320:
therapeutic effects. This method is to first discover the in vivo or in vitro functional activity of drugs (such as extract drugs or natural products), and then perform target identification. Phenotypic discovery uses a practical and target-independent approach to generate initial leads, aiming to discover pharmacologically active compounds and therapeutics that operate through novel drug mechanisms. This method allows the exploration of disease phenotypes to find potential treatments for conditions with unknown, complex, or multifactorial origins, where the understanding of molecular targets is insufficient for effective intervention.
1356:. Phenotypic screening is characterized by the process of screening drugs using cellular or animal disease models to identify compounds that alter the phenotype and produce beneficial disease-related effects. Emerging technologies in high-throughput screening substantially enhance processing speed and decrease the required detection volume. Virtual screening is completed by computer, enabling a large number of molecules can be screened with a short cycle and low cost. Virtual screening uses a range of computational methods that empower chemists to reduce extensive virtual libraries into more manageable sizes.
332:) begins with a hypothesis that modulation of a specific biological target may have therapeutic value. In order for a biomolecule to be selected as a drug target, two essential pieces of information are required. The first is evidence that modulation of the target will be disease modifying. This knowledge may come from, for example, disease linkage studies that show an association between mutations in the biological target and certain disease states. The second is that the target is capable of binding to a small molecule and that its activity can be modulated by the small molecule.
490:
35:
531:
1196:, the number of hydrogen bonds formed, etc. In practice, the components of the master equation are fit to experimental data using multiple linear regression. This can be done with a diverse training set including many types of ligands and receptors to produce a less accurate but more general "global" model or a more restricted set of ligands and receptors to produce a more accurate but less general "local" model.
1150:
895:
948:
1204:
A particular example of rational drug design involves the use of three-dimensional information about biomolecules obtained from such techniques as X-ray crystallography and NMR spectroscopy. Computer-aided drug design in particular becomes much more tractable when there is a high-resolution structure
1179:
The basic idea is that the overall binding free energy can be decomposed into independent components that are known to be important for the binding process. Each component reflects a certain kind of free energy alteration during the binding process between a ligand and its target receptor. The Master
1187:
Various computational methods are used to estimate each of the components of the master equation. For example, the change in polar surface area upon ligand binding can be used to estimate the desolvation energy. The number of rotatable bonds frozen upon ligand binding is proportional to the motion
558:
model that defines the minimum necessary structural characteristics a molecule must possess in order to bind to the target. A model of the biological target may be built based on the knowledge of what binds to it, and this model in turn may be used to design new molecular entities that interact with
701:
457:
Ideally, the computational method will be able to predict affinity before a compound is synthesized and hence in theory only one compound needs to be synthesized, saving enormous time and cost. The reality is that present computational methods are imperfect and provide, at best, only qualitatively
622:
A second category is de novo design of new ligands. In this method, ligand molecules are built up within the constraints of the binding pocket by assembling small pieces in a stepwise manner. These pieces can be either individual atoms or molecular fragments. The key advantage of such a method is
610:
Current methods for structure-based drug design can be divided roughly into three main categories. The first method is identification of new ligands for a given receptor by searching large databases of 3D structures of small molecules to find those fitting the binding pocket of the receptor using
302:
technologies may have therapeutic applications. For example, nanomedicines based on mRNA can streamline and expedite the drug development process, enabling transient and localized expression of immunostimulatory molecules. In vitro transcribed (IVT) mRNA allows for delivery to various accessible
497:
In order to overcome the insufficient prediction of binding affinity calculated by recent scoring functions, the protein-ligand interaction and compound 3D structure information are used for analysis. For structure-based drug design, several post-screening analyses focusing on protein-ligand
319:
is a traditional drug discovery method, also known as forward pharmacology or classical pharmacology. It uses the process of phenotypic screening on collections of synthetic small molecules, natural products, or extracts within chemical libraries to pinpoint substances exhibiting beneficial
458:
accurate estimates of affinity. In practice, it requires several iterations of design, synthesis, and testing before an optimal drug is discovered. Computational methods have accelerated discovery by reducing the number of iterations required and have often provided novel structures.
2693:
Becker OM, Dhanoa DS, Marantz Y, Chen D, Shacham S, Cheruku S, et al. (June 2006). "An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression".
1145:{\displaystyle {\begin{array}{lll}\Delta G_{\text{bind}}=-RT\ln K_{\text{d}}\\K_{\text{d}}={\dfrac {}{}}\\\Delta G_{\text{bind}}=\Delta G_{\text{desolvation}}+\Delta G_{\text{motion}}+\Delta G_{\text{configuration}}+\Delta G_{\text{interaction}}\end{array}}}
646:(protein without ligand) structures are available and the reliable identification of unoccupied sites that have the potential to bind ligands with high affinity is non-trivial. In brief, binding site identification usually relies on identification of
2848:
Amari S, Aizawa M, Zhang J, Fukuzawa K, Mochizuki Y, Iwasawa Y, et al. (2006). "VISCANA: visualized cluster analysis of protein-ligand interaction based on the ab initio fragment molecular orbital method for virtual ligand screening".
354:
The search for small molecules that bind to the target is begun by screening libraries of potential drug compounds. This may be done by using the screening assay (a "wet screen"). In addition, if the structure of the target is available, a
890:{\displaystyle \Delta G_{\text{bind}}=\Delta G_{\text{0}}+\Delta G_{\text{hb}}\Sigma _{h-bonds}+\Delta G_{\text{ionic}}\Sigma _{ionic-int}+\Delta G_{\text{lipophilic}}\left\vert A\right\vert +\Delta G_{\text{rot}}{\mathit {NROT}}}
2657:
Singh J, Chuaqui CE, Boriack-Sjodin PA, Lee WC, Pontz T, Corbley MJ, et al. (December 2003). "Successful shape-based virtual screening: the discovery of a potent inhibitor of the type I TGFbeta receptor kinase (TbetaRI)".
3940:
1421:
3885:
303:
cell types via the blood or alternative pathways. The use of IVT mRNA serves to convey specific genetic information into a person's cells, with the primary objective of preventing or altering a particular disease.
3841:
453:
or other statistical techniques to derive predictive binding affinity equations by fitting experimental affinities to computationally derived interaction energies between the small molecule and the target.
141:
design (i.e., design of a molecule that will bind tightly to its target). Although design techniques for prediction of binding affinity are reasonably successful, there are many other properties, such as
3930:
1416:
688:
drugs with fewer side effects. Thus, one of the most important principles for designing or obtaining potential new ligands is to predict the binding affinity of a certain ligand to its target (and known
638:
is determined in the presence of a bound ligand, then the ligand should be observable in the structure in which case location of the binding site is trivial. However, there may be unoccupied
1205:
of a target protein bound to a potent ligand. This approach to drug discovery is sometimes referred to as structure-based drug design. The first unequivocal example of the application of
129:
are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.
