166:
95:
275:
1687:
1699:
466:
tools calculate match scores using only the best scoring alignment, HMMER3 calculates match scores by integrating across all possible alignments, to account for uncertainty in which alignment is best. HMMER sequence alignments are accompanied by posterior probability annotations, indicating which portions of the alignment have been assigned high confidence and which are more uncertain.
27:
286:
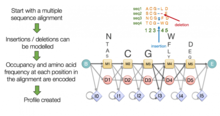
that residue has been observed in that column of the alignment, but also incorporates prior information on patterns of residues that tend to co-occur in the same columns of sequence alignments. This string of match states emitting amino acids at particular frequencies is analogous to position specific score matrices or weight matrices.
482:
While HMMER2 could perform local alignment (align a complete model to a subsequence of the target) and global alignment (align a complete model to a complete target sequence), HMMER3 only performs local alignment. This restriction is due to the difficulty in calculating the significance of hits when
465:
The major advance in speed was made possible by the development of an approach for calculating the significance of results integrated over a range of possible alignments. In discovering remote homologs, alignments between query and hit proteins are often very uncertain. While most sequence alignment
293:
The HMMER2 and HMMER3 releases used an architecture for building profile HMMs called the Plan 7 architecture, named after the seven states captured by the model. In addition to the three major states (M, I and D), six additional states capture non-homologous flanking sequence in the alignment. These
289:
A profile HMM takes this modelling of sequence alignments further by modelling insertions and deletions, using I and D states, respectively. D states do not emit a residue, while I state do emit a residue. Multiple I state can occur consecutively, corresponding to multiple residues between consensus
285:
Profile HMMs center around a linear set of match (M) states, with one state corresponding to each consensus column in a sequence alignment. Each M state emits a single residue (amino acid or nucleotide). The probability of emitting a particular residue is determined largely by the frequency at which
266:
A profile HMM is a variant of an HMM relating specifically to biological sequences. Profile HMMs turn a multiple sequence alignment into a position-specific scoring system, which can be used to align sequences and search databases for remotely homologous sequences. They capitalise on the fact that
203:
constructed explicitly for a particular search) to either a single sequence or a database of sequences. Sequences that score significantly better to the profile-HMM compared to a null model are considered to be homologous to the sequences that were used to construct the profile-HMM. Profile-HMMs
411:
in the United
Kingdom, while development of the algorithm is still performed by Sean Eddy's team in the United States. Major reasons for relocating the web service were to leverage the computing infrastructure at the EBI, and to cross-link HMMER searches with relevant databases that are also
267:
certain positions in a sequence alignment tend to have biases in which residues are most likely to occur, and are likely to differ in their probability of containing an insertion or a deletion. Capturing this information gives them a better ability to detect true homologs than traditional
429:
A major aim of the HMMER3 project, started in 2004 was to improve the speed of HMMER searches. While profile HMM-based homology searches were more accurate than BLAST-based approaches, their slower speed limited their applicability. The main performance gain is due to a
1542:
294:
6 states collectively are important for controlling how sequences are aligned to the model, e.g. whether a sequence can have multiple consecutive hits to the same model (in the case of sequences with multiple instances of the same domain).
290:
columns in an alignment. M, I and D states are connected by state transition probabilities, which also vary by position in the sequence alignment, to reflect the different frequencies of insertions and deletions across sequence alignments.
143:
420:
The latest stable release of HMMER is version 3.0. HMMER3 is complete rewrite of the earlier HMMER2 package, with the aim of improving the speed of profile-HMM searches. Major changes are outlined below:
368:
In addition to the software package, the HMMER search function is available in the form of a web server. The service facilitates searches across a range of databases, including sequence databases such as
1487:
1451:
457:
HMMER still lags behind BLAST in speed of DNA-based searches; however, DNA-based searches can be tuned such that an improvement in speed comes at the expense of accuracy.
1390:
1285:
1469:
882:
Finn, Robert D.; Clements, Jody; Arndt, William; Miller, Benjamin L.; Wheeler, Travis J.; Schreiber, Fabian; Bateman, Alex; Eddy, Sean R. (1 July 2015).
