867:. As mentioned, the T3SS is closely related to the bacterial flagellum. There are three competing hypotheses: first, that the flagellum evolved first and the T3SS is derived from that structure, second, that the T3SS evolved first and the flagellum is derived from it, and third, that the two structures are derived from a common ancestor. There was some controversy about the different scenarios, since they all explain protein homology between the two structures, as well as their functional diversity. Yet, recent phylogenomic evidence favours the hypothesis that the T3SS derived from the flagellum by a process involving initial gene loss and then gene acquisition. A key step of the latter process was the recruitment of secretins to the T3SS, an event that occurred at least three times from other membrane-associated systems.
837:. The bacterium must know when the time is right to secrete effectors. Unnecessary secretion, when no host cell is in vicinity, is wasteful for the bacterium in terms of energy and resources. The bacterium is somehow able to recognize contact of the needle with the host cell. How this is done is still being researched, and the method may well be dependent on the pathogen. Some theories postulate a delicate conformational change in the structure of the needle upon contact with the host cell; this change perhaps serves as a signal for the base to commence secretion. One method of recognition has been discovered in
469:
786:
877:
315:
777:. When injected into plants, these proteins can enter the nucleus of the plant cell, bind plant promoter sequences, and activate transcription of plant genes that aid in bacterial infection. TAL effector-DNA recognition has recently been demonstrated to comprise a simple code and this has greatly improved the understanding of how these proteins can alter the transcription of genes in the host plant cells.
831:. As mentioned above, the existence of a secretion signal in effector proteins is known. The signal allows the system to distinguish T3SS-transported proteins from any other protein. Its nature, requirements and the mechanism of recognition are poorly understood, but methods for predicting which bacterial proteins can be transported by the Type III secretion system have recently been developed.
31:
815:. It is not known how the bacterium "knows" when a new needle has reached its proper length. Several theories exist, among them the existence of a "ruler protein" that somehow connects the tip and the base of the needle. Addition of new monomers to the tip of the needle should stretch the ruler protein and thereby signal the needle length to the base.
284:, for instance). Technically speaking, type III secretion is used both for secreting infection-related proteins and flagellar components. However, the term "type III secretion" is used mainly in relation to the infection apparatus. The bacterial flagellum shares a common ancestor with the type III secretion system.
885:
Since the beginning of the 1990s new T3SS proteins are being found in different bacterial species at a steady rate. Abbreviations have been given independently for each series of proteins in each organism, and the names usually do not reveal much about the protein's function. Some proteins discovered
716:
T3SS effectors manipulate host cells in several ways. The most striking effect is the promoting of uptake of the bacterium by the host cell. Many bacteria possessing T3SSs must enter host cells in order to replicate and propagate infection. The effectors they inject into the host cell induce the host
728:
and it also participates in motility and in changes in cell shape. Through its T3SS effectors the bacterium is able to utilize the host cell's own machinery for its own benefit. Once the bacterium has entered the cell it is able to secrete other effectors more easily and it can penetrate neighboring
407:
separate the two cytoplasms: the double membranes (inner and outer membranes) of the Gram-negative bacterium and the eukaryotic membrane. The needle provides a smooth passage through those highly selective and almost impermeable membranes. A single bacterium can have several hundred needle complexes
1637:. The structural components of the NC can be separated from each other (the needle part from the base part, for instance), and by analyzing those fractions the proteins participating in each one can be deduced. Alternatively, isolated NCs can be directly analyzed by mass spectrometry, without prior
695:
T3SS effectors enter the needle complex at the base and make their way inside the needle towards the host cell. The exact way in which effectors enter the host is mostly unknown. It has been previously suggested that the needle itself is capable of puncturing a hole in the host cell membrane; this
682:
of T3SS genes are known. Some of the chaperones that bind T3SS effectors also act as transcription factors. A feedback mechanism has been suggested: when the bacterium does not secrete, its effector proteins are bound to chaperones and float in the cytoplasm. When secretion starts, the chaperones
427:
The base is composed of several circular rings and is the first structure that is built in a new needle complex. Once the base is completed, it serves as a secretion machine for the outer proteins (the needle). Once the whole complex is completed the system switches to secreting proteins that are
287:
T3SSs are essential for the pathogenicity (the ability to infect) of many pathogenic bacteria. Defects in the T3SS may render a bacterium non-pathogenic. It has been suggested that some non-invasive strains of gram-negative bacteria have lost the T3SS because the energetically costly system is no
1713:
The ability of a T3SS to secrete a specific protein or to secrete at all. In order to assay this, secretion is induced in bacteria growing in liquid medium. The bacteria and medium are then separated by centrifugation, and the medium fraction (the supernatant) is then assayed for the presence of
423:
connects the needle to the base. The needle itself, although the biggest and most prominent part of the T3SS, is made out of many units of a single protein. The majority of the different T3SS proteins are therefore those that build the base and those that are secreted into the host. As mentioned
424:
above, the needle complex shares similarities with bacterial flagella. More specifically, the base of the needle complex is structurally very similar to the flagellar base; the needle itself is analogous to the flagellar hook, a structure connecting the base to the flagellar filament.
1589:
Numerous T3SS proteins have been crystallized over the years. These include structural proteins of the NC, effectors and chaperones. The first structure of a needle-complex monomer was NMR structure of BsaL from "Burkholderia pseudomallei" and later the crystal structure of MixH from
708:
bacteria that lack translocators are able to secrete proteins but are not able to deliver them into host cells. In general each T3SS includes three translocators. Some translocators serve a double role; after they participate in pore formation they enter the cell and act as
1714:
secreted proteins. In order to prevent a normally secreted protein from being secreted, a large molecule can be artificially attached to it. If the then non-secreted protein stays "stuck" at the bottom of the needle complex, the secretion is effectively blocked.
537:, on the other hand, has a large virulence plasmid on which all T3SS genes reside. It is important to note that many pathogenicity islands and plasmids contain elements that allow for frequent horizontal gene transfer of the island/plasmid to a new species.
683:
detach from the effectors and the latter are secreted and leave the cell. The lone chaperones then act as transcription factors, binding to the genes encoding their effectors and inducing their transcription and thereby the production of more effectors.
808:
methods. The rest, being perhaps rare, have proven difficult to detect and they remain theoretical (although genetic rather than biochemical studies have been performed on many T3SS genes/proteins). The localization of each protein is also not entirely
686:
Structures similar to Type3SS injectisomes have been proposed to rivet gram negative bacterial outer and inner membranes to help release outer membrane vesicles targeted to deliver bacterial secretions to eukaryotic host or other target cells in vivo.
1182:
Several key elements appear in all T3SSs: the needle monomer, the inner rod of the needle, the ring proteins, the two translocators, the needle-tip protein, the ruler protein (which is thought to determine the needle's length; see above) and the
1522:). The tagged protein is retained in the column, and with it the entire needle complex. High degrees of purity can be achieved using such methods. This purity is essential for many delicate assays that have been used for NC characterization.
451:
layer, for instance) do not interfere with secretion. The hole of the needle has a 3 nm diameter. Most folded effector proteins are too large to pass through the needle opening, so most secreted proteins must pass through the needle
1533:
characterize the three-dimensional structure of the NC in detail, and through this to draw conclusions regarding the mechanism of secretion (for example, that the narrow width of the needle requires unfolding of effectors prior to
754:. It serves a double role, both as a translocator, creating a pore in the host cell membrane, and as an effector, exerting multiple detrimental effects on the host cell. It had been demonstrated that IpaB induces apoptosis in
880:
Flagellum of Gram-negative bacteria. The rings of the base are very similar to needle-complex rings, although the existence of a C-ring in the needle complex has not been proven. The flagellar hook is homologous to the T3SS
110:. Many animal and plant associated bacteria possess similar T3SSs. These T3SSs are similar as a result of convergent evolution and phylogenetic analysis supports a model in which gram-negative bacteria can transfer the T3SS
1653:
The T3SS in many bacteria has been manipulated by researchers. Observing the influence of individual manipulations can be used to draw insights into the role of each component of the system. Examples of manipulations are:
1423:
membrane structures from cells has constituted a challenge for many years. By the end of the 1990s, however, several approaches have been developed for the isolation of T3SS NCs. In 1998 the first NCs were isolated from
87:, many argue that the injectisome is only part of the type III secretion system, which also include structures like the flagellar export apparatus. The T3SS is a needle-like protein complex found in several species of
549:) of the protein (usually within the first 20 amino acids), that the needle complex is able to recognize. Unlike other secretion systems, the secretion signal of T3SS proteins is never cleaved off the protein.