2030:
Scomparin A, Polyak D, Krivitsky A, Satchi-Fainaro R (November 2015). "Achieving successful delivery of oligonucleotides--From physico-chemical characterization to in vivo evaluation".
1759:
Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, et al. (July 2015). "An analysis of the attrition of drug candidates from four major pharmaceutical companies".
623:
that novel structures, not contained in any database, can be suggested. A third method is the optimization of known ligands by evaluating proposed analogs within the binding cavity.
696:
One early general-purposed empirical scoring function to describe the binding energy of ligands to receptors was developed by Böhm. This empirical scoring function took the form:
3236:
Böhm HJ (June 1994). "The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure".
3985:
3980:
3910:
154:, that first must be optimized before a ligand can become a safe and effictive drug. These other characteristics are often difficult to predict with rational design techniques.
3995:
2813:
Deng Z, Chuaqui C, Singh J (January 2004). "Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein-ligand binding interactions".
2562:
Lewis RA (2011). "Chapter 4: The
Development of Molecular Modelling Programs: The Use and Limitations of Physical Models". In Gramatica P, Livingstone DJ, Davis AM (eds.).
430:
are often used to provide optimized parameters for the molecular mechanics calculations and also provide an estimate of the electronic properties (electrostatic potential,
3915:
595:
of the target based on the experimental structure of a related protein. Using the structure of the biological target, candidate drugs that are predicted to bind with high
2778:
Oda A, Tsuchida K, Takakura T, Yamaotsu N, Hirono S (2006). "Comparison of consensus scoring strategies for evaluating computational models of protein-ligand complexes".
4000:
3945:
1426:
3394:
Greer J, Erickson JW, Baldwin JJ, Varney MD (April 1994). "Application of the three-dimensional structures of protein target molecules in structure-based drug design".
3831:
1376:
4814:
382:
techniques are sometimes employed. Finally because of the limitations in the current methods for prediction of activity, drug design is still very much reliant on
4009:
3960:
3865:
1396:
3875:
2370:
3821:
3990:
3925:
3900:
1406:
1180:
Equation is the linear combination of these components. According to Gibbs free energy equation, the relation between dissociation equilibrium constant, K
3950:
1431:
169:
properties are predicted to result in fewer complications during development and hence more likely to lead to an approved, marketed drug. Furthermore,
2509:"Bounded Rationality and the Search for Organizational Architecture: An Evolutionary Perspective on the Design of Organizations and Their Evolvability"
3497:
3905:
3880:
2595:
Rajamani R, Good AC (May 2007). "Ranking poses in structure-based lead discovery and optimization: current trends in scoring function development".
6064:
3870:
1401:
3975:
588:
5495:
3970:
3454:
Capdeville R, Buchdunger E, Zimmermann J, Matter A (July 2002). "Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug".
1446:
5943:
4014:
1436:
907:– empirically derived offset that in part corresponds to the overall loss of translational and rotational entropy of the ligand upon binding.
1192:
calculations. Finally the interaction energy can be estimated using methods such as the change in non polar surface, statistically derived
232:
will be designed to enhance or inhibit the target function in the specific disease modifying pathway. Small molecules (for example receptor
3935:
3920:
560:
476:
2299:
Yuan Y, Pei J, Lai L (Dec 2013). "Binding site detection and druggability prediction of protein targets for structure-based drug design".
3855:
3846:
3826:
672:
Structure-based drug design attempts to use the structure of proteins as a basis for designing new ligands by applying the principles of
268:
of target. Small molecules (drugs) can be designed so as not to affect any other important "off-target" molecules (often referred to as
105:
to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on
3836:
554:) relies on knowledge of other molecules that bind to the biological target of interest. These other molecules may be used to derive a
367:, adequate chemical and metabolic stability, and minimal toxic effects. Several methods are available to estimate druglikeness such as
2266:
437:
Molecular mechanics methods may also be used to provide semi-quantitative prediction of the binding affinity. Also, knowledge-based
3358:
5564:
1681:
Shirai H, Prades C, Vita R, Marcatili P, Popovic B, Xu J, et al. (November 2014). "Antibody informatics for drug discovery".
3895:
3890:
419:
3203:
Warren GL, Warren SD (2011). "Chapter 16: Scoring Drug-Receptor
Interactions". In Gramatica P, Livingstone DJ, Davis AM (eds.).
1411:
1319:
398:
The most fundamental goal in drug design is to predict whether a given molecule will bind to a target and if so how strongly.
1463:
It has been argued that the highly rigid and focused nature of rational drug design suppresses serendipity in drug discovery.
113:. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as
5569:
3965:
3751:
3438:
2947:
2893:
2407:
2382:
2014:
1802:
Yu H, Adedoyin A (September 2003). "ADME-Tox in drug discovery: integration of experimental and computational technologies".
1547:
1441:
1386:
418:
of the small molecule and to model conformational changes in the target that may occur when the small molecule binds to it.
34:
4350:
3955:
228:. Potential drug targets are not necessarily disease causing but must by definition be disease modifying. In some cases,
2910:
600:
423:
5513:
4308:
3378:
3220:
2922:
2579:
1577:
634:
identification is the first step in structure based design. If the structure of the target or a sufficiently similar
5936:
5523:
2278:
1381:
584:
5754:
5373:
4047:
1716:
Tollenaere JP (April 1996). "The role of structure-based ligand design and molecular modelling in drug discovery".
1498:
534:
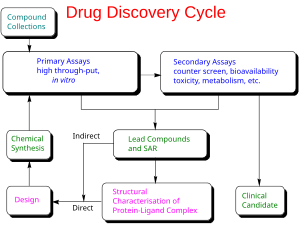
Drug discovery cycle highlighting both ligand-based (indirect) and structure-based (direct) drug design strategies.
2519:(3). Sage Publications, Inc. on behalf of the Johnson Graduate School of Management, Cornell University: 404–437.
1366:
3322:
Murcko MA (December 1995). "Computational methods to predict binding free energy in ligand-receptor complexes".
6085:
5518:
4876:
1886:
Imming P, Sinning C, Meyer A (October 2006). "Drugs, their targets and the nature and number of drug targets".