638:
Krogh A, Brown M, Mian IS, Sjölander K, Haussler D (February 1994). "Hidden Markov models in computational biology. Applications to protein modeling".
434:
that finds high-scoring un-gapped matches within database sequences to a query profile. This heuristic results in a computation time comparable to
1481:
1548:
1495:
1396:
1524:
513:
Several implementations of profile HMM methods and related position-specific scoring matrix methods are available. Some are listed below:
302:
The HMMER package consists of a collection of programs for performing functions using profile hidden Markov models. The programs include:
1559:
474:
A major improvement in HMMER3 was the inclusion of DNA/DNA comparison tools. HMMER2 only had functionality to compare protein sequences.
1536:
1530:
1117:
1090:
271:-based approaches, which penalise substitutions, insertions and deletions equally, regardless of where in an alignment they occur.
1735:
1518:
1295:
522:
212:
program. The profile-HMM implementation used in the HMMER software was based on the work of Krogh and colleagues. HMMER is a
1745:
1475:
1373:
537:
1231:
254:
to increase computational speed. This work is based upon an earlier publication showing a significant acceleration of the
1501:
1553:
1457:
1424:
1407:
1367:
622:
408:
1725:
1355:
1343:
125:
1703:
1185:
1740:
1730:
1402:
1322:
1239:
1207:
1652:
1171:
1110:
74:
1280:
1203:
1145:
542:
497:
205:
48:
165:
1260:
993:"A probabilistic model of local sequence alignment that simplifies statistical significance estimation"
557:
1430:
1384:
789:
547:
1543:
International
Conference on Computational Intelligence Methods for Bioinformatics and Biostatistics
1413:
255:
108:
1691:
1657:
1300:
1153:
1103:
397:). The search function accepts single sequences as well as sequence alignments or profile HMMs.
1621:
1361:
1149:
784:
587:
213:
1642:
1255:
527:
439:
435:
404:
organisation of the hits. Search results can then be filtered according to either parameter.
268:
1626:
1004:
776:
675:"Striped Smith-Waterman speeds database searches six times over other SIMD implementations"
833:
8:
1616:
1435:
1223:
1051:
854:
492:
431:
200:
94:
1163:
1008:
780:
1672:
1576:
1379:
1027:
992:
965:
932:
908:
883:
809:
764:
451:
185:
1350:
Microsoft
Research - University of Trento Centre for Computational and Systems Biology
737:
720:
374:
1667:
1606:
1181:
1032:
970:
952:
913:
814:
742:
696:
655:
618:
447:
378:
225:
1167:
691:
674:
580:
393:. The four search types phmmer, hmmsearch, hmmscan and jackhmmer are supported (see
1647:
1611:
1419:
1085:
1022:
1012:
960:
944:
903:
895:
804:
794:
732:
686:
647:
567:
562:
400:
The search results are accompanied by a report on the taxonomic breakdown, and the
217:
195:
sequences, and to perform sequence alignments. It detects homology by comparing a
138:
118:
1338:
1017:
799:
615:
Biological
Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids
1126:
401:
130:
1091:
A blog posting on HMMER policy on trademark, copyright, patents, and licensing
608:
1719:
1601:
1197:
956:
353:
177:
1591:
1586:
1581:
1036:
974:
917:
818:
700:
651:
610:
438:
with little impact on accuracy. Further gains in performance are due to a
746:
659:
948:
899:
517:
390:
83:
1662:
1596:
1215:
1177:
274:
192:
34:
181:
180:
and commonly used software package for sequence analysis written by
1290:
1227:
1219:
1189:
386:
333:
nhmmscan – search nucleotide sequences against a nucleotide profile
330:
nhmmer – search DNA/RNA queries against a DNA/RNA sequence database
327:
jackhmmer – iteratively search sequences against a protein database
311:
hmmbuild – construct profile HMMs from multiple sequence alignments
240:
1265:
1211:
1159:
1141:
1095:
443:
370:
321:
hmmscan – search protein sequences against a profile HMM database
188:
235:
HMMER is the core utility that protein family databases such as
1463:
1317:
1270:
1235:
229:
1349:
933:"HMMER web server: interactive sequence similarity searching"
931:
Finn, Robert D.; Clements, Jody; Eddy, Sean R. (2011-07-01).
