525:
1521:
1516:
1511:
1507:
1503:
1500:(1): 96–107.
1499:
1495:
1491:
1484:
1476:
1472:
1468:
1464:
1460:
1456:
1453:(7): 786–93.
1452:
1448:
1441:
1434:
1426:
1422:
1418:
1414:
1410:
1406:
1402:
1398:
1390:
1382:
1378:
1374:
1370:
1366:
1362:
1358:
1354:
1347:
1345:
1336:
1332:
1327:
1322:
1318:
1314:
1310:
1303:
1301:
1292:
1288:
1283:
1278:
1274:
1270:
1266:
1262:
1259:(6040): 307.
1258:
1254:
1250:
1243:
1241:
1232:
1228:
1223:
1218:
1214:
1210:
1206:
1202:
1198:
1191:
1189:
1187:
1178:
1174:
1170:
1166:
1162:
1158:
1154:
1150:
1142:
1140:
1138:
1136:
1134:
1125:
1121:
1116:
1111:
1107:
1103:
1099:
1095:
1091:
1084:
1082:
1080:
1071:
1067:
1062:
1057:
1052:
1047:
1043:
1039:
1036:(6): 2623–8.
1035:
1031:
1027:
1020:
1012:
1008:
1003:
998:
994:
990:
987:(1): 359–72.
986:
982:
978:
971:
963:
959:
954:
949:
945:
941:
938:(2): 757–61.
937:
933:
929:
922:
914:
910:
905:
900:
896:
892:
888:
884:
880:
876:
872:
865:
857:
853:
849:
845:
841:
837:
833:
829:
825:
821:
814:
806:
802:
798:
794:
790:
786:
782:
778:
774:
770:
763:
755:
751:
747:
743:
739:
735:
728:
720:
716:
712:
708:
704:
700:
696:
692:
685:
683:
678:
669:
666:
664:
661:
659:
656:
654:
651:
650:
644:
635:
632:
628:
623:
621:
620:meganucleases
616:
614:
608:
606:
602:
598:
594:
589:
587:
583:
582:knockout rats
579:
578:
573:
569:
565:
554:
552:
548:
545:
543:
539:
535:
522:
519:
515:
511:
496:
492:
490:
485:
481:
477:
473:
463:
460:
456:
446:
443:
439:
435:
434:infect plants
431:
427:
426:
421:
420:TAL effectors
412:
410:
406:
402:
398:
394:
393:
388:
384:
379:
375:
372:
368:
364:
360:
349:
344:
342:
337:
335:
330:
329:
327:
326:
321:
318:
316:
313:
311:
308:
306:
303:
301:
298:
296:
293:
292:
291:
290:
287:Controversies
286:
285:
280:
279:Designer baby
277:
275:
272:
268:
265:
264:
263:
260:
259:
258:
257:
253:
252:
245:
242:
240:
237:
236:
235:
232:
228:
225:
223:
220:
218:
215:
214:
213:
212:Gene delivery
210:
206:
203:
202:
201:
198:
196:
193:
192:
191:
190:
186:
185:
178:
175:
173:
170:
169:
168:
165:
164:
161:
158:
157:
156:
155:
151:
150:
143:
140:
138:
135:
133:
130:
128:
125:
124:
123:
120:
119:
114:
111:
109:
106:
104:
101:
100:
99:
96:
95:
92:
89:
87:
84:
83:
82:
81:
78:
75:
74:
70:
69:
65:
61:
60:
57:
56:
52:
51:
47:
43:
42:
35:
31:
27:
21:
2665:
2656:
2647:
2612:
2608:
2598:
2571:
2567:
2556:
2521:
2517:
2507:
2472:
2468:
2458:
2413:
2409:
2399:
2374:
2370:
2364:
2329:
2325:
2265:
2261:
2251:
2206:
2202:
2192:
2160:(8): 697–8.
2157:
2153:
2143:
2098:
2094:
2084:
2049:
2045:
1997:
1993:
1983:
1956:
1952:
1942:
1907:
1903:
1893:
1848:
1844:
1834:
1789:
1785:
1775:
1740:
1736:
1726:
1691:
1687:
1677:
1642:
1636:
1601:
1597:
1549:
1545:
1497:
1493:
1483:
1450:
1446:
1433:
1400:
1396:
1389:
1356:
1352:
1319:(8): 695–6.
1316:
1312:
1256:
1252:
1207:(8): 731–4.
1204:
1200:
1155:(2): 143–8.
1152:
1148:
1097:
1093:
1033:
1029:
1019:
984:
980:
970:
935:
931:
921:
878:
874:
864:
823:
819:
813:
772:
768:
762:
737:
733:
727:
697:(2): 135–6.