1717:
The ability of the bacteria to assemble an intact needle complex. NCs can be isolated from manipulated bacteria and examined microscopically. Minor changes, however cannot always be detected by microscopy.
540:
Effector proteins that are to be secreted through the needle need to be recognized by the system, since they float in the cytoplasm together with thousands of other proteins. Recognition is done through a
296:
strains constantly emerge. Understanding the way the T3SS works and developing drugs targeting it specifically have become an important goal of many research groups around the world since the late 1990s.
1515:) into the researched bacteria. After initial NC isolation, as described above, the lysate is passed through a column coated with particles with high affinity to the tag (in the case of histidine tags:
1529:
and T3SS proteins led researchers to suspects the existence of an outer T3SS structure similar to flagella. The identification and subsequent isolation of the needle structure enabled researchers to:
557:
Contact of the needle with a host cell triggers the T3SS to start secreting; not much is known about this trigger mechanism (see below). Secretion can also be induced by lowering the concentration of
1680:
The introduction of a gene or a protein from one species of bacteria into another (cross-complementation assay). This is done in order to check for differences and similarities between two T3SSs.
1171:
Following those abbreviations is a letter or a number. Letters usually denote a "serial number", either the chronological order of discovery or the physical order of appearance of the gene in an
3125:
Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, et al. (April 1998). "Supramolecular structure of the
Salmonella typhimurium type III protein secretion system".
905:
Below is a summary of the most common protein-series names in several T3SS-containing species. Note that these names include proteins that form the T3SS machinery as well as the secreted
861:. Although much was revealed since the beginning of the 21st century about the ways in which T3SS effectors manipulate the host, the majority of effects and pathways remains unknown.
651:, while the cecum does not. The bacteria sense these molecules, determine that they are at the ileum and activate their secretion machinery. Molecules present in the cecum, such as
1617:. The model also revealed an extended amino-terminal domain that is positioned on the surface of the needle, while the highly conserved carboxy terminus points towards the lumen.
3367:
Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN (June 2006). "Solution structure of monomeric BsaL, the type III secretion needle protein of
Burkholderia pseudomallei".
432:
protein pile upon each other, so that the unit at the tip of the needle is the last one added. The needle subunit is one of the smallest T3SS proteins, measuring at around 9 k
1557:. The first images of NCs (1998) showed needle structures protruding from the cell wall of live bacteria and flat, two-dimensional isolated NCs. In 2001 images of NCs from
1525:
Type III effectors were known since the beginning of the 1990s, but the way in which they are delivered into host cells was a complete mystery. The homology between many
264:
The T3SS is composed of approximately 30 different proteins, making it one of the most complex secretion systems. Its structure shows many similarities with bacterial
1481:. This treatment enriches large macromolecular structures and discards smaller cell components. Optionally, the final lysate is subjected to further purification by
2656:
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. (December 2009). "Breaking the code of DNA binding specificity of TAL-type III effectors".
1607:
T3SS needle. It was shown that the 80-residue PrgI subunits form a right-handed helical assembly with roughly 11 subunits per two turns, similar to that of the
825:
is associated with the base of the T3SS and participates in directing proteins into the needle; but whether it supplies the energy for transport is not clear.
2465:"Eucaryotic cell intoxication by gram-negative pathogens: A novel bacterial outermembrane-bound nanovesicular exocytosis model for Type III secretion system"
411:
The needle complex starts at the cytoplasm of the bacterium, crosses the two membranes and protrudes from the cell. The part anchored in the membrane is the
2486:
Zychlinsky A, Kenny B, Ménard R, Prévost MC, Holland IB, Sansonetti PJ (February 1994). "IpaB mediates macrophage apoptosis induced by
Shigella flexneri".
2208:"Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases"
890:; the historical names, however, have mostly been kept, a fact that might cause confusion. For example, the proteins SicA, IpgC and SycD are homologs from
1783:, an antibiotic capable of inhibiting the translation of T3SS proteins has been shown to able to prevent T3SS effectors in vitro and in animal models
1625:
Several methods have been employed in order to identify the array of proteins that comprise the T3SS. Isolated needle complexes can be separated with
2033:
Nguyen L, Paulsen IT, Tchieu J, Hueck CJ, Saier MH (April 2000). "Phylogenetic analyses of the constituents of Type III protein secretion systems".
1586:
was published. Recent advances and approaches have allowed high-resolution 3D images of the NC, further clarifying the complex structure of the NC.
3753:
1677:
Point or regional changes in T3SS genes or proteins. This is done in order to define the function of specific amino acids or regions in a protein.
1684:
Manipulation of T3SS components can have influence on several aspects of bacterial function and pathogenicity. Examples of possible influences:
362:
350:
1998:
Gophna U, Ron EZ, Graur D (July 2003). "Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events".
797:
Hundreds of articles on T3SS have been published since the mid-nineties. However, numerous issues regarding the system remain unresolved:
2062:"Differential expression of Salmonella type III secretion system factors InvJ, PrgJ, SipC, SipD, SopA and SopB in cultures and in mice"
1601:, electron microscopy and Rosetta modeling revealed the supramolecular interfaces and ultimately the complete atomic structure of the
3703:"A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium"
408:
spread across its membrane. It has been proposed that the needle complex is a universal feature of all T3SSs of pathogenic bacteria.
3320:"Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout"
3014:"The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems"
17:
1466:
443:
in length and 8 nm in external width. It needs to have a minimal length so that other extracellular bacterial structures (
1187:, which supplies energy for secretion. The following table shows some of these key proteins in four T3SS-containing bacteria:
3463:"The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system"
1862:"Characterization of the Mode of Action of Aurodox, a Type III Secretion System Inhibitor from Streptomyces goldiniensis"
643:. The bacteria are able to know where they are thanks to the different ions present in these regions; the ileum contains
1688:
The ability of the bacteria to invade host cells, in the case of intracellular pathogens. This can be measured using an
717:
to engulf the bacterium and to practically "eat" it. In order for this to happen the bacterial effectors manipulate the
3744:
102:
The term Type III secretion system was coined in 1993. This secretion system is distinguished from at least five other
1726:
to be able to infect host cells, their ability to sustain an infection in a live organism cannot be taken for granted.
1563:
were digitally analyzed and averaged to obtain a first semi-3D structure of the NC. The helical structure of NCs from
276:
sequence homology to flagellar proteins. Some of the bacteria possessing a T3SS have flagella as well and are motile (
35:
3662:"Development and validation of a high-content screening assay for inhibitors of enteropathogenic E. coli adhesion"
2707:
Schraidt O, Lefebre MD, Brunner MJ, Schmied WH, Schmidt A, Radics J, et al. (April 2010). Stebbins CE (ed.).
1960:
Salmond GP, Reeves PJ (January 1993). "Membrane traffic wardens and protein secretion in gram-negative bacteria".
428:
intended to be delivered into host cells. The needle is presumed to be built from bottom to top; units of needle
2155:
Galán JE, Wolf-Watz H (November 2006). "Protein delivery into eukaryotic cells by type III secretion machines".
1537:
analyze the protein components of the NC, this by subjecting isolated needles to proteomic analysis (see below),
2876:"Protein homology network families reveal step-wise diversification of Type III and Type IV secretion systems"
1702:
The ability of the bacteria to kill host cells. This can be measured by several methods, for instance by the
602:), for instance. These methods and other are used in laboratories to artificially induce type III secretion.
207:
674:
The external cues listed above either regulate secretion directly or through a genetic mechanism. Several
2353:
Akeda Y, Galán JE (October 2005). "Chaperone release and unfolding of substrates in type III secretion".
1693:
3268:"Structural organization of the needle complex of the type III secretion apparatus of Shigella flexneri"
1544:
T3SS genes, isolating NCs from the mutated bacteria and examining the changes that the mutations caused.
804:. Of the approximately 30 T3SS proteins less than 10 in each organism have been directly detected using
3748:
906:
490:
72:
60:
3295:
2709:"Topology and organization of the Salmonella typhimurium type III secretion needle complex components"
2116:"Structure and composition of the Shigella flexneri "needle complex", a part of its type III secreton"
3570:
Holmes TC, May AE, Zaleta-Rivera K, Ruby JG, Skewes-Cox P, Fischbach MA, et al. (October 2012).
2605:
Moscou MJ, Bogdanove AJ (December 2009). "A simple cipher governs DNA recognition by TAL effectors".
1450:
1629:. The bands that appear after staining can be individually excised from the gel and analyzed using
1512:
114:
2406:"Contribution of Salmonella typhimurium type III secretion components to needle complex formation"
1720:
The ability of bacteria to infect live animals or plants. Even if manipulated bacteria are shown
1492:
679:
3701:
Kimura K, Iwatsuki M, Nagai T, Matsumoto A, Takahashi Y, Shiomi K, et al. (February 2011).