507:
244:
221:
1452:
African
Society for Bioinformatics and Computational Biology
552:
483:
performing local/global alignments using the new algorithm.
460:
336:
phmmer – search protein sequences against a protein database
26:
1391:
Max Planck
Institute of Molecular Cell Biology and Genetics
1193:
502:
382:
360:
The package contains numerous other specialised functions.
324:
hmmsearch – search profile HMMs against a sequence database
251:
236:
1049:
852:
1470:
International
Nucleotide Sequence Database Collaboration
243:
are based upon. Some other bioinformatics tools such as
881:
637:
763:
Eddy, Sean R.; Pearson, William R. (20 October 2011).
532:
349:
hmmemit – produce sample sequences from a profile HMM
169:
A profile HMM modelling a multiple sequence alignment
1078:
154:
1162:, database of protein sequences grouping together
477:
442:model that requires no calibration for estimating
394:
39:Sean Eddy, Travis Wheeler, HMMER development team
1717:
1397:US National Center for Biotechnology Information
930:
297:
1482:International Society for Computational Biology
986:
984:
450:to be used for computing the significance of a
278:The core profile HMM architecture used by HMMER
1549:ISCB Africa ASBCB Conference on Bioinformatics
848:
846:
1496:Institute of Genomics and Integrative Biology
1111:
1525:European Conference on Computational Biology
981:
407:The web service is currently run out of the
1560:Research in Computational Molecular Biology
843:
762:
666:
346:hmmalign – align sequences to a profile HMM
1537:International Conference on Bioinformatics
1118:
1104:
877:
875:
758:
756:
714:
712:
710:
631:
469:
93:
25:
1531:Intelligent Systems for Molecular Biology
1026:
1016:
964:
907:
808:
798:
788:
736:
690:
461:Improvements in remote homology searching
672:
424:
273:
164:
1519:Basel Computational Biology Conference
990:
872:
753:
707:
409:European Bioinformatics Institute (EBI)
363:
305:
1718:
991:Eddy SR (2008). Rost, Burkhard (ed.).
16:Software package for sequence analysis
1476:International Society for Biocuration
1374:European Molecular Biology Laboratory
1099:
415:
315:
1698:
718:
1502:Japanese Society for Bioinformatics
250:HMMER3 also makes extensive use of
184:. Its general usage is to identify
13:
1464:European Molecular Biology network
1125:
765:"Accelerated Profile HMM Searches"
340:
220:, including different versions of
14:
1757:
1554:Pacific Symposium on Biocomputing
1458:Australia Bioinformatics Resource
1425:Swiss Institute of Bioinformatics
1408:Netherlands Bioinformatics Centre
1368:European Bioinformatics Institute
1070:
1050:Sean R. Eddy; Travis J. Wheeler.
853:Sean R. Eddy; Travis J. Wheeler.
1697:
1686:
1685:
1356:Database Center for Life Science
1344:Computational Biology Department
1232:Arabidopsis Information Resource
1054:. and the HMMER development team
860:. and the HMMER development team
831:
1202:Specialised genomic databases:
1043:
924:
884:"HMMER web server: 2015 update"
738:10.1093/bioinformatics/14.9.755
609:Durbin, Richard; Sean R. Eddy;
478:Restriction to local alignments
446:, and allows the more accurate
261:
208:in the HMMER package using the
1403:Japanese Institute of Genetics
825:
721:"Profile hidden Markov models"
617:. Cambridge University Press.
602:
352:hmmlogo – produce data for an
216:utility ported to every major
1:
1736:Free software programmed in C
1323:Rosalind (education platform)
1240:Zebrafish Information Network
1208:Saccharomyces Genome Database
943:(Web Server issue): W29–W37.