694:
690:
663:Meganuclease
641:
624:
617:
612:
609:
592:
590:
575:
560:
557:Applications
549:
546:
536:
533:
507:
504:Transfection
469:
452:
423:
418:
399:. Alongside
390:
383:gene editing
371:TAL effector
362:
358:
357:
274:Gene therapy
254:Applications
227:Transduction
222:Transfection
53:
2691:E-TALEN.org
1910:(1): 20–7.
1403:(1): 74–9.
1100:(12): e82.
514:transfected
489:zinc finger
425:Xanthomonas
405:CRISPR/Cas9
2707:Categories
740:: 419–36.
674:References
577:C. elegans
530:Mechanisms
442:nucleotide
438:amino acid
432:when they
195:Techniques
167:Regulation
127:Maize/corn
2332:(3): e3.
805:206522347
586:zebrafish
518:expressed
476:annealing
459:nucleases
2639:24285304
2590:26183927
2548:26061646
2499:23546300
2450:24236034
2410:PLOS ONE
2391:25245091
2356:23344620
2302:24523878
2262:PLOS ONE
2243:25733846
2184:21822241
2135:23555929
2095:PLOS ONE
2076:25971621
2052:: 7085.
2024:24871200
2000:: 3831.
1975:24851712
1934:23124327
1885:21625552
1845:PLOS ONE
1826:21625585
1786:PLOS ONE
1767:21459844
1718:21421566
1669:22328436
1628:21248753
1576:21813459
1524:20447404
1475:22079561
1467:17603476
1425:14334237
1417:21131970
1381:28802632
1373:21822242
1335:21822240
1291:21700836
1231:21738127
1177:53549397
1169:21179091
1124:21493687
1070:21262818
1011:20699274
962:20660643
932:Genetics
913:25632877
881:: 8150.
848:19933106
797:19933107
754:19400638
711:21301438
647:See also
593:in vitro
564:biofuels
510:plasmids
389:editing
378:nuclease
86:Bacteria
46:a series
44:Part of
2630:3936731
2539:4817890
2490:3677309
2441:3827243
2418:Bibcode
2347:3381595
2293:3921256
2270:Bibcode
2234:4386332
2211:Bibcode
2175:3154023
2126:3610929
2103:Bibcode
2054:Bibcode
2002:Bibcode
1925:3532252
1876:3098256
1853:Bibcode
1817:3098229
1794:Bibcode
1758:3152341
1709:3141260
1619:3084533
1567:3241638
1515:2885538
1282:3489282
1261:Bibcode
1253:Science
1222:3152587
1115:3130291
1061:3038751
1038:Bibcode
1002:3017587
953:2942870
904:4311247
883:Bibcode
856:6648530
828:Bibcode
820:Science
777:Bibcode
769:Science
613:in situ
392:in situ
385:or for
187:Process
160:History
137:Soybean
113:Insects
103:Mammals
98:Animals
91:Viruses
2637:
2627:
2588:
2546:
2536:
2497:
2487:
2448:
2438:
2389:
2354:
2344:
2300:
2290:
2241:
2231:
2182:
2172:
2133:
2123:
2074:
2022:
1973:
1932:
1922:
1883:
1873:
1824:
1814:
1765:
1755:
1716:
1706:
1667:
1657:
1626:
1616:
1574:
1564:
1522:
1512:
1473:
1465:
1423:
1415:
1379:
1371:
1333:
1289:
1279:
1229:
1219:
1175:
1167:
1122:
1112:
1068:
1058:
1009:
999:
960:
950:
911:
901:
854:
846:
803:
795:
752:
719:304571
717:
709:
668:CRISPR
631:CRISPR
603:, and
542:indels
387:genome
365:) are
244:CRISPR
142:Potato
122:Plants
71:
2675:(PDF)
1471:S2CID
1443:(PDF)
1421:S2CID
1377:S2CID
1173:S2CID
852:S2CID
801:S2CID
715:S2CID
363:TALEN
239:TALEN
2635:PMID
2586:PMID
2544:PMID
2495:PMID
2446:PMID
2387:PMID
2352:PMID
2298:PMID
2239:PMID
2180:PMID
2131:PMID
2072:PMID
2020:PMID
1971:PMID
1930:PMID
1881:PMID
1822:PMID
1763:PMID
1714:PMID
1665:PMID
1655:ISBN
1624:PMID
1572:PMID
1520:PMID
1463:PMID
1413:PMID
1369:PMID
1331:PMID
1287:PMID
1227:PMID
1165:PMID
1120:PMID
1066:PMID
1007:PMID
958:PMID
909:PMID
844:PMID
793:PMID
750:PMID
707:PMID
627:ZFNs
570:and
455:FokI
403:and
267:food
132:Rice
108:Fish
34:3UGM
30:1FOK
2718:DNA
2625:PMC
2617:doi
2576:doi
2534:PMC
2526:doi
2485:PMC
2477:doi
2436:PMC
2426:doi
2379:doi
2342:PMC
2334:doi
2288:PMC
2278:doi
2229:PMC
2219:doi
2207:112
2170:PMC
2162:doi
2121:PMC
2111:doi
2062:doi
2010:doi
1961:doi
1920:PMC
1912:doi
1908:161
1871:PMC
1861:doi
1812:PMC
1802:doi
1753:PMC
1745:doi
1704:PMC
1696:doi
1647:doi
1614:PMC
1606:doi
1562:PMC
1554:doi
1510:PMC
1502:doi
1498:400
1455:doi
1405:doi
1361:doi
1321:doi
1277:PMC
1269:doi
1257:333
1217:PMC
1209:doi
1157:doi
1110:PMC
1102:doi
1056:PMC
1046:doi
1034:108
997:PMC
989:doi
948:PMC
940:doi
936:186
899:PMC
891:doi
836:doi
824:326
785:doi
773:326
742:doi
699:doi
484:PCR
26:PDB
2709::
2655:.