563:(QSAR), in which a correlation between calculated properties of molecules and their experimentally determined
5589:
5142:
4665:
4242:
4232:
667:
650:
surfaces on the protein that can accommodate drug sized molecules that also possess appropriate "hot spots" (
438:
378:
Due to the large number of drug properties that must be simultaneously optimized during the design process,
6095:
5929:
5395:
5378:
5100:
4940:
4375:
4291:
4264:
2622:
de
Azevedo WF, Dias R (December 2008). "Computational methods for calculation of ligand-binding affinity".
1237:
379:
245:
4692:
4254:
3799:
607:. Alternatively, various automated computational procedures may be used to suggest new drug candidates.
5887:
6019:
5759:
5621:
5533:
4903:
4442:
3613:
Hertzberg RP, Pope AJ (August 2000). "High-throughput screening: new technology for the 21st century".
1518:
1241:
567:, may be derived. These QSAR relationships in turn may be used to predict the activity of new analogs.
472:
407:
375:. Several methods for predicting drug metabolism have also been proposed in the scientific literature.
368:
498:
interaction have been developed for improving enrichment and effectively mining potential candidates:
165:, more attention is being focused early in the drug design process on selecting candidate drugs whose
5716:
5385:
5000:
4995:
4925:
4670:
4655:
4466:
4382:
3744:
1508:
1349:
953:
427:
3036:
Wang R, Gao Y, Lai L (2000). "LigBuilder: A Multi-Purpose
Program for Structure-Based Drug Design".
2271:
Introduction to
Biological and Small Molecule Drug Research and Development: theory and case studies
1172:
configuration – conformational strain energy required to put the ligand in its "active" conformation
348:
5721:
5478:
5390:
5152:
4866:
4839:
4769:
4630:
4486:
4397:
4271:
3716:
1391:
415:
1252:
simply target rapidly dividing cells, not differentiating between cancer cells and other tissues.
97:. In the most basic sense, drug design involves the design of molecules that are complementary in
6090:
5873:
5686:
5651:
5579:
5538:
5490:
5348:
5167:
5125:
5090:
5070:
4834:
3789:
1229:
1193:
681:
5901:
1640:
Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, Kmiecik S (August 2018).
6004:
5206:
5075:
5040:
4491:
4392:
4365:
4303:
1513:
1270:
1261:
316:
126:
3168:
Leis S, Schneider S, Zacharias M (2010). "In silico prediction of binding sites on proteins".
3071:
Schneider G, Fechner U (August 2005). "Computer-based de novo design of drug-like molecules".
2423:
Nicolaou CA, Brown N (September 2013). "Multi-objective optimization methods in drug design".
6024:
5671:
5554:
5368:
5174:
5147:
5137:
5105:
5095:
5060:
4975:
4920:
4915:
4898:
4844:
4682:
4635:
4496:
4360:
4281:
4137:
2548:
673:
596:
508:
May lose the relationship between protein-ligand structural information and scoring criterion
372:
151:
138:
2731:"Consensus scoring for enriching near-native structures from protein-protein docking decoys"
591:. If an experimental structure of a target is not available, it may be possible to create a
294:) produced through biological processes are becoming increasingly more common. In addition,
5841:
5696:
5656:
5410:
5363:
5184:
5132:
5055:
5045:
4945:
4908:
4888:
4819:
4809:
4217:
4207:
4164:
4142:
4097:
4040:
3737:
3245:
3123:
2963:
Mauser H, Guba W (May 2008). "Recent developments in de novo design and scaffold hopping".
1345:
639:
364:
344:
329:
147:
3515:
Prior M, Chiruta C, Currais A, Goldberg J, Ramsey J, Dargusch R, et al. (July 2014).
2267:"The small molecule drug discovery process — from target selection to candidate selection"
8:
6039:
6029:
5953:
5636:
5559:
5468:
5353:
5201:
5179:
5085:
5065:
4930:
4789:
4774:
4687:
4454:
4407:
4188:
1503:
1189:
677:
612:
604:
564:
461:
Computer-aided drug design may be used at any of the following stages of drug discovery:
399:
387:
237:
210:
194:
106:
3249:
3127:
2334:
Rishton GM (January 2003). "Nonleadlikeness and leadlikeness in biochemical screening".
2093:
2066:
1970:
Recanatini M, Bottegoni G, Cavalli A (December 2004). "In silico antitarget screening".
5749:
5701:
5676:
5115:
4950:
4935:
4794:
4610:
4481:
4427:
4159:
3712:
3595:
3541:
3516:
3479:
3269:
3147:
3096:
3053:
3018:
2755:
2730:
2536:
2528:
2484:
2459:
2247:
2193:
2166:
2147:
1911:
1863:
1838:
1784:
1741:
1296:
580:
403:
340:
166:
90:
3661:
3626:
1815:
383:
359:
may be performed of candidate drugs. Ideally, the candidate drug compounds should be "
6014:
5856:
5826:
5631:
5584:
5440:
5430:
5251:
5246:
5241:
5211:
5050:
5035:
4990:
4980:
4965:
4960:
4893:
4829:
4824:
4727:
4562:
4459:
4437:
4387:
4345:
4335:
4249:
4127:
4067:
3692:
3630:
3587:
3546:
3498:"AutoDock's role in Developing the First Clinically-Approved HIV Integrase Inhibitor"
3471:
3434:
3411:
3374:
3339:
3304:
3261:
3216:
3185:
3139:
3088:
3010:
2972:
2943:
2918:
2889:
2866:
2830:
2795:
2760:
2711:
2675:
2639:
2604:
2575:
2544:
2540:
2489:
2440:
2403:
2378:
2351:
2316:
2274:
2239:
2198:
2139:
2098:
2047:
2043:
2010:
1987:
1952:
1903:
1868:
1819:
1776:
1733:
1698:
1663:
1619:
1573:
1543:
1371:
1353:
616:
592:
466:
442:
356:
336:
291:
280:
have the highest chance of cross reactivity and hence highest side effect potential.
277:
206:
118:
67:
3599:
3057:
3022:
2218:"Opportunities and challenges in phenotypic drug discovery: an industry perspective"
2151:
1788:
1745:
6049:
6044:
6009:
5994:
5831:
5796:
5771:
5484:
5458:
5420:
5226:
5162:
5110:
5015:
5005:
4985:
4871:
4861:
4779:
4732:
4702:
4677:
4645:
4605:
4545:
4476:
4315:
3860:
3784:
3723:
3684:
3657:
3622:
3577:
3536:
3528:
3483:
3463:
3403:
3366:
3331:
3296:
3273:
3253:
3208:
3177:
3151:
3131:
3100:
3080:
3045:
3002:
2858:
2822:
2787:
2750:
2742:
2703:
2667:
2631:
2567:
2520:
2479:
2471:
2432:
2343:
2308:
2251:
2229:
2188:
2183:
2178:
2129:
2088:
2078:
2039:
1979:
1947:
1942:
1930:
1915:
1895:
1858:
1850:
1811:
1768:
1725:
1690:
1653:
1609:
1488:
1451:
655:
635:
482:
446:
284:
276:. Due to similarities in binding sites, closely related targets identified through
253:
249:
162:
75:
2475:
2402:. Wiley's Methods and Principles in Medicinal Chemistry. Vol. 63. Wiley-VCH.