692:10.1093/bioinformatics/btl582
573:
298:Programs in the HMMER package
1746:Free bioinformatics software
1653:List of biological databases
1172:Protein Information Resource
1018:10.1371/journal.pcbi.1000069
800:10.1371/journal.pcbi.1002195
381:, and HMM databases such as
258:for aligning two sequences.
7:
1146:European Nucleotide Archive
613:; Graeme Mitchison (1998).
498:Sequence alignment software
486:
206:multiple sequence alignment
10:
1762:
1052:"HMMER3.1b2 Release Notes"
769:PLOS Computational Biology
1681:
1635:
1569:
1511:
1444:
1431:Wellcome Sanger Institute
1385:J. Craig Venter Institute
1331:
1309:
1248:
1133:
673:Farrar M (January 2007).
149:
137:
124:
114:
104:
73:
69:
47:
43:
33:
24:
1414:Philippine Genome Center
256:Smith-Waterman algorithm
1726:Bioinformatics software
1658:Molecular phylogenetics
1154:China National GeneBank
470:DNA sequence comparison
412:maintained by the EBI.
204:are constructed from a
56:; 13 months ago
54:3.4 / 15 August 2023
1362:DNA Data Bank of Japan
1150:DNA Data Bank of Japan
937:Nucleic Acids Research
888:Nucleic Acids Research
652:10.1006/jmbi.1994.1104
282:
170:
1741:Computational science
1731:Free science software
1643:Computational biology
1158:Secondary databases:
834:"HMMER2 User's Guide"
425:Improvements in speed
277:
168:
1140:Sequence databases:
855:"HMMER User's Guide"
364:The HMMER web server
306:Profile HMM building
1436:Whitehead Institute
1224:Rat Genome Database
1086:HMMER3 announcement
1009:2008PLSCB...4E0069E
781:2011PLSCB...7E2195E
493:Hidden Markov model
252:vector instructions
201:Hidden Markov model
21:
1673:Sequence alignment
1380:Flatiron Institute
949:10.1093/nar/gkr367
900:10.1093/nar/gkv397
416:The HMMER3 release
316:Homology searching
283:
171:
19:
1713:
1712:
1668:Sequence database
1182:Protein Data Bank
1176:Other databases:
719:Eddy, SR (1998).
379:Protein Data Bank
163:
162:
1753:
1701:
1700:
1689:
1688:
1648:List of biobanks
1612:Stockholm format
1420:Scripps Research
1120:
1113:
1106:
1097:
1096:
1082:
1081:
1079:Official website
1064:
1063:
1061:
1059:
1047:
1041:
1040:
1030:
1020:
997:PLOS Comput Biol
988:
979:
978:
968:
928:
922:
921:
911:
879:
870:
869:
867:
865:
859:
850:
841:
840:
838:
829:
823:
822:
812:
802:
792:
775:(10): e1002195.
760:
751:
750:
740:
716:
705:
704:
694:
670:
664:
663:
635:
629:
628:
606:
600:
599:
597:
595:
590:. 15 August 2023
584:
432:heuristic filter
356:from an HMM file
247:also use HMMER.
218:operating system
159:
156:
97:
92:
89:
87:
85:
64:
62:
57:
29:
22:
18:
1761:
1760:
1756:
1755:
1754:
1752:
1751:
1750:
1716:
1715:
1714:
1709:
1677:
1631:
1565:
1507:
1488:Student Council
1440:
1339:Broad Institute
1327:
1305:
1244:
1129:
1124:
1077:
1076:
1073:
1068:
1067:
1057:
1055:
1048:
1044:
1003:(5): e1000069.
989:
982:
929:
925:
894:(W1): W30–W38.