3572:"Molecular insights into the biosynthesis of guadinomine: a type III secretion system inhibitor"
2570:
Boch J, Bonas U (2010). "Xanthomonas AvrBs3 family-type III effectors: discovery and function".
3318:
Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, da
Fonseca PC, et al. (May 2009).
2649:
1762:
1613:
1603:
1426:
769:
Another well characterized class of T3SS effectors are
Transcription Activator-like effectors (
107:
91:
40:
1477:. After several rounds of lysis and washing, the opened bacteria are subjected to a series of
821:. The force that drives the passage of proteins inside the needle is not completely known. An
762:—after being engulfed by them. It was later shown that IpaB achieves this by interacting with
520:, for instance, has a chromosomal region in which most T3SS genes are gathered, the so-called
1776:
1746:
1703:
852:
845:
through the pathogenicity island 2-encoded T3SS in order to switch on secretion of effectors.
399:
is excluded; see below). Bacterial proteins that need to be secreted pass from the bacterial
202:
3402:
Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, et al. (August 2006).
2973:"Evolution of the type III secretion system and its effectors in plant-microbe interactions"
902:, respectively, but the last letter (the "serial number") in their name does not show that.
605:
Induction of secretion by external cues other than contact with host cells also takes place
3526:
3415:
3404:"Molecular model of a type III secretion system needle: Implications for host-cell sensing"
3222:
3134:
3076:
2887:
2822:
2665:
2614:
2417:
2362:
2309:
2164:
675:
512:. These operons are located on the bacterial chromosome in some species and on a dedicated
293:
3267:
2114:
Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, et al. (February 2001).
8:
3777:
3513:
Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, et al. (May 2012).
1554:
1478:
127:
88:
3530:
3419:
3226:
3138:
3080:
2891:
2826:
2669:
2618:
2583:
2421:
2366:
2313:
2168:
30:
3782:
3596:
3571:
3547:
3514:
3487:
3462:
3438:
3403:
3344:
3319:
3243:
3210:
3170:"Helical structure of the needle of the type III secretion system of Shigella flexneri"
3040:
3013:
2910:
2875:
2851:
2810:
2786:
2759:
2735:
2708:
2689:
2638:
2529:
Hilbi H, Moss JE, Hersh D, Chen Y, Arondel J, Banerjee S, et al. (December 1998).
2511:
2499:
2386:
2335:
2273:
2256:
2232:
2207:
2188:
2088:
2061:
1937:
1910:
1886:
1861:
1837:
1812:
1630:
448:
355:
2011:
494:: get secreted into the host cell and promote infection / suppress host cell defences.
3724:
3683:
3642:
3601:
3552:
3492:
3443:
3384:
3349:
3300:
3248:
3191:
3150:
3104:
3099:
3064:
3045:
2994:
2989:
2972:
2953:
2915:
2856:
2791:
2740:
2693:
2681:
2630:
2587:
2552:
2503:
2464:
2445:
2440:
2405:
2378:
2327:
2278:
2237:
2180:
2137:
2132:
2115:
2093:
2042:
2015:
1977:
1973:
1942:
1891:
1842:
1828:
1634:
1575:
1559:
1508:
750:
444:
3168:
Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM (May 2003).
2936:
Saier MH (March 2004). "Evolution of bacterial type III protein secretion systems".
2515:
2339:
1710:
LDH, which leaks from dead cells, is identified by measuring its enzymatic activity.
468:
83:. While the type III secretion system has been widely regarded as equivalent to the
3714:
3673:
3632:
3591:
3583:
3542:
3534:
3482:
3474:
3433:
3423:
3376:
3339:
3331:
3290:
3282:
3238:
3230:
3181:
3142:
3094:
3084:
3035:
3025:
2984:
2945:
2905:
2895:
2846:
2838:
2830:
2781:
2771:
2730:
2720:
2673:
2642:
2622:
2579:
2542:
2495:
2435:
2425:
2390:
2370:
2317:
2268:
2227:
2219:
2192:
2172:
2127:
2083:
2073:
2007:
1969:
1932:
1922:
1881:
1873:
1832:
1824:
1813:"The Injectisome, a Complex Nanomachine for Protein Injection into Mammalian Cells"
1729:
The expression levels of other genes. This can be assayed in several ways, notably
1485:
1438:
887:
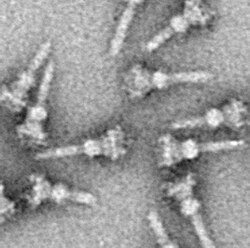
730:
433:
195:
146:
785:
367:
343:
3286:
3209:
Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM (November 2004).
3146:
3030:
2900:
2776:
2725:
1665:
1638:
1598:
1553:
As with almost all proteins, the visualization of T3SS NCs is only possible with
1496:
696:
theory has been refuted. It is now clear that some effectors, collectively named
585:
453:
233:
167:
3678:
3661:
3211:"Structural insights into the assembly of the type III secretion needle complex"
851:. It is not known when chaperones bind their effectors (whether during or after
3637:
3620:
3408:
Proceedings of the
National Academy of Sciences of the United States of America
3069:
Proceedings of the
National Academy of Sciences of the United States of America
2410:
Proceedings of the
National Academy of Sciences of the United States of America
1689:
721:
627:
levels, and use them to "decide" whether to activate their T3SS. For instance,
500:: bind effectors in the bacterial cytoplasm, protect them from aggregation and
155:
3478:
3380:
2949:
2322:
2297:
3771:
3266:
Sani M, Allaoui A, Fusetti F, Oostergetel GT, Keegstra W, Boekema EJ (2007).
2547:
2530:
1911:"Protein Export via the Type III Secretion System of the Bacterial Flagellum"
1730:
1659:
1541:
1504:
1434:
759:
565:
404:
139:
111:
3428:
3234:
2834:
2677:
2626:
876:
3728:
3687:
3646:
3605:
3556:
3496:
3447:
3388:
3353:
3304:
3252:
3195:
3186:
3169:
3108:
3089:
3049:
2998:
2957:
2919:
2860:
2842:
2795:
2744:
2685:
2634:
2591:
2449:
2430:
2382:
2331:
2282:
2241:
2184:
2141:
2097:
2078:
2046:
2019:
1946:
1895:
1846:
1771:
1470:
1454:
805:
770:
725:
178:
80:
3461:
Galkin VE, Schmied WH, Schraidt O, Marlovits TC, Egelman EH (March 2010).
3154:
2556:
2531:"Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB"
2507:
1981:
117:
to other species. Some of the most researched T3SSs are from species of:
2223:
1927:
1877:
1860:
McHugh RE, O'Boyle N, Connolly JP, Hoskisson PA, Roe AJ (February 2019).
1766:
1500:
1446:
1420:
1175:. Numbers, the rarer case, denote the molecular weight of the protein in
774:
660:
612:
501:
289:
250:
214:
3719:
3702:
3538:
2374:
2176:
667:
found in most eukaryotic cell membranes, is able to induce secretion in
2811:"pH sensing by intracellular Salmonella induces effector translocation"
1742:
1442:
755:
737:
652:
620:
546:
273:
134:
3587:
3335:
704:) in the host cell membrane, through which other effectors may enter.
314:
1608:
1526:
1474:
1462:
763:
741:
640:
595:
440:
400:
256:
244:
151:
103:
76:
68:
2298:"Evolution: reducible complexity -- the case for bacterial flagella"
1699:
The ability of intracellular bacteria to migrate between host cells.
1722:
1642:
1626:
1597:
In 2012, a combination of recombinant wild-type needle production,
1579:
1568:
1458:
705:
656:
589:
577:
269:
265:
190:
183:
171:
122:
64:
3621:"Non-traditional Antibacterial Therapeutic Options and Challenges"
1859:
1548:
1433:
For the isolation, bacteria are grown in a large volume of liquid
1780:
1670:
648:
644:
607:
558:
513:
429:
419:) of the T3SS. The extracellular part is the needle. A so-called
238:
886:
independently in different bacteria have later been shown to be
855:) and how they dissociate from their effectors before secretion.
659:, provide a negative cue to the bacteria and inhibit secretion.
1738:
1734:
1707:
1516:
1184:
1172:
822:
624:
545:—a short sequence of amino acids located at the beginning (the
509:
472:
Diagram of individual substructures of the needle complex from
457:
396:
223:
162:
3460:
3124:
1761:
A few compounds have been discovered that inhibit the T3SS in
736:
T3SS effectors have also been shown to tamper with the host's
3265:
2809:
Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW (May 2010).