5969:
5821:
5801:
5781:
5776:
5766:
5661:
5296:
5286:
5271:
5231:
5196:
5189:
5120:
5080:
4849:
4722:
4712:
4620:
4577:
4557:
4286:
4237:
4033:
1694:
1658:
1641:
1614:
1597:
1483:
1221:
261:
241:
173:
experiments complemented with computation methods are increasingly used in early
143:
102:
55:
5921:
3370:
3212:
2571:
2436:
2312:
1983:
643:
603:
to the target may be designed using interactive graphics and the intuition of a
351:. In addition, the three-dimensional structure of the target may be determined.
6034:
5999:
5846:
5791:
5744:
5726:
5528:
5276:
5266:
5261:
5236:
5030:
5010:
4883:
4854:
4804:
4737:
4707:
4697:
4660:
4582:
4540:
4535:
4417:
4412:
4402:
4320:
4298:
4132:
4122:
3794:
3774:
3648:
Walters WP, Stahl MT, Murcko MA (April 1998). "Virtual screening—an overview".
3181:
2671:
2635:
2067:"Enabling mRNA Therapeutics: Current Landscape and Challenges in Manufacturing"
1854:
1493:
1478:
1332:
1323:
1281:
1169:
penalty for reducing the degrees of freedom when a ligand binds to its receptor
431:
411:
299:
290:
produced through chemical synthesis, but biopolymer-based drugs (also known as
287:
273:
257:
229:
174:
158:
78:
5908:
3114:
Jorgensen WL (March 2004). "The many roles of computation in drug discovery".
2508:
2347:
6079:
6059:
5984:
5974:
5851:
5836:
5816:
5739:
5641:
5473:
5463:
5291:
5281:
5256:
5221:
4784:
4717:
4640:
4615:
4567:
4552:
4259:
4179:
3591:
3431:
Structure-based Ligand Design (Methods and
Principles in Medicinal Chemistry)
2917:(1st ed.). Cambridge, UK: Cambridge University Press. pp. 151–164.
1313:
939:– entropy penalty due to freezing a rotatable in the ligand bond upon binding
555:
222:
182:
20:
5894:
5880:
3688:
3135:
272:) since drug interactions with off-target molecules may lead to undesirable
6054:
5734:
5711:
5681:
5666:
4955:
4572:
4513:
4325:
4276:
4227:
4202:
4154:
4147:
3696:
3634:
3582:
3565:
3550:
3475:
3308:
3287:
Liu J, Wang R (March 2015). "Classification of current scoring functions".
3189:
3143:
3092:
3049:
3014:
2976:
2870:
2834:
2799:
2764:
2715:
2679:
2643:
2608:
2493:
2444:
2355:
2320:
2243:
2202:
2143:
2102:
2051:
1991:
1956:
1907:
1872:
1823:
1780:
1702:
1667:
1623:
1249:
1175:
interaction – enthalpic gain for "resolvating" the ligand with its receptor
631:
516:
Represent and cluster candidates according to protein-ligand 3D information
360:
265:
202:
3415:
3343:
3265:
3006:
1737:
647:
5811:
5806:
5786:
5706:
5626:
5435:
5340:
4650:
4597:
4526:
4471:
4449:
4197:
4174:
4169:
4112:
4107:
3779:
2234:
2217:
2083:
1473:
1309:
1290:
1210:
651:
530:
218:
82:
63:
3675:
Klein DF (March 2008). "The loss of serendipity in psychopharmacology".
3407:
3335:
493:
Flowchart of a Usual
Clustering Analysis for Structure-Based Drug Design
363:", that is they should possess properties that are predicted to lead to
5979:
5425:
5020:
4799:
4521:
4501:
4355:
4193:
3257:
2746:
2532:
1729:
1266:
932:| is surface area of lipophilic contact between the ligand and receptor
690:
538:
There are two major types of drug design. The first is referred to as
450:
414:
and its biological target. These methods are also used to predict the
269:
3532:
3300:
2993:
Klebe G (2000). "Recent developments in structure-based drug design".
2862:
2826:
2791:
2707:
2029:
441:
may be used to provide binding affinity estimates. These methods use
16:
Invention of new medications based on knowledge of a biological target
5691:
5415:
5400:
5358:
4970:
4530:
4370:
2065:
Youssef M, Hitti C, Puppin Chaves Fulber J, Kamen AA (October 2023).
2064:
1328:
1304:
214:
59:
3453:
3207:. RSC Drug Discovery. Royal Society of Chemistry. pp. 440–457.
3084:
2524:
2134:
2117:
1899:
1772:
684:
binding to the target is generally desirable since it leads to more
434:, etc.) of the drug candidate that will influence binding affinity.
5025:
4072:
3467:
2566:. RSC Drug Discovery. Royal Society of Chemistry. pp. 88–107.
1300:
1233:
1217:
1159:
685:
335:
Once a suitable target has been identified, the target is normally
225:
170:
3429:
Timmerman H, Gubernator K, Böhm HJ, Mannhold R, Kubinyi H (1998).
1188:
term. The configurational or strain energy can be estimated using
489:
475:
optimization of affinity and selectivity (structure-based design,
5646:
5574:
5314:
4184:
1166:
233:
198:
122:
94:
86:
3490:
2216:
Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M (August 2017).
1537:
1209:
leading to an approved drug is the carbonic anhydrase inhibitor
542:-based drug design and the second, structure-based drug design.
213:
pathway that is associated with a specific disease condition or
5405:
4056:
3428:
3365:. RSC Drug Discovery. Royal Society of Chemistry. p. 466.
1245:
1244:). Imatinib is substantially different from previous drugs for
693:) and use the predicted affinity as a criterion for selection.
539:
519:
Needs meaningful representation of protein-ligand interactions.