2633:.
2623:.
2613:42
2611:.
2607:.
2584:.
2572:75
2570:.
2566:.
2542:.
2532:.
2522:23
2520:.
2516:.
2493:.
2483:.
2473:21
2471:.
2467:.
2444:.
2434:.
2424:.
2412:.
2408:.
2385:.
2375:14
2373:.
2350:.
2340:.
2328:.
2324:.
2310:^
2296:.
2286:.
2276:.
2264:.
2260:.
2237:.
2227:.
2217:.
2205:.
2201:.
2178:.
2168:.
2158:29
2156:.
2152:.
2129:.
2119:.
2109:.
2097:.
2093:.
2070:.
2060:.
2048:.
2044:.
2032:^
2018:.
2008:.
1996:.
1992:.
1969:.
1957:12
1955:.
1951:.
1928:.
1918:.
1906:.
1902:.
1879:.
1869:.
1859:.
1847:.
1843:.
1820:.
1810:.
1800:.
1788:.
1784:.
1761:.
1751:.
1741:39
1739:.
1735:.
1712:.
1702:.
1692:39
1690:.
1686:.
1663:.
1653:.
1622:.
1612:.
1602:29
1600:.
1596:.
1584:^
1570:.
1560:.
1550:39
1548:.
1544:.
1532:^
1518:.
1508:.
1496:.
1492:.
1469:.
1461:.
1451:25
1449:.
1445:.
1419:.
1411:.
1399:.
1375:.
1367:.
1357:29
1355:.
1343:^
1329:.
1317:29
1315:.
1311:.
1299:^
1285:.
1275:.
1267:.
1255:.
1251:.
1239:^
1225:.
1215:.
1205:29
1203:.
1199:.
1185:^
1171:.
1163:.
1153:29
1151:.
1132:^
1118:.
1108:.
1098:39
1096:.
1092:.
1078:^
1064:.
1054:.
1044:.
1032:.
1028:.
1005:.
995:.
985:39
983:.
979:.
956:.
946:.
934:.
930:.
907:.
897:.
889:.
877:.
873:.
850:.
842:.
834:.
822:.
799:.
791:.
783:.
771:.
748:.
738:48
736:.
713:.
705:.
695:29
693:.
681:^
599:,
580:,
411:.
48:on
32:,
28::
2677:.
2641:.
2619::
2592:.
2578::
2550:.
2528::
2501:.
2479::
2452:.
2428::
2420::
2414:8
2393:.
2381::
2358:.
2336::
2330:1
2304:.
2280::
2272::
2266:9
2245:.
2221::
2213::
2186:.
2164::
2137:.
2113::
2105::
2099:8
2078:.
2064::
2056::
2050:6
2026:.
2012::
2004::
1998:5
1977:.
1963::
1936:.
1914::
1887:.
1863::
1855::
1849:6
1828:.
1804::
1796::
1790:6
1769:.
1747::
1720:.
1698::
1671:.
1649::
1630:.
1608::
1578:.
1556::
1526:.
1504::
1477:.
1457::
1427:.
1407::
1401:8
1383:.
1363::
1337:.
1323::
1293:.
1271::
1263::
1233:.
1211::
1179:.
1159::
1126:.
1104::
1072:.
1048::
1040::
1013:.
991::
964:.
942::
915:.
893::
885::
879:5
858:.
838::
830::
807:.
787::
779::
756:.
744::
721:.
701::
361:(
347:e
340:t
333:v
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.