2706:
1572:
1179:. Examples: IpaA, IpaB, IpaC; MxiH, MxiG, MxiM; Spa9, Spa47.
718:
664:
636:
632:
227:
219:
3700:
3317:
2655:
2598:
2485:
1792:
1749:
and regulatory networks were discovered using these methods.
3208:
2113:
1786:
1482:
581:
403:
through the needle directly into the host cytoplasm. Three
338:
3569:
3512:
2060:
Gong H, Vu GP, Bai Y, Yang E, Liu F, Lu S (January 2010).
3401:
3167:
1519:
1176:
611:, in infected organisms. The bacteria sense such cues as
592:
561:
2032:
724:
machinery of the host cell. Actin is a component of the
700:, are secreted first and produce a pore or a channel (a
479:
The T3SS proteins can be grouped into three categories:
375:
The hallmark of T3SS is the needle (more generally, the
3366:
842:
616:
3515:"Atomic model of the type III secretion system needle"
2758:
Grynberg M, Godzik A (April 2009). Stebbins CE (ed.).
1668:
of one or more T3SS genes (in other words: production
1125:: Hypersensitive response conserved (or Hrp conserved)
3618:
2808:
2563:
1491:
An additional approach for further purification uses
1414:
3764:
and tomato plant leading to bacterial speck disease.
3747:
outlining the chemistry of the injectisome from the
2295:
1409:
272:). Some of the proteins participating in T3SS share
2873:
2257:"Bacterial flagella and type III secretion systems"
2205:
2035:
Journal of
Molecular Microbiology and Biotechnology
1674:
of a T3SS protein in quantities larger than usual).
292:were effective against these bacteria in the past,
3659:
2528:
1641:, in order to obtain a complete picture of the NC
766:, a major regulatory protein in eukaryotic cells.
1648:
3769:
1810:
871:
268:(long, rigid, extracellular structures used for
2874:Medini D, Covacci A, Donati C (December 2006).
2604:
2403:
2206:Pallen MJ, Bailey CM, Beatson SA (April 2006).
1779:have been developed that inhibit the T3SS too.
1549:Microscopy, crystallography and solid-state NMR
1540:assign roles to various NC components, this by
486:: build the base, the inner rod and the needle.
3120:
3118:
2757:
2154:
2109:
2107:
1997:
841:, which relies on sensing host cell cytosolic
744:. One of the most researched T3SS effector is
2970:
2059:
1959:
38:image of isolated T3SS needle complexes from
3612:
2964:
2867:
2751:
2346:
1993:
1991:
1908:
3115:
2700:
2104:
2053:
1119:: Hypersensitive response and pathogenicity
504:and direct them towards the needle complex.
3619:Theuretzbacher U, Piddock LJ (July 2019).
3563:
3296:11370/9ee8c380-a931-4313-89cf-d9faa49cdf3b
2404:Kimbrough TG, Miller SI (September 2000).
2352:
1804:
690:
3718:
3677:
3660:Pylkkö T, Ilina P, Tammela P (May 2021).
3636:
3595:
3546:
3508:
3506:
3486:
3437:
3427:
3343:
3324:Nature Structural & Molecular Biology
3294:
3242:
3185:
3098:
3088:
3039:
3029:
3011:
2988:
2909:
2899:
2850:
2785:
2775:
2734:
2724:
2569:
2546:
2439:
2429:
2321:
2296:Doolittle WF, Zhaxybayeva O (July 2007).
2272:
2231:
2131:
2087:
2077:
1988:
1936:
1926:
1885:
1836:
1756:
552:
436:. 100−150 subunits comprise each needle.
3576:Journal of the American Chemical Society
2931:
2929:
1787:Type III signal peptide prediction tools
875:
784:
467:
29:
2462:
631:can replicate and invade better in the
14:
3770:
3503:
2254:
1811:Lara-Tejero M, Galán JE (March 2019).
1737:. The expression levels of the entire
439:The T3SS needle measures around 60−80
3065:"Bacterial menageries inside insects"
3062:
2935:
2926:
789:The topology and organization of the
3012:Abby SS, Rocha EP (September 2012).
1658:Deletion of one or more T3SS genes (
1594:, which were both resolved in 2006.
780:
740:and some of them are able to induce
288:longer of use. Although traditional
3174:The Journal of Biological Chemistry
2584:10.1146/annurev-phyto-080508-081936
2535:The Journal of Biological Chemistry
1909:Halte M, Erhardt M (January 2021).
1752:The growth and fitness of bacteria.
1567:was resolved at a resolution of 16
1453:(the bacteria) is resuspended in a
1314:Chaperone for the two translocators
729:cells and quickly infect the whole
24:
3738:
3666:Journal of Microbiological Methods
2500:10.1111/j.1365-2958.1994.tb00341.x
2274:10.1111/j.1574-6968.2001.tb10797.x
1853:
1449:(the medium) is discarded and the
1415:Isolation of T3SS needle complexes
280:, for instance), and some do not (
25:
3794:
2760:"The signal for signaling, found"
1419:The isolation of large, fragile,
1410:Methods employed in T3SS research
1043:: Surface presentation of antigen
958:: Surface presentation of antigen
75:into the host's cells to promote
2990:10.1111/j.1469-8137.2007.02293.x
2133:10.1046/j.1365-2958.2001.02200.x
1829:10.1128/ecosalplus.ESP-0039-2018
1769:which are naturally produced by
1507:, for instance) are produced by
1473:. This buffer disintegrates the
508:Most T3SS genes are laid out in
463:
313:
36:transmission electron microscope
3694:
3653:
3454:
3395:
3360:
3311:
3259:
3202:
3161:
3056:
3005:
2802:
2572:Annual Review of Phytopathology
2522:
2479:
2456:
2397:
2289:
2248:
2199:
1578:in 2003, and a year later a 17-
1066:: Translocated intimin receptor
2971:McCann HC, Guttman DS (2008).
2148:
2026:
1962:Trends in Biochemical Sciences
1953:
1902:
1649:Genetic and functional studies
460:at the base of the structure.
13:
1:
2012:10.1016/S0378-1119(03)00612-7
1798:
1706:-release assay, in which the
1620:
872:Nomenclature of T3SS proteins
3467:Journal of Molecular Biology
3369:Journal of Molecular Biology
3287:10.1016/j.micron.2006.04.007
3147:10.1126/science.280.5363.602
3031:10.1371/journal.pgen.1002983
2901:10.1371/journal.pcbi.0020173
2777:10.1371/journal.ppat.1000398
2726:10.1371/journal.ppat.1000824
1974:10.1016/0968-0004(93)90080-7
1164:"Protochlamydia amoebophila"
1037:: Membrane expression of Ipa
456:, a task carried out by the
300:
208:sexually transmitted disease
7:
3679:10.1016/j.mimet.2021.106201
1694:gentamicin protection assay
1499:T3SS proteins that carry a
1189:
97:
61:bacterial secretion systems
10:
3799:
3749:Royal Society of Chemistry
3707:The Journal of Antibiotics
3638:10.1016/j.chom.2019.06.004
3063:Moran NA (February 2001).
2880:PLOS Computational Biology
1031:: Invasion plasmid antigen
1012:: Invasion-associated gene
598:to the growth medium (for
27:Bacterial virulence factor
3754:Host-Pathogen Interaction
3479:10.1016/j.jmb.2010.01.001
3381:10.1016/j.jmb.2006.03.028
2950:10.1016/j.tim.2004.01.003
2323:10.1016/j.cub.2007.05.003
2261:FEMS Microbiology Letters
2255:Aizawa SI (August 2001).