3729:
943:
A more general thermodynamic "master" equation is as follows:
109:
techniques. This type of modeling is sometimes referred to as
4117:
2886:
Pharmacophore
Perception, Development, and use in Drug Design
2656:
1683:
Biochimica et
Biophysica Acta (BBA) - Proteins and Proteomics
1285:
485:
of other pharmaceutical properties while maintaining affinity
98:
1639:
1598:"Peptide therapeutics: current status and future directions"
505:
Selecting candidates by voting of multiple scoring functions
5964:
5157:
3514:
2777:
1758:
1206:
642:
that may be of interest. Furthermore, it may be that only
295:
178:
71:
3941:
Nucleoside and nucleotide reverse-transcriptase inhibitors
3393:
2118:"mRNA-based therapeutics--developing a new class of drugs"
1969:
1422:
Nucleoside and nucleotide reverse transcriptase inhibitors
583:
of the biological target obtained through methods such as
181:(absorption, distribution, metabolism, and excretion) and
4025:
1680:
1572:(1 ed.). Cambridge, UK: Cambridge University Press.
882:
879:
876:
2847:
1277:
from which the later members of the class were developed
2729:
Liang S, Meroueh SO, Wang G, Qiu C, Zhou Y (May 2009).
2692:
2264:
2009:(2nd ed.). Totowa, NJ Humana Press: Humana Press.
1839:"Identifying druggable disease-modifying gene products"
1642:"Protein-peptide docking: opportunities and challenges"
3167:
1322:(selective serotonin reuptake inhibitors), a class of
3886:
Dual serotonin and norepinephrine reuptake inhibitors
1015:
951:
704:
2215:
928:– contribution from lipophilic interactions where |A
25:
3361:. In Gramatica P, Livingstone DJ, Davis AM (eds.).
3359:"Chapter 17: Modeling Chemicals in the Environment"
2965:
Current
Opinion in Drug Discovery & Development
2915:
Drug Design: Structure- and Ligand-Based Approaches
2728:
2597:
Current Opinion in Drug Discovery & Development
1591:
1589:
1570:
Drug Design: Structure- and Ligand-Based Approaches
1538:Madsen U, Krogsgaard-Larsen P, Liljefors T (2002).
406:is most often used to estimate the strength of the
347:. The purified protein is then used to establish a
3647:
2888:. La Jolla, Calif: International University Line.
2004:
1885:
1144:
889:
5951:
3422:
2931:
2877:
2115:
1567:
264:) will be designed that are complementary to the
6077:
3724:https://www.drugdesign.org/chapters/drug-design/
3070:
2812:
2506:
2377:(3 ed.). Academic Press. pp. 521–527.
1595:
1586:
3931:Non-nucleoside reverse-transcriptase inhibitors
3363:Drug Design Strategies: Quantitative Approaches
3205:Drug Design Strategies: Quantitative Approaches
2564:Drug Design Strategies: Quantitative Approaches
1563:
1561:
1559:
1417:Non-nucleoside reverse transcriptase inhibitors
1184:, and the components of free energy was built.
157:Due to high attrition rates, especially during
3517:"Back to the future with phenotypic screening"
2621:
2167:"Recent advances in phenotypic drug discovery"
1836:
1216:Another case study in rational drug design is
626:
5937:
4041:
3745:
3612:
3504:. The Scripps Research Institute. 2007-12-17.
2422:
2007:Biopharmaceutical drug design and development
393:
205:) is a key molecule involved in a particular
81:that activates or inhibits the function of a
6065:Quantitative structure–activity relationship
3668:
3289:Journal of Chemical Information and Modeling
3202:
3163:
3161:
3107:
2988:
2986:
2902:
2851:Journal of Chemical Information and Modeling
2841:
2806:
2780:Journal of Chemical Information and Modeling
2771:
2722:
2686:
2660:Bioorganic & Medicinal Chemistry Letters
2650:
2615:
2594:
2588:
2294:
2292:
2290:
2265:Ganellin CR, Jefferis R, Roberts SM (2013).
2116:Sahin U, Karikó K, Türeci Ö (October 2014).
1931:"The process of structure-based drug design"
1635:
1633:
1568:Reynolds CH, Merz KM, Ringe D, eds. (2010).
1556:
1531:
1162:penalty for removing the ligand from solvent
561:quantitative structure-activity relationship
311:
3387:
3229:
3196:
2937:
2913:. In Reynolds CH, Merz KM, Ringe D (eds.).
2460:"The role of serendipity in drug discovery"
1830:
1795:
1752:
5944:
5930:
4048:
4034:
3752:
3738:
3238:Journal of Computer-Aided Molecular Design
3064:
3035:
3029:
2962:
2555:
2298:
2164:
1801:
1715:
1709:
1542:. Washington, D.C.: Taylor & Francis.
1228:fusion protein that is characteristic for
323:
3715:at the U.S. National Library of Medicine
3581:
3540:
3356:
3315:
3158:
3113:
2983:
2754:
2483:
2397:
2287:
2233:
2192:
2182:
2133:
2092:
2082:
1946:
1862:
1657:
1630:
1613:
5565:Good Design Award (Museum of Modern Art)
3280:
1928:
1837:Dixon SJ, Stockwell BR (December 2009).
1224:inhibitor designed specifically for the
658:sites, etc.) that drive ligand binding.
529:
488:
177:to select compounds with more favorable
3891:Selective serotonin reuptake inhibitors
3286:
2908:
2368:
2333:
1596:Fosgerau K, Hoffmann T (January 2015).
371:and a range of scoring methods such as
137:The phrase "drug design" is similar to
6078:
3321:
921:– contribution from ionic interactions
5925:
5610:
5570:Good Design Award (Chicago Athenaeum)
5313:
4758:
4085:
4029:
3733:
3674:
2992:
2883:
2561:
1540:Textbook of Drug Design and Discovery
3563:
3235:
2507:Ethiraj SK, Levinthal D (Sep 2004).
914:– contribution from hydrogen bonding
661:
3615:Current Opinion in Chemical Biology
2457:
2375:The Practice of Medicinal Chemistry
2371:"Chapter 25: Pharmacological space"
1843:Current Opinion in Chemical Biology
469:(structure- or ligand-based design)
424:ab initio quantum chemistry methods
13:
5611:
5514:American Institute of Graphic Arts
3981:Bcr-Abl tyrosine-kinase inhibitors
2464:Dialogues in Clinical Neuroscience
2425:Drug Discovery Today. Technologies
1972:Drug Discovery Today. Technologies
1382:Bcr-Abl tyrosine-kinase inhibitors
1125:
1109:
1093:
1077:
1061:
956:
873:
858:
831:
795:
781:
751:
737:
721:
705:
615:programs. This method is known as
570:
328:Rational drug design (also called
117:. In addition to small molecules,
14:
6107:
5524:Design and Industries Association
3996:Neurokinin 1 receptor antagonists
3871:Dipeptidyl peptidase-4 inhibitors
3706:
2005:Wu-Pong S, Rojanasakul Y (2008).