880:
873:
863:
861:
857:
851:
844:
836:
830:
826:
790:10.1.1.290.1476
761:
754:
717:
708:
671:
667:
636:
632:
625:
607:
603:
593:
591:
586:
585:
581:
576:
489:
480:
472:
463:
427:
418:
366:
343:
341:Other functions
318:
308:
300:
264:
153:
100:
82:
65:
60:
58:
55:
17:
12:
11:
5:
1759:
1749:
1748:
1743:
1738:
1733:
1728:
1711:
1710:
1708:
1707:
1695:
1682:
1679:
1678:
1676:
1675:
1670:
1665:
1660:
1655:
1650:
1645:
1639:
1637:
1636:Related topics
1633:
1632:
1630:
1629:
1624:
1619:
1614:
1609:
1604:
1599:
1594:
1589:
1584:
1579:
1573:
1571:
1567:
1566:
1564:
1563:
1557:
1551:
1546:
1540:
1534:
1528:
1522:
1515:
1513:
1509:
1508:
1506:
1505:
1499:
1493:
1492:
1491:
1479:
1473:
1467:
1461:
1455:
1448:
1446:
1442:
1441:
1439:
1438:
1433:
1428:
1422:
1417:
1411:
1405:
1400:
1394:
1388:
1382:
1377:
1371:
1365:
1359:
1353:
1347:
1341:
1335:
1333:
1329:
1328:
1326:
1325:
1320:
1313:
1311:
1307:
1306:
1304:
1303:
1298:
1293:
1288:
1283:
1278:
1273:
1268:
1263:
1258:
1252:
1250:
1246:
1245:
1243:
1242:
1200:
1174:
1156:
1137:
1135:
1131:
1130:
1127:Bioinformatics
1123:
1122:
1115:
1108:
1100:
1094:
1093:
1088:
1083:
1072:
1071:External links
1069:
1066:
1065:
1042:
980:
923:
871:
842:
824:
752:
725:Bioinformatics
706:
679:Bioinformatics
665:
646:(5): 1501–31.
630:
623:
601:
578:
577:
575:
572:
571:
570:
565:
560:
555:
550:
545:
540:
535:
530:
525:
520:
511:
510:
505:
500:
495:
488:
485:
479:
476:
471:
468:
462:
459:
448:forward scores
440:log-likelihood
426:
423:
417:
414:
365:
362:
358:
357:
350:
347:
342:
339:
338:
337:
334:
331:
328:
325:
322:
317:
314:
313:
312:
307:
304:
299:
296:
263:
260:
161:
160:
151:
147:
146:
141:
135:
134:
131:Bioinformatics
128:
122:
121:
116:
112:
111:
106:
102:
101:
99:
98:
79:
77:
71:
70:
67:
66:
61:15 August 2023
53:
51:
49:Stable release
45:
44:
41:
40:
37:
31:
30:
15:
9:
6:
4:
3:
2:
1758:
1747:
1744:
1742:
1739:
1737:
1734:
1732:
1729:
1727:
1724:
1723:
1721:
1706:
1705:
1696:
1694:
1693:
1684:
1683:
1680:
1674:
1671:
1669:
1666:
1664:
1661:
1659:
1656:
1654:
1651:
1649:
1646:
1644:
1641:
1640:
1638:
1634:
1628:
1625:
1623:
1620:
1618:
1615:
1613:
1610:
1608:
1605:
1603:
1602:Pileup format
1600:
1598:
1595:
1593:
1590:
1588:
1585:
1583:
1580:
1578:
1575:
1574:
1572:
1568:
1561:
1558:
1555:
1552:
1550:
1547:
1544:
1541:
1538:
1535:
1532:
1529:
1526:
1523:
1520:
1517:
1516:
1514:
1510:
1503:
1500:
1497:
1494:
1489:
1486:
1485:
1483:
1480:
1477:
1474:
1471:
1468:
1465:
1462:
1459:
1456:
1453:
1450:
1449:
1447:
1445:Organizations
1443:
1437:
1434:
1432:
1429:
1426:
1423:
1421:
1418:
1415:
1412:
1409:
1406:
1404:
1401:
1398:
1395:
1392:
1389:
1386:
1383:
1381:
1378:
1375:
1372:
1369:
1366:
1363:
1360:
1357:
1354:
1351:
1348:
1345:
1342:
1340:
1337:
1336:
1334:
1330:
1324:
1321:
1319:
1315:
1314:
1312:
1308:
1302:
1299:
1297:
1294:
1292:
1289:
1287:
1284:
1282:
1279:
1277:
1274:
1272:
1269:
1267:
1264:
1262:
1259:
1257:
1254:
1253:
1251:
1247:
1241:
1237:
1233:
1229:
1225:
1221:
1217:
1213:
1209:
1205:
1201:
1199:
1198:Gene Ontology
1195:
1191:
1187:
1183:
1179:
1175:
1173:
1169:
1165:
1161:
1157:
1155:
1151:
1147:
1143:
1139:
1138:
1136:
1132:
1128:
1121:
1116:
1114:
1109:
1107:
1102:
1101:
1098:
1092:
1089:
1087:
1084:
1080:
1075:
1074:
1053:
1046:
1038:
1034:
1029:
1024:
1019:
1014:
1010:
1006:
1002:
998:
994:
987:
985:
976:
972:
967:
962:
958:
954:
950:
946:
942:
938:
934:
927:
919:
915:
910:
905:
901:
897:
893:
889:
885:
878:
876:
856:
849:
847:
835:
828:
820:
816:
811:
806:
801:
796:
791:
786:
782:
778:
774:
770:
766:
759:
757:
748:
744:
739:
734:
731:(9): 755–63.
730:
726:
722:
715:
713:
711:
702:
698:
693:
688:
685:(2): 156–61.
684:
680:
676:
669:
661:
657:
653:
649:
645:
641:
634:
626:
624:0-521-62971-3
620:
616:
612:
605:
589:
588:"Release 3.4"
583:
579:
569:
566:
564:
561:
559:
556:
554:
551:
549:
546:
544:
541:
539:
536:
534:
531:
529:
526:
524:
521:
519:
516:
515:
514:
509:
506:
504:
501:
499:
496:
494:
491:
490:
484:
475:
467:
458:
455:
453:
449:
445:
441:
437:
433:
422:
413:
410:
405:
403:
398:
396:
392:
388:
384:
380:
376:
372:
361:
355:
351:
348:
345:
344:
335:
332:
329:
326:
323:
320:
319:
310:
309:
303:
295:
291:
287:
281:
276:
272:
270:
259:
257:
253:
248:
246:
242:
238:
233:
231:
227:
223:
219:
215:
211:
207:
202:
198:
194:
190:
187:
183:
179:
175:
167:
158:
152:
148:
145:
142:
140:
136:
132:
129:
127:
123:
120:
117:
113:
110:
107:
103:
96:
91:
88:/EddyRivasLab
81:
80:
78:
76:
72:
68:
52:
50:
46:
42:
38:
36:
32:
28:
23:
1702:
1690:
1597:Nexus format
1592:NeXML format
1587:FASTQ format
1582:FASTA format
1570:File formats
1332:Institutions
1275:
1056:. Retrieved
1045:
1000:
996:
940:
936:
926:
891:
887:
862:. Retrieved
832:Eddy, Sean.
827:
772:
768:
728:
724:
682:
678:
668:
643:
640:J. Mol. Biol
639:
633:
614:
611:Anders Krogh
604:
594:18 September
592:. Retrieved
582:
512:
481:
473:
464:
456:
428:
419:
406:
399:
367:
359:
301:
292:
288:
284:
279:
265:
262:Profile HMMs
249:
234:
209:
196:
173:
172:
115:Available in
35:Developer(s)
1577:CRAM format
1498:(CSIR-IGIB)
568:DeCypherHMM
391:SUPERFAMILY
197:profile-HMM
1720:Categories
1663:Sequencing
1627:GTF format
1622:GFF format
1617:VCF format
1607:SAM format
1370:(EMBL-EBI)
1296:SOAP suite
1216:VectorBase
1178:BioNumbers
1164:Swiss-Prot
574:References
454:sequence.