1582:3D structure of NCs from
361:
349:
337:
329:
324:
312:
308:Type III secretion system
307:
254:, and the plant symbiont
49:type III secretion system
2548:10.1074/jbc.273.49.32895
2469:Toxicology International
813:The length of the needle
3625:Cell Host & Microbe
3429:10.1073/pnas.0602689103
3235:10.1126/science.1102610
2835:10.1126/science.1189000
2678:10.1126/science.1178811
2627:10.1126/science.1178817
1493:affinity chromatography
1025:: Invasion plasmid gene
996:: Oxygen-regulated gene
835:Activation of secretion
691:T3SS-mediated infection
319:The T3SS needle complex
18:Type 3 secretion system
3187:10.1074/jbc.M300091200
3090:10.1073/pnas.98.4.1338
2938:Trends in Microbiology
2488:Molecular Microbiology
2431:10.1073/pnas.200209497
2120:Molecular Microbiology
2079:10.1099/mic.0.032318-0
1866:Infection and Immunity
1763:gram-negative bacteria
1757:Inhibitors of the T3SS
1614:Salmonella typhimurium
1604:Salmonella typhimurium
1584:Salmonella typhimurium
1427:Salmonella typhimurium
1167:"Sodalis glossinidius"
882:
794:
553:Induction of secretion
476:
474:Salmonella typhimurium
108:gram-negative bacteria
92:gram-negative bacteria
44:
41:Salmonella typhimurium
1777:Monoclonal antibodies
1747:transcription factors
1511:and then introduced (
1457:typically containing
1193:↓ Function / Genus →
1086:secretion (component)
984:: PhoP-repressed gene
935:secretion (component)
879:
849:Binding of chaperones
788:
758:—cells of the animal
676:transcription factors
471:
154:, some strains cause
33:
3758:Pseudomonas syringae
2224:10.1110/ps.051958806
1928:10.3390/biom11020186
1878:10.1128/IAI.00595-18
1479:ultracentrifugations
1153:In several species:
1138:: Nodulation protein
588:) and by adding the
527:pathogenicity island
294:antibiotic-resistant
3720:10.1038/ja.2010.155
3582:(42): 17797–17806.
3539:10.1038/nature11079
3531:2012Natur.486..276L
3420:2006PNAS..10312529D
3414:(33): 12529–12533.
3227:2004Sci...306.1040M
3221:(5698): 1040–1042.
3180:(19): 17103–17107.
3139:1998Sci...280..602K
3081:2001PNAS...98.1338M
2977:The New Phytologist
2892:2006PLSCB...2..173M
2827:2010Sci...328.1040Y
2821:(5981): 1040–1043.
2670:2009Sci...326.1509B
2664:(5959): 1509–1512.
2619:2009Sci...326.1501M
2541:(49): 32895–32900.
2463:YashRoy RC (2003).
2422:2000PNAS...9711008K
2416:(20): 11008–11013.
2375:10.1038/nature03992
2367:2005Natur.437..911A
2314:2007CBio...17.R510D
2177:10.1038/nature05272
2169:2006Natur.444..567G
1555:electron microscopy
859:Effector mechanisms
635:rather than in the
576:; done by adding a
484:Structural proteins
128:bacillary dysentery
1741:can be assayed by
1631:protein sequencing
1257:Needle-tip protein
968:invasion chaperone
883:
795:
678:that regulate the
516:in other species.
477:
449:lipopolysaccharide
45:
3588:10.1021/ja308622d
3525:(7402): 276–279.
3336:10.1038/nsmb.1599
3133:(5363): 602–605.
2361:(7060): 911–915.
2308:(13): R510–R512.
2163:(7119): 567–573.
2072:(Pt 1): 116–127.
1635:mass spectrometry
1592:Shigella flexneri
1576:fiber diffraction
1565:Shigella flexneri
1560:Shigella flexneri
1509:molecular cloning
1437:until they reach
1407:
1406:
1096:secretion protein
1006:-secreted protein
907:effector proteins
781:Unresolved issues
751:Shigella flexneri
491:Effector proteins
373:
372:
104:secretion systems
73:effector proteins
16:(Redirected from
3790:
3733:
3732:
3722:
3698:
3692:
3691:
3681:
3657:
3651:
3650:
3640:
3616:
3610:
3609:
3599:
3567:
3561:
3560:
3550:
3510:
3501:
3500:
3490:
3473:(5): 1392–1397.
3458:
3452:
3451:
3441:
3431:
3399:
3393:
3392:
3364:
3358:
3357:
3347:
3315:
3309:
3308:
3298:
3272:
3263:
3257:
3256:
3246:
3206:
3200:
3199:
3189:
3165:
3159:
3158:
3122:
3113:
3112:
3102:
3092:
3075:(4): 1338–1340.
3060:
3054:
3053:
3043:
3033:
3009:
3003:
3002:
2992:
2968:
2962:
2961:
2933:
2924:
2923:
2913:
2903:
2871:
2865:
2864:
2854:
2806:
2800:
2799:
2789:
2779:
2755:
2749:
2748:
2738:
2728:
2704:
2698:
2697:
2653:
2647:
2646:
2602:
2596:
2595:
2567:
2561:
2560:
2550:
2526:
2520:
2519:
2483:
2477:
2476:
2460:
2454:
2453:
2443:
2433:
2401:
2395:
2394:
2350:
2344:
2343:
2325:
2293:
2287:
2286:
2276:
2252:
2246:
2245:
2235:
2203:
2197:
2196:
2152:
2146:
2145:
2135:
2111:
2102:
2101:
2091:
2081:
2057:
2051:
2050:
2030:
2024:
2023:
1995:
1986:
1985:
1957:
1951:
1950:
1940:
1930:
1906:
1900:
1899:
1889:
1872:(2): e00595–18.
1857:
1851:
1850:
1840:
1808:
1765:, including the
1745:. Many type III
1486:density gradient
1461:and sometimes a
1441:. They are then
1190:
978:invasion protein
829:Secretion signal
543:secretion signal
317:
305:
304:
147:Escherichia coli
59:) is one of the
21:
3798:
3797:
3793:
3792:
3791:
3789:
3788:
3787:
3768:
3767:
3745:Instant insight
3741:
3739:Further reading
3736:
3699:
3695:
3658:
3654:
3617:
3613:
3568:
3564:
3511:
3504:
3459:
3455:
3400:
3396:
3365:
3361:
3316:
3312:
3270:
3264:
3260:
3207:
3203:
3166:
3162:
3123:
3116:
3061:
3057:
3024:(9): e1002983.
3010:
3006:
2969:
2965:
2934:
2927:
2872:
2868:
2807:
2803:
2770:(4): e1000398.
2756:
2752:
2719:(4): e1000824.
2705:
2701:
2654:
2650:
2603:
2599:
2568:
2564:
2527:
2523:
2484:
2480:
2461:
2457:
2402:
2398:
2351:
2347:
2302:Current Biology
2294:
2290:
2253:
2249:
2212:Protein Science
2204:
2200:
2153:
2149:
2112:
2105:
2058:
2054:
2031:
2027:
1996:
1989:
1958:
1954:
1907:
1903:
1858:
1854:
1809:
1805:
1801:
1789:
1759:
1651:
1639:electrophoresis
1623:
1599:solid-state NMR
1551:
1417:
1412:
1102:: Chaperone of
1072:: Secretion of
874:
793:needle complex.
783:
693:
555:
466:
391:); also called
351:OPM superfamily
320:
303:
234:Plant pathogens
168:gastroenteritis
100:
28:
23:
22:
15:
12:
11:
5:
3796:
3786:
3785:
3780:
3766:
3765:
3751:
3740:
3737:
3735:
3734:
3713:(2): 197–203.
3693:
3652:
3611:
3562:
3502:
3453:
3394:
3375:(2): 322–330.
3359:
3330:(5): 477–485.
3310:
3281:(3): 291–301.
3258:
3201:
3160:
3114:
3055:
3004:
2963:
2944:(3): 113–115.
2925:
2866:
2801:
2764:PLOS Pathogens
2750:
2713:PLOS Pathogens
2699:
2648:
2613:(5959): 1501.
2597:
2562:
2521:
2494:(4): 619–627.
2478:
2455:
2396:
2345:
2288:
2267:(2): 157–164.
2247:
2218:(4): 935–941.
2198:
2147:
2126:(3): 652–663.
2103:
2052:
2041:(2): 125–144.