1402:Dipeptidyl peptidase-4 inhibitors
1339:
306:
3986:Cannabinoid receptor antagonists
3566:"Phenotypic screening, take two"
2513:Administrative Science Quarterly
2044:10.1016/j.biotechadv.2015.04.008
1499:List of pharmaceutical companies
1387:Cannabinoid receptor antagonists
1377:Angiotensin receptor antagonists
1344:Types of drug screening include
575:Structure-based drug design (or
33:
4086:
3815:and development of drug classes
3759:
3641:
3606:
3557:
3508:
3447:
3350:
2956:
2500:
2451:
2416:
2391:
2362:
2327:
2258:
2209:
2158:
2109:
2058:
2023:
1998:
1963:
1922:
1879:
1372:Acetylcholine receptor agonists
1359:
1293:, a peptide HIV entry inhibitor
545:
188:
5519:Chartered Society of Designers
4759:
3456:Nature Reviews. Drug Discovery
3396:Journal of Medicinal Chemistry
3324:Journal of Medicinal Chemistry
3073:Nature Reviews. Drug Discovery
2940:Structure-based Drug Discovery
2815:Journal of Medicinal Chemistry
2696:Journal of Medicinal Chemistry
2222:Nature Reviews. Drug Discovery
2184:10.12688/f1000research.25813.1
2122:Nature Reviews. Drug Discovery
1948:10.1016/j.chembiol.2003.09.002
1929:Anderson AC (September 2003).
1888:Nature Reviews. Drug Discovery
1761:Nature Reviews. Drug Discovery
1674:
1213:, which was approved in 1995.
1050:
1042:
1037:
1029:
1026:
1018:
1:
5590:Prince Philip Designers Prize
4233:Architectural lighting design
3832:Angiotensin receptor blockers
3662:10.1016/S1359-6446(97)01163-X
3627:10.1016/S1367-5931(00)00110-1
3038:Journal of Molecular Modeling
2995:Journal of Molecular Medicine
2301:Current Pharmaceutical Design
1816:10.1016/S1359-6446(03)02828-9
1525:
1255:Additional examples include:
668:Scoring functions for docking
579:) relies on knowledge of the
559:the target. Alternatively, a
550:Ligand-based drug design (or
132:
89:, which in turn results in a
5396:Electronic design automation
5379:Virtual home design software
4351:Automotive suspension design
1718:Pharmacy World & Science
1695:10.1016/j.bbapap.2014.07.006
1659:10.1016/j.drudis.2018.05.006
1615:10.1016/j.drudis.2014.10.003
1458:
1238:chronic myelogenous leukemia
380:multi-objective optimization
66:based on the knowledge of a
7:
4255:Environmental impact design
4010:Melatonin receptor agonists
3961:Thalidomide and its analogs
3916:Memantine and related drugs
3866:Cyclooxygenase 2 inhibitors
3371:10.1039/9781849733410-00458
3213:10.1039/9781849733410-00440
3170:Current Medicinal Chemistry
2938:Leach AR, Harren J (2007).
2572:10.1039/9781849733410-00088
2476:10.31887/DCNS.2006.8.3/tban
2437:10.1016/j.ddtec.2013.02.001
2313:10.2174/1381612811319120019
2165:Swinney DC, Lee JA (2020).
1984:10.1016/j.ddtec.2004.10.004
1466:
1397:Cyclooxygenase 2 inhibitors
1207:structure-based drug design
1199:
627:Binding site identification
581:three dimensional structure
394:Computer-aided drug design
115:structure-based drug design
10:
6112:
5534:International Forum Design
4904:Engineering design process
4055:
3876:Direct thrombin inhibitors
3813:Case studies of discovery
3182:10.2174/092986710790979944
2672:10.1016/j.bmcl.2003.09.028
2636:10.2174/138945008786949405
2400:Drug Metabolism Prediction
1855:10.1016/j.cbpa.2009.08.003
1519:Retrometabolic drug design
1242:acute lymphocytic leukemia
665:
408:intermolecular interaction
111:computer-aided drug design
18:
5960:
5869:
5617:
5606:
5547:
5506:
5449:
5339:
5335:
5309:
4765:
4754:
4656:Integrated circuit design
4591:
4578:Stage/set lighting design
4510:
4467:Hardware interface design
4426:
4383:Hardware interface design
4334:
4216:
4096:
4092:
4081:
4063:
3991:CCR5 receptor antagonists
3812:
3767:
3570:Science-Business EXchange
3521:ACS Chemical Neuroscience
2348:10.1016/s1359644602025722
2177:: F1000 Faculty Rev–944.
1509:Molecular design software
1392:CCR5 receptor antagonists
1350:high-throughput screening
465:hit identification using
428:density functional theory
317:Phenotypic drug discovery
312:Phenotypic drug discovery
283:Most commonly, drugs are
5491:Industrial design rights
5479:Fashion design copyright
5391:Design quality indicator
4840:Creative problem-solving
4631:Electrical system design
4487:Sonic interaction design
4398:Photographic lens design
4272:Healthy community design
3926:Neuraminidase inhibitors
3717:Medical Subject Headings
2911:"QSAR in Drug Discovery"
1412:NK1 receptor antagonists
1194:potentials of mean force
640:allosteric binding sites
525:
19:Not to be confused with
6020:Lipinski's rule of five
5687:New product development
5652:Enterprise architecture
5580:IF Product Design Award
5539:Design Research Society
5091:Reliability engineering
3901:HIV-protease inhibitors
3822:5α-Reductase inhibitors
3689:10.1001/jama.299.9.1063
3433:. Weinheim: Wiley-VCH.
3136:10.1126/science.1096361
2373:. In Wermuth CG (ed.).
1935:Chemistry & Biology
1407:HIV protease inhibitors
1262:atypical antipsychotics
1230:Philadelphia chromosome
369:Lipinski's Rule of Five
324:Rational drug discovery
62:process of finding new
50:, often referred to as
5143:Top-down and bottom-up
4492:User experience design
4393:Packaging and labeling
4366:Electric guitar design
4304:Landscape architecture
3951:Proton pump inhibitors
3583:10.1038/scibx.2012.380
3050:10.1007/s0089400060498
2032:Biotechnology Advances
1514:Molecular modification
1432:Proton pump inhibitors
1146:
891:
535:
494:
127:therapeutic antibodies
6086:Design of experiments
6025:Lipophilic efficiency
5672:Innovation management
5555:European Design Award
5321:Intellectual property
5138:Theory of constraints
5101:Responsibility-driven
4941:For manufacturability
4845:Creativity techniques
4683:Nuclear weapon design
4497:User interface design
4361:Corrugated box design
4282:Interior architecture
3564:Kotz J (April 2012).