452:homologous
377:, and the
193:nucleotide
186:homologous
105:Written in
75:Repository
1490:(ISCB-SC)
1460:(EMBL-AR)
1393:(MPI-CBG)
1134:Databases
957:0305-1048
785:CiteSeerX
563:GPU-HMMER
553:META-MEME
528:PSI-BLAST
375:SwissProt
182:Sean Eddy
1692:Category
1562:(RECOMB)
1512:Meetings
1466:(EMBnet)
1316:Server:
1291:SAMtools
1286:PANGOLIN
1249:Software
1228:PHI-base
1220:WormBase
1190:InterPro
1037:18516236
975:21593126
918:25943547
819:22039361
701:17110365
543:GENEWISE
518:HH-suite
487:See also
444:E-values
395:Programs
387:TIGRFAMs
354:HMM logo
241:InterPro
210:hmmbuild
1704:Commons
1539:(InCoB)
1484:(ISCB)
1472:(INSDC)
1454:(ASBCB)
1358:(DBCLS)
1352:(COSBI)
1266:Clustal
1212:FlyBase
1186:Ensembl
1160:UniProt
1142:GenBank
1058:23 July
1028:2396288
1005:Bibcode
966:3125773
909:4489315
864:23 July
810:3197634
777:Bibcode
747:9918945
660:8107089
538:PFTOOLS
533:MMseqs2
371:UniProt
226:Windows
214:console
189:protein
150:Website
139:License
119:English
59: (
1545:(CIBB)
1533:(ISMB)
1527:(ECCB)
1504:(JSBi)
1410:(NBIC)
1399:(NCBI)
1387:(JCVI)
1376:(EMBL)
1364:(DDBJ)
1318:ExPASy
1301:TopHat
1281:MUSCLE
1271:EMBOSS
1261:Bowtie
1236:GISAID
1196:, and
1168:TrEMBL
1035:
1025:
973:
963:
955:
916:
906:
817:
807:
787:
745:
699:
658:
621:
558:BLOCKS
402:domain
228:, and
90:/hmmer
84:github
1556:(PSB)
1478:(ISB)
1427:(SIB)
1416:(PGC)
1346:(CBD)
1310:Other
1276:HMMER
1256:BLAST
858:(PDF)
837:(PDF)
548:PROBE
508:UGENE
436:BLAST
269:BLAST
245:UGENE
230:macOS
222:Linux
176:is a
174:HMMER
155:hmmer
144:BSD-3
20:HMMER
1238:and
1204:BOLD
1194:KEGG
1170:and
1152:and
1060:2017
1033:PMID
971:PMID
953:ISSN
914:PMID
866:2017
815:PMID
743:PMID
697:PMID
656:PMID
619:ISBN
596:2023
503:Pfam
389:and
383:Pfam
239:and
237:Pfam
178:free
157:.org
133:tool
126:Type
86:.com
1023:PMC
1013:doi
961:PMC
945:doi
904:PMC
896:doi
805:PMC
795:doi
733:doi
687:doi
648:doi
644:235
523:SAM
199:(a
191:or
1722::
1521:()
1234:,
1230:,
1226:,
1222:,
1218:,
1214:,
1210:,
1206:,
1192:,
1188:,
1184:,
1180:,
1166:,
1148:,
1144:,
1031:.
1021:.
1011:.
999:.
995:.
983:^
969:.
959:.
951:.
941:39
939:.
935:.
912:.
902:.
892:43
890:.
886:.
874:^
845:^
813:.
803:.
793:.
783:.
771:.
767:.
755:^
741:.
729:14
727:.
723:.
709:^
695:.
683:23
681:.
677:.
654:.
642:.
385:,
373:,
232:.
224:,
1119:e
1112:t
1105:v
1062:.
1039:.
1015::
1007::
1001:4
977:.
947::
920:.
898::
868:.
839:.
821:.
797::
779::
773:7
749:.
735::
703:.
689::
662:.
650::
627:.
598:.
280:.
109:C
63:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.