2025:
1987:
1952:
1901:
1852:
1802:
1800:
1797:
1796:
1795:
1788:
1785:
1758:
1755:
1754:
1753:
1750:
1727:
1718:
1715:
1711:
1700:
1697:
1690:invasion assay
1682:
1681:
1678:
1675:
1666:Overexpression
1663:
1650:
1647:
1622:
1619:
1550:
1547:
1546:
1545:
1538:
1535:
1416:
1413:
1411:
1408:
1405:
1404:
1401:
1398:
1395:
1392:
1386:
1385:
1382:
1379:
1376:
1373:
1367:
1366:
1363:
1360:
1357:
1354:
1348:
1347:
1344:
1341:
1338:
1335:
1329:
1328:
1325:
1322:
1319:
1316:
1310:
1309:
1306:
1303:
1300:
1297:
1291:
1290:
1287:
1284:
1281:
1278:
1272:
1271:
1268:
1265:
1262:
1259:
1253:
1252:
1249:
1246:
1243:
1240:
1234:
1233:
1230:
1227:
1224:
1221:
1219:Needle monomer
1215:
1214:
1209:
1204:
1199:
1194:
1169:
1168:
1165:
1162:
1161:
1160:
1151:
1150:
1149:
1139:
1128:
1127:
1126:
1120:
1109:
1108:
1107:
1097:
1087:
1077:
1067:
1056:
1055:
1054:
1044:
1038:
1032:
1026:
1015:
1014:
1013:
1007:
997:
991:
985:
979:
969:
959:
948:
947:
946:
945:protein kinase
936:
926:
873:
870:
869:
868:
862:
856:
846:
832:
826:
816:
810:
782:
779:
722:polymerization
692:
689:
554:
551:
506:
505:
495:
487:
465:
462:
385:T3SS apparatus
377:needle complex
371:
370:
365:
359:
358:
353:
347:
346:
341:
335:
334:
331:
327:
326:
322:
321:
318:
310:
309:
302:
299:
262:
261:
231:
211:
199:
187:
175:
159:
156:food poisoning
143:
131:
99:
96:
26:
9:
6:
4:
3:
2:
3795:
3784:
3781:
3779:
3776:
3775:
3773:
3763:
3759:
3755:
3752:
3750:
3746:
3743:
3742:
3730:
3726:
3721:
3716:
3712:
3708:
3704:
3697:
3689:
3685:
3680:
3675:
3671:
3667:
3663:
3656:
3648:
3644:
3639:
3634:
3630:
3626:
3622:
3615:
3607:
3603:
3598:
3593:
3589:
3585:
3581:
3577:
3573:
3566:
3558:
3554:
3549:
3544:
3540:
3536:
3532:
3528:
3524:
3520:
3516:
3509:
3507:
3498:
3494:
3489:
3484:
3480:
3476:
3472:
3468:
3464:
3457:
3449:
3445:
3440:
3435:
3430:
3425:
3421:
3417:
3413:
3409:
3405:
3398:
3390:
3386:
3382:
3378:
3374:
3370:
3363:
3355:
3351:
3346:
3341:
3337:
3333:
3329:
3325:
3321:
3314:
3306:
3302:
3297:
3292:
3288:
3284:
3280:
3276:
3269:
3262:
3254:
3250:
3245:
3240:
3236:
3232:
3228:
3224:
3220:
3216:
3212:
3205:
3197:
3193:
3188:
3183:
3179:
3175:
3171:
3164:
3156:
3152:
3148:
3144:
3140:
3136:
3132:
3128:
3121:
3119:
3110:
3106:
3101:
3096:
3091:
3086:
3082:
3078:
3074:
3070:
3066:
3059:
3051:
3047:
3042:
3037:
3032:
3027:
3023:
3019:
3018:PLOS Genetics
3015:
3008:
3000:
2996:
2991:
2986:
2982:
2978:
2974:
2967:
2959:
2955:
2951:
2947:
2943:
2939:
2932:
2930:
2921:
2917:
2912:
2907:
2902:
2897:
2893:
2889:
2885:
2881:
2877:
2870:
2862:
2858:
2853:
2848:
2844:
2843:10044/1/19679
2840:
2836:
2832:
2828:
2824:
2820:
2816:
2812:
2805:
2797:
2793:
2788:
2783:
2778:
2773:
2769:
2765:
2761:
2754:
2746:
2742:
2737:
2732:
2727:
2722:
2718:
2714:
2710:
2703:
2695:
2691:
2687:
2683:
2679:
2675:
2671:
2667:
2663:
2659:
2652:
2644:
2640:
2636:
2632:
2628:
2624:
2620:
2616:
2612:
2608:
2601:
2593:
2589:
2585:
2581:
2577:
2573:
2566:
2558:
2554:
2549:
2544:
2540:
2536:
2532:
2525:
2517:
2513:
2509:
2505:
2501:
2497:
2493:
2489:
2482:
2474:
2470:
2466:
2459:
2451:
2447:
2442:
2437:
2432:
2427:
2423:
2419:
2415:
2411:
2407:
2400:
2392:
2388:
2384:
2380:
2376:
2372:
2368:
2364:
2360:
2356:
2349:
2341:
2337:
2333:
2329:
2324:
2319:
2315:
2311:
2307:
2303:
2299:
2292:
2284:
2280:
2275:
2270:
2266:
2262:
2258:
2251:
2243:
2239:
2234:
2229:
2225:
2221:
2217:
2213:
2209:
2202:
2194:
2190:
2186:
2182:
2178:
2174:
2170:
2166:
2162:
2158:
2151:
2143:
2139:
2134:
2129:
2125:
2121:
2117:
2110:
2108:
2099:
2095:
2090:
2085:
2080:
2075:
2071:
2067:
2063:
2056:
2048:
2044:
2040:
2036:
2029:
2021:
2017:
2013:
2009:
2005:
2001:
1994:
1992:
1983:
1979:
1975:
1971:
1967:
1963:
1956:
1948:
1944:
1939:
1934:
1929:
1924:
1920:
1916:
1912:
1905:
1897:
1893:
1888:
1883:
1879:
1875:
1871:
1867:
1863:
1856:
1848:
1844:
1839:
1834:
1830:
1826:
1822:
1818:
1814:
1807:
1803:
1794:
1791:
1790:
1784:
1782:
1778:
1774:
1773:
1768:
1764:
1751:
1748:
1744:
1740:
1736:
1732:
1731:northern blot
1728:
1725:
1724:
1719:
1716:
1712:
1709:
1705:
1701:
1698:
1695:
1691:
1687:
1686:
1685:
1679:
1676:
1673:
1672:
1667:
1664:
1661:
1660:gene knockout
1657:
1656:
1655:
1646:
1644:
1640:
1636:
1632:
1628:
1618:
1616:
1615:
1610:
1606:
1605:
1600:
1595:
1593:
1587:
1585:
1581:
1577:
1574:
1570:
1566:
1562:
1561:
1556:
1543:
1539:
1536:
1532:
1531:
1530:
1528:
1523:
1521:
1518:
1514:
1510:
1506:
1505:histidine tag
1502:
1498:
1494:
1489:
1487:
1484:
1480:
1476:
1472:
1468:
1464:
1460:
1456:
1452:
1448:
1444:
1440:
1436:
1435:growth medium
1431:
1429:
1428:
1422:
1402:
1399:
1396:
1393:
1391:
1388:
1387:
1383:
1380:
1377:
1374:
1372:
1369:
1368:
1364:
1361:
1358:
1355:
1353:
1352:Ruler protein
1350:
1349:
1345:
1342:
1339:
1336:
1334:
1331:
1330:
1326:
1323:
1320:
1317:
1315:
1312:
1311:
1307:
1304:
1301:
1298:
1296:
1293:
1292:
1288:
1285:
1282:
1279:
1277:
1274:
1273:
1269:
1266:
1263:
1260:
1258:
1255:
1254:
1250:
1247:
1244:
1241:
1239:
1236:
1235:
1231:
1228:
1225:
1222:
1220:
1217:
1216:
1213:
1210:
1208:
1205:
1203:
1200:
1198:
1195:
1192:
1191:
1188:
1186:
1180:
1178:
1174:
1166:
1163:
1158:
1155:
1154:
1152:
1147:
1143:
1140:
1137:
1134:
1133:
1132:
1129:
1124:
1121:
1118:
1115:
1114:
1113:
1110:
1105:
1101:
1098:
1095:
1091:
1088:
1085:
1081:
1078:
1075:
1071:
1068:
1065:
1062:
1061:
1060:
1057:
1052:
1048:
1045:
1042:
1039:
1036:
1033:
1030:
1027:
1024:
1021:
1020:
1019:
1016:
1011:
1008:
1005:
1001:
998:
995:
992:
989:
986:
983:
980:
977:
973:
970:
967:
963:
960:
957:
954:
953:
952:
949:
944:
940:
937:
934:
930:
927:
925:outer protein
924:
920:
917:
916:
915:
912:
911:
910:
908:
903:
901:
897:
893:
889:
878:
866:
863:
860:
857:
854:
850:
847:
844:
840:
836:
833:
830:
827:
824:
820:
817:
814:
811:
807:
803:
802:T3SS proteins
800:
799:
798:
792:
787:
778:
776:
772:
771:TAL effectors
767:
765:
761:
760:immune system
757:
753:
752:
747:
743:
739:
734:
732:
727:
723:
720:
714:
712:
707:
703:
699:
698:translocators
688:
684:
681:
677:
672:
670:
666:
662:
658:
654:
650:
646:
642:
638:
634:
630:
626:
622:
618:
614:
610:
609:
603:
601:
597:
594:
591:
587:
583:
579:
575:
571:
567:
566:growth medium
563:
560:
550:
548:
544:
538:
536:
532:
528:
525:
524:
519:
515:
511:
503:
499:
496:
493:
492:
488:
485:
482:
481:
480:
475:
470:
464:T3SS proteins
461:
459:
455:
450:
446:
442:
437:
435:
431:
425:
422:
418:
414:
409:
406:
402:
398:
394:
390:
386:
382:
378:
369:
366:
364:
360:
357:
354:
352:
348:
345:
342:
340:
336:
332:
328:
323:
316:
311:
306:
298:
295:
291:
285:
283:
279:
275:
271:
267:
259:
258:
253:
252:
247:
246:
241:
240:
235:
232:
229:
225:
221:
217:
216:
212:
209:
205:
204:
200:
197:
193:
192:
188:
185:
181:
180:
176:
173:
169:
165:
164:
160:
157:
153:
149:
148:
144:
141:
140:typhoid fever
137:
136:
132:
129:
125:
124:
120:
119:
118:
116:
113:
112:gene cassette
109:
105:
95:
93:
90:
86:
82:
78:
74:
70:
66:
62:
58:
54:
50:
43:
42:
37:
32:
19:
3761:
3757:
3710:
3706:
3696:
3669:
3665:
3655:
3631:(1): 61–72.