3007:10.1007/s001090000084
2038:(6 Pt 3): 1294–1309.
1147:
892:
674:molecular recognition
585:x-ray crystallography
533:
492:
373:lipophilic efficiency
5717:Unintelligent design
5697:Philosophy of design
5411:Design specification
5364:Comprehensive layout
4936:For behaviour change
4909:Probabilistic design
4671:Power network design
4208:Visual merchandising
4165:Instructional design
4143:Postage stamp design
3906:Integrase inhibitors
3881:Direct Xa inhibitors
3650:Drug Discovery Today
3357:Gramatica P (2011).
2942:. Berlin: Springer.
2624:Current Drug Targets
2398:Kirchmair J (2014).
2336:Drug Discovery Today
2235:10.1038/nrd.2017.111
2084:10.3390/biom13101497
1804:Drug Discovery Today
1646:Drug Discovery Today
1602:Drug Discovery Today
1346:phenotypic screening
1275:-receptor antagonist
1248:, as most agents of
949:
702:
552:indirect drug design
365:oral bioavailability
330:reverse pharmacology
74:is most commonly an
52:rational drug design
6096:Medicinal chemistry
6040:New chemical entity
6030:Mechanism of action
5954:medicinal chemistry
5637:Creative industries
5560:German Design Award
5469:Design infringement
5354:Architectural model
4693:Organization design
4688:Nucleic acid design
4636:Experimental design
4189:Traffic sign design
3408:10.1021/jm00034a001
3336:10.1021/jm00026a001
3250:1994JCAMD...8..243B
3128:2004Sci...303.1813J
3122:(5665): 1813–1818.
2369:Hopkins AL (2011).
1504:Medicinal chemistry
1269:, the prototypical
1190:molecular mechanics
565:biological activity
400:Molecular mechanics
388:bounded rationality
195:biomolecular target
148:metabolic half-life
5702:Process simulation
5677:Intelligent design
5001:Intelligence-based
4996:Integrated topside
4926:Framework-oriented
4611:Behavioural design
4482:Information design
4160:Information design
3976:Tubulin inhibitors
3258:10.1007/BF00126743
2909:Tropsha A (2010).
2747:10.1002/prot.22252
1730:10.1007/BF00579706
1297:Nonbenzodiazepines
1142:
1140:
1055:
887:
577:direct drug design
536:
502:Consensus scoring
495:
404:molecular dynamics
292:biopharmaceuticals
119:biopharmaceuticals
6073:
6072:
6015:Ligand efficiency
5919:
5918:
5865:
5864:
5632:Conceptual design
5602:
5601:
5598:
5597:
5585:James Dyson Award
5441:Website wireframe
5431:Technical drawing
5305:
5304:
5153:Transgenerational
4894:Ecological design
4770:Activity-centered
4750:
4749:
4746:
4745:
4728:Spacecraft design
4522:Public art design
4460:Video game design
4438:Experience design
4408:Production design
4388:Motorcycle design
4346:Automotive design
4250:Ecological design
4128:Film title design
4023:
4022:
3971:TRPV1 antagonists
3911:Lipase inhibitors
3533:10.1021/cn500051h
3440:978-3-527-29343-8
3330:(26): 4953–4967.
3301:10.1021/ci500731a
3176:(15): 1550–1562.
2949:978-1-4020-4406-9
2895:978-0-9636817-6-8
2884:Guner OF (2000).
2863:10.1021/ci050262q
2827:10.1021/jm030331x
2792:10.1021/ci050283k
2708:10.1021/jm0508641
2702:(11): 3116–3135.
2666:(24): 4355–4359.
2630:(12): 1031–1039.
2409:978-3-527-67301-8
2384:978-0-12-374194-3
2307:(12): 2326–2333.
2016:978-1-59745-532-9
1689:(11): 2002–2015.
1549:978-0-415-28288-8
1447:TRPV1 antagonists
1367:5-HT3 antagonists
1354:virtual screening
1240:and occasionally
1135:
1119:
1103:
1087:
1071:
1054:
1048:
1035:
1024:
1008:
994:
966:
868:
841:
791:
747:
731:
715:
662:Scoring functions
617:virtual screening
611:fast approximate
605:medicinal chemist
513:Cluster analysis
483:lead optimization
467:virtual screening
443:linear regression
278:sequence homology
256:; or ion channel
221:or survival of a
197:(most commonly a
107:computer modeling
68:biological target
45:
44:
6103:
6050:Pharmacokinetics
6045:Pharmacodynamics
6010:Enzyme inhibitor
5995:Drug development
5946:
5939:
5932:
5923:
5922:
5911:
5904:
5897:
5890:
5883:
5876:
5608:
5607:
5485:Geschmacksmuster
5459:Community design
5337:
5336:
5311:
5310:
5071:Process-centered
4867:Design–bid–build
4835:Cradle-to-cradle
4815:Concept-oriented
4756:
4755:
4733:Strategic design
4703:Processor design
4678:Mechanism design
4646:Geometric design
4606:Algorithm design
4546:Jewellery design
4477:Immersive design
4371:Furniture design
4316:Landscape design
4094:
4093:
4083:
4082:
4050:
4043:
4036:
4027:
4026:
4015:Renin inhibitors
3861:c-Met inhibitors
3785:Drug development
3754:
3747:
3740:
3731:
3730:
3701:
3700:
3683:(9): 1063–1065.
3672:
3666:
3665:
3645:
3639:
3638:
3610:
3604:
3603:
3585:
3561:
3555:
3554:
3544:
3512:
3506:
3505:
3494:
3488:
3487:
3451:
3445:
3444:
3426:
3420:
3419:
3402:(8): 1035–1054.
3391:
3385:
3384:
3354:
3348:
3347:
3319:
3313:
3312:
3284:
3278:
3277:
3233:
3227:
3226:
3200:
3194:
3193:
3165:
3156:
3155:
3111:
3105:
3104:
3068:
3062:
3061:
3044:(7–8): 498–516.