3628:
3624:
3614:
3579:
3575:
3565:
3522:
3518:
3470:
3466:
3456:
3411:
3407:
3397:
3372:
3368:
3362:
3327:
3323:
3313:
3278:
3274:
3261:
3218:
3214:
3204:
3177:
3173:
3163:
3130:
3126:
3072:
3068:
3058:
3021:
3017:
3007:
2983:(1): 33–47.
2980:
2976:
2966:
2941:
2937:
2886:(12): e173.
2883:
2879:
2869:
2818:
2814:
2804:
2767:
2763:
2753:
2716:
2712:
2702:
2661:
2657:
2651:
2610:
2606:
2600:
2575:
2571:
2565:
2538:
2534:
2524:
2491:
2487:
2481:
2472:
2468:
2458:
2413:
2409:
2399:
2358:
2354:
2348:
2305:
2301:
2291:
2264:
2260:
2250:
2215:
2211:
2201:
2160:
2156:
2150:
2123:
2119:
2069:
2066:Microbiology
2065:
2055:
2038:
2034:
2028:
2003:
1999:
1965:
1961:
1955:
1918:
1915:Biomolecules
1914:
1904:
1869:
1865:
1855:
1820:
1816:
1806:
1772:Streptomyces
1770:
1767:guadinomines
1760:
1721:
1683:
1669:
1652:
1624:
1612:
1602:
1596:
1591:
1588:
1583:
1564:
1558:
1552:
1542:knocking out
1524:
1490:
1471:Triton X-100
1455:lysis buffer
1432:
1425:
1418:
1400:YopN (TyeA)
1389:
1370:
1351:
1346:SepB (EscN)
1332:
1313:
1295:Translocator
1294:
1276:Translocator
1275:
1256:
1237:
1218:
1211:
1206:
1201:
1196:
1181:
1170:
1156:
1145:
1141:
1135:
1130:
1122:
1116:
1111:
1103:
1099:
1093:
1089:
1083:
1079:
1073:
1069:
1063:
1058:
1050:
1046:
1040:
1034:
1028:
1022:
1017:
1009:
1003:
999:
993:
987:
981:
975:
971:
965:
961:
955:
950:
942:
938:
932:
928:
922:
918:
913:
904:
899:
895:
891:
884:
864:
858:
848:
838:
834:
828:
818:
812:
801:
796:
790:
768:
749:
745:
735:
726:cytoskeleton
715:
710:
701:
697:
694:
685:
673:
668:
628:
606:
604:
599:
573:
569:
556:
542:
539:
534:
530:
526:
522:
521:
517:
507:
497:
489:
483:
478:
473:
438:
426:
420:
416:
412:
410:
392:
388:
384:
380:
376:
374:
286:
281:
277:
263:
255:
249:
243:
237:
213:
201:
189:
179:Burkholderia
177:
161:
145:
133:
121:
115:horizontally
101:
84:
81:colonisation
56:
52:
48:
46:
39:
2578:: 419–436.
2006:: 151–163.
1968:(1): 7–12.
1817:EcoSal Plus
1793:EffectiveT3
1534:secretion),
1513:transformed
1501:protein tag
1497:Recombinant
1447:supernatant
1443:centrifuged
1421:hydrophobic
1212:Escherichia
1159:: Virulence
1112:Pseudomonas
1094:Escherichia
1084:Escherichia
1059:Escherichia
853:translation
806:biochemical
775:Xanthomonas
756:macrophages
713:effectors.
661:Cholesterol
613:temperature
574:Pseudomonas
502:degradation
393:injectisome
363:OPM protein
325:Identifiers
290:antibiotics
251:Xanthomonas
215:Pseudomonas
85:injectisome
3778:Organelles
3772:Categories
3672:: 106201.
1921:(2): 186.
1799:References
1775:species.
1743:microarray
1621:Proteomics
1390:Gatekeeper
1202:Salmonella
1004:Salmonella
990:: Invasion
976:Salmonella
966:Salmonella
951:Salmonella
892:Salmonella
888:homologous
839:Salmonella
819:Energetics
791:Salmonella
738:cell cycle
702:translocon
680:expression
653:propionate
639:of animal
629:Salmonella
621:osmolarity
547:N-terminus
523:Salmonella
518:Salmonella
498:Chaperones
417:basal body
278:Salmonella
274:amino-acid
135:Salmonella
89:pathogenic
3783:Secretion
2694:206522347
2475:(1): 1–9.
1609:flagellum
1527:flagellar
1475:cell wall
1463:detergent
1439:log phase
1238:Inner rod
1148:conserved
1146:Rhizobium
1131:Rhizobium
1106:secretion
865:Evolution
764:caspase 1
742:apoptosis
711:bona fide
641:intestine
596:Congo red
421:inner rod
405:membranes
401:cytoplasm
395:when the
383:) or the
301:Structure
257:Rhizobium
245:Ralstonia
230:) and the
218:(infects
203:Chlamydia
152:Gut flora
106:found in
77:virulence
3729:21139624
3688:33713725
3647:31295426
3606:23030602
3557:22699623
3497:20060835
3448:16888041
3389:16631790
3354:19396171
3305:16920362
3253:15528446
3196:12571230
3109:11171951
3050:23028376
2999:18078471
2958:15001186
2920:17140285
2861:20395475
2796:19390616
2745:20368966
2686:19933107
2635:19933106
2592:19400638
2516:40167923
2450:10984518
2383:16208377
2340:17452659
2332:17610831
2283:11520608
2242:16522800
2185:17136086
2142:11169106
2098:19762438
2047:10939240
2020:12909351
1947:33572887
1896:30455200
1847:30942149
1723:in vitro
1643:proteome
1627:SDS-PAGE
1465:such as
1459:lysozyme
1207:Yersinia
1197:Shigella
1076:proteins
1051:Shigella
1049:: Outer
1018:Shigella
943:Yersinia
933:Yersinia
923:Yersinia
914:Yersinia
900:Yersinia
896:Shigella
669:Shigella
657:butyrate
600:Shigella
590:aromatic
580:such as
578:chelator
570:Yersinia
535:Shigella
454:unfolded
447:and the
445:adhesins
282:Shigella
270:motility
266:flagella
236:such as
191:Yersinia
184:glanders
172:diarrhea
126:(causes
123:Shigella
98:Overview
65:bacteria
63:used by
3597:3483642
3548:3598588
3527:Bibcode
3488:2823972
3439:1567912
3416:Bibcode
3345:2681179
3244:1459965
3223:Bibcode
3215:Science
3155:9554854
3135:Bibcode
3127:Science
3077:Bibcode
3041:3459982
2911:1676029
2888:Bibcode
2852:6485629
2823:Bibcode
2815:Science
2787:2668190
2736:2848554
2666:Bibcode
2658:Science
2643:6648530
2615:Bibcode
2607:Science
2557:9830039
2508:8196540
2418:Bibcode
2391:4355750
2363:Bibcode
2310:Bibcode
2233:2242474
2193:4411244
2165:Bibcode
2089:2889428
1982:8438237
1938:7911332
1887:6346137
1838:6450406
1781:Aurodox
1671:in vivo
1104:E. coli
1074:E. coli
1053:protein
773:) from
706:Mutated
649:acetate
645:formate
608:in vivo
564:in the
559:calcium
514:plasmid
510:operons
430:monomer
239:Erwinia
224:animals
69:secrete
3762:tomato
3727:
3686:
3645:
3604:
3594:
3555:
3545:
3519:Nature
3495:
3485:
3446:
3436:
3387:
3352:
3342:
3303:
3275:Micron
3251:
3241:
3194:
3153:
3107:
3097:
3048:
3038:
2997:
2956:
2918:
2908:
2859:
2849:
2794:
2784:
2743:
2733:
2692:
2684:
2641:
2633:
2590:
2555:
2514:
2506:
2448:
2438:
2389:
2381:
2355:Nature
2338:
2330:
2281:
2240:
2230:
2191:
2183:
2157:Nature
2140:
2096:
2086:
2045:
2018:
1980:
1945:
1935:
1894:
1884:
1845:
1835:
1739:genome
1735:RT-PCR
1708:enzyme
1571:using
1517:nickel
1451:pellet
1445:; the
1375:Spa40
1371:Switch
1365:Orf16
1356:Spa32
1337:Spa47
1333:ATPase
1185:ATPase
1173:operon
881:needle
823:ATPase
809:known.
731:tissue
625:oxygen
458:ATPase
397:ATPase
344:1.B.22
330:Symbol
228:plants
220:humans
196:plague
163:Vibrio
71:their
3271:(PDF)
3100:33380
2690:S2CID
2639:S2CID
2512:S2CID
2441:27139
2387:S2CID
2336:S2CID
2189:S2CID
1823:(2).
1573:X-ray
1403:SepL
1397:InvE
1394:MxiC
1384:EscU
1381:YscU
1378:SpaS
1362:YscP
1359:InvJ
1343:YscN
1340:InvC
1327:CesD
1324:SycD
1321:SicA
1318:IpgC
1308:EspB
1305:YopD
1302:SipC
1299:IpaC
1289:EspD
1286:YopB
1283:SipB
1280:IpaB
1270:EspA
1267:LcrV
1264:SipD
1261:IpaD
1251:EscI
1248:YscI
1245:PrgJ
1242:MxiI
1232:EscF
1229:YscF
1226:PrgI
1223:MxiH
748:from
719:actin
665:lipid
637:cecum
633:ileum
568:(for
3760:pv.
3725:PMID
3684:PMID
3643:PMID
3602:PMID
3553:PMID
3493:PMID
3444:PMID
3385:PMID
3350:PMID
3301:PMID
3249:PMID
3192:PMID
3151:PMID
3105:PMID
3046:PMID
2995:PMID
2954:PMID
2916:PMID
2857:PMID
2792:PMID
2741:PMID
2682:PMID
2631:PMID
2588:PMID
2553:PMID
2504:PMID
2446:PMID
2379:PMID
2328:PMID
2279:PMID
2238:PMID
2181:PMID
2138:PMID
2094:PMID
2043:PMID
2016:PMID
2000:Gene
1978:PMID
1943:PMID
1892:PMID
1843:PMID
1733:and
1633:and
1520:ions
1483:CsCl
1467:LDAO
898:and
746:IpaB
663:, a
655:and
647:and
623:and
586:EGTA
582:EDTA
572:and
562:ions
415:(or
413:base
389:T3SA
368:5tcq
339:TCDB
333:T3SS
248:and
226:and
170:and
79:and
57:TTSS
53:T3SS
47:The
3756:in
3715:doi
3674:doi
3670:184
3633:doi
3592:PMC
3584:doi
3580:134
3543:PMC
3535:doi
3523:486
3483:PMC
3475:doi
3471:396
3434:PMC
3424:doi
3412:103
3377:doi
3373:359
3340:PMC
3332:doi
3291:hdl
3283:doi
3239:PMC
3231:doi
3219:306
3182:doi
3178:278
3143:doi
3131:280
3095:PMC
3085:doi
3036:PMC
3026:doi
2985:doi
2981:177
2946:doi
2906:PMC
2896:doi
2847:PMC
2839:hdl
2831:doi
2819:328
2782:PMC
2772:doi
2731:PMC
2721:doi
2674:doi
2662:326
2623:doi
2611:326
2580:doi
2543:doi
2539:273
2496:doi
2436:PMC
2426:doi
2371:doi
2359:437
2318:doi
2269:doi
2265:202
2228:PMC
2220:doi
2173:doi
2161:444
2128:doi
2084:PMC
2074:doi
2070:156
2008:doi
2004:312
1970:doi
1933:PMC
1923:doi
1882:PMC
1874:doi
1833:PMC
1825:doi
1704:LDH
1611:of
1503:(a
1469:or
1177:kDa
1157:Vir
1142:Rhc
1136:Nop
1123:Hrc
1117:Hrp
1100:Ces
1090:Esp
1080:Esc
1070:Sep
1064:Tir
1047:Osp
1041:Spa
1035:Mxi
1029:Ipa
1023:Ipg
1010:Iag
1000:Ssp
994:Org
988:Inv
982:Prg
972:Sip
962:Sic
956:Spa
939:Ypk
929:Ysc
919:Yop
593:dye
584:or
533:).
531:SPI
356:348
67:to
55:or
3774::
3723:.
3711:64
3709:.
3705:.
3682:.
3668:.
3664:.
3641:.
3629:26
3627:.
3623:.
3600:.
3590:.
3578:.
3574:.
3551:.
3541:.
3533:.
3521:.
3517:.
3505:^
3491:.
3481:.
3469:.
3465:.
3442:.
3432:.
3422:.
3410:.
3406:.
3383:.
3371:.
3348:.
3338:.
3328:16
3326:.
3322:.
3299:.
3289:.
3279:38
3277:.
3273:.
3247:.
3237:.
3229:.
3217:.
3213:.
3190:.
3176:.
3172:.
3149:.
3141:.
3129:.
3117:^
3103:.
3093:.
3083:.
3073:98
3071:.
3067:.
3044:.
3034:.
3020:.
3016:.
2993:.
2979:.
2975:.
2952:.
2942:12
2940:.
2928:^
2914:.
2904:.
2894:.
2882:.
2878:.
2855:.
2845:.
2837:.
2829:.
2817:.
2813:.
2790:.
2780:.
2766:.
2762:.
2739:.
2729:.
2715:.
2711:.
2688:.
2680:.
2672:.
2660:.
2637:.
2629:.
2621:.
2609:.
2586:.
2576:48
2574:.
2551:.
2537:.
2533:.
2510:.
2502:.
2492:11
2490:.
2473:10
2471:.
2467:.
2444:.
2434:.
2424:.
2414:97
2412:.
2408:.
2385:.
2377:.
2369:.
2357:.
2334:.
2326:.
2316:.
2306:17
2304:.
2300:.
2277:.
2263:.
2259:.
2236:.
2226:.
2216:15
2214:.
2210:.
2187:.
2179:.
2171:.
2159:.
2136:.
2124:39
2122:.
2118:.
2106:^
2092:.
2082:.
2068:.
2064:.
2037:.
2014:.
2002:.
1990:^
1976:.
1966:18
1964:.
1941:.
1931:.
1919:11
1917:.
1913:.
1890:.
1880:.
1870:87
1868:.
1864:.
1841:.
1831:.
1819:.
1815:.
1696:).
1662:).
1645:.
1495:.
1488:.
1430:.
1144::
1092::
1082::
1002::
974::
964::
941::
931::
921::
909::
894:,
843:pH
733:.
671:.
619:,
617:pH
615:,
441:nm
434:Da
381:NC
242:,
222:,
210:),
198:),
186:),
174:),
158:),
142:),
130:),
94:.
34:A
3731:.
3717::
3690:.
3676::
3649:.
3635::
3608:.
3586::
3559:.
3537::
3529::
3499:.
3477::
3450:.
3426::
3418::
3391:.
3379::
3356:.
3334::
3307:.
3293::
3285::
3255:.
3233::
3225::
3198:.
3184::
3157:.
3145::
3137::
3111:.
3087::
3079::
3052:.
3028::
3022:8
3001:.
2987::
2960:.
2948::
2922:.
2898::
2890::
2884:2
2863:.
2841::
2833::
2825::
2798:.
2774::
2768:5
2747:.
2723::
2717:6
2696:.
2676::
2668::
2645:.
2625::
2617::
2594:.
2582::
2559:.
2545::
2518:.
2498::
2452:.
2428::
2420::
2393:.
2373::
2365::
2342:.
2320::
2312::
2285:.
2271::
2244:.
2222::
2195:.
2175::
2167::
2144:.
2130::
2100:.
2076::
2049:.
2039:2
2022:.
2010::
1984:.
1972::
1949:.
1925::
1898:.
1876::
1849:.
1827::
1821:8
1692:(
1580:Å
1569:Å
529:(
387:(
379:(
260:.
206:(
194:(
182:(
166:(
150:(
138:(
51:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.