3033:
3027:
3026:
2990:
2981:
2980:
2960:
2954:
2953:
2935:
2929:
2928:
2906:
2900:
2899:
2881:
2875:
2874:
2845:
2839:
2838:
2810:
2804:
2803:
2775:
2769:
2768:
2758:
2726:
2720:
2719:
2690:
2684:
2683:
2654:
2648:
2647:
2619:
2613:
2612:
2592:
2586:
2585:
2559:
2553:
2552:
2504:
2498:
2497:
2487:
2455:
2449:
2448:
2431:(3): e427–e435.
2420:
2414:
2413:
2395:
2389:
2388:
2366:
2360:
2359:
2331:
2325:
2324:
2296:
2285:
2284:
2262:
2256:
2255:
2237:
2213:
2207:
2206:
2196:
2186:
2162:
2156:
2155:
2137:
2113:
2107:
2106:
2096:
2086:
2062:
2056:
2055:
2027:
2021:
2020:
2002:
1996:
1995:
1967:
1961:
1960:
1950:
1926:
1920:
1919:
1883:
1877:
1876:
1866:
1849:(5–6): 549–555.
1834:
1828:
1827:
1799:
1793:
1792:
1756:
1750:
1749:
1713:
1707:
1706:
1678:
1672:
1671:
1661:
1652:(8): 1530–1537.
1637:
1628:
1627:
1617:
1593:
1584:
1583:
1565:
1554:
1553:
1535:
1489:Drug development
1452:c-Met inhibitors
1437:Renin inhibitors
1151:
1149:
1148:
1143:
1141:
1137:
1136:
1133:
1121:
1120:
1117:
1105:
1104:
1101:
1089:
1088:
1085:
1073:
1072:
1069:
1056:
1053:
1049:
1046:
1040:
1036:
1033:
1025:
1022:
1016:
1010:
1009:
1006:
996:
995:
992:
968:
967:
964:
896:
894:
893:
888:
886:
885:
870:
869:
866:
854:
843:
842:
839:
827:
826:
793:
792:
789:
777:
776:
749:
748:
745:
733:
732:
729:
717:
716:
713:
656:hydrogen bonding
589:NMR spectroscopy
447:machine learning
439:scoring function
242:inverse agonists
163:drug development
37:
26:
6111:
6110:
6106:
6105:
6104:
6102:
6101:
6100:
6076:
6075:
6074:
6069:
5970:Bioavailability
5956:
5950:
5920:
5915:
5909:
5902:
5895:
5888:
5881:
5874:
5861:
5662:Futures studies
5613:
5594:
5543:
5502:
5451:
5445:
5331:
5330:
5301:
5207:Value sensitive
5197:User innovation
5076:Public interest
5041:Object-oriented
4761:
4742:
4723:Software design
4713:Research design
4666:Physical design
4621:Database design
4595:
4593:
4587:
4563:Property design
4558:Game art design
4512:
4506:
4429:
4422:
4337:
4330:
4287:Interior design
4238:Building design
4219:
4212:
4099:
4088:
4077:
4059:
4054:
4024:
4019:
4004:
3946:PDE5 inhibitors
3936:NS5A inhibitors
3921:mTOR inhibitors
3850:
3814:
3808:
3768:Steps in design
3763:
3758:
3709:
3704:
3673:
3669:
3646:
3642:
3611:
3607:
3562:
3558:
3513:
3509:
3496:
3495:
3491:
3452:
3448:
3441:
3427:
3423:
3392:
3388:
3381:
3355:
3351:
3320:
3316:
3285:
3281:
3234:
3230:
3223:
3201:
3197:
3166:
3159:
3112:
3108:
3085:10.1038/nrd1799
3069:
3065:
3034:
3030:
2991:
2984:
2961:
2957:
2950:
2936:
2932:
2925:
2907:
2903:
2896:
2882:
2878:
2846:
2842:
2811:
2807:
2776:
2772:
2727:
2723:
2691:
2687:
2655:
2651:
2620:
2616:
2593:
2589:
2582:
2560:
2556:
2525:10.2307/4131441
2505:
2501:
2458:Ban TA (2006).
2456:
2452:
2421:
2417:
2410:
2396:
2392:
2385:
2367:
2363:
2332:
2328:
2297:
2288:
2281:
2263:
2259:
2214:
2210:
2163:
2159:
2135:10.1038/nrd4278
2128:(10): 759–780.
2114:
2110:
2063:
2059:
2028:
2024:
2017:
2003:
1999:
1968:
1964:
1927:
1923:
1900:10.1038/nrd2132
1894:(10): 821–834.
1884:
1880:
1835:
1831:
1810:(18): 852–861.
1800:
1796:
1773:10.1038/nrd4609
1757:
1753:
1714:
1710:
1679:
1675:
1638:
1631:
1594:
1587:
1580:
1566:
1557:
1550:
1536:
1532:
1528:
1523:
1484:Cheminformatics
1469:
1461:
1456:
1427:PDE5 inhibitors
1362:
1342:
1337:
1324:antidepressants
1274:
1222:tyrosine kinase
1202:
1183:
1139:
1138:
1132:
1128:
1116:
1112:
1100:
1096:
1084:
1080:
1068:
1064:
1058:
1057:
1045:
1041:
1032:
1021:
1017:
1014:
1005:
1001:
998:
997:
991:
987:
963:
959:
952:
950:
947:
946:
938:
931:
927:
920:
913:
906:
872:
871:
865:
861:
844:
838:
834:
798:
794:
788:
784:
754:
750:
744:
740:
728:
724:
712:
708:
703:
700:
699:
670:
664:
629:
573:
571:Structure-based
548:
528:
396:
349:screening assay
326:
314:
309:
288:small molecules
230:small molecules
191:
167:physicochemical
159:clinical phases
144:bioavailability
135:
125:and especially
93:benefit to the
56:rational design
41:
40:
39:
38:
24:
17:
12:
11:
5:
6109:
6099:
6098:
6093:
6091:Drug discovery
6088:
6071:
6070:
6068:
6067:
6062:
6057:
6052:
6047:
6042:
6037:
6035:Mode of action
6032:
6027:
6022:
6017:
6012:
6007:
6005:Drug targeting
6002:
6000:Drug discovery
5997:
5992:
5987:
5982:
5977:
5972:
5967:
5961:
5958:
5957:
5949:
5948:
5941:
5934:
5926:
5917:
5916:
5914:
5913:
5906:
5899:
5892:
5885:
5878:
5870:
5867:
5866:
5863:
5862:
5860:
5859:
5854:
5849:
5844:
5839:
5834:
5829:
5824:
5819:
5814:
5809:
5804:
5799:
5794:
5789:
5784:
5779:
5774:
5769:
5764:
5763:
5762:
5757:
5747:
5742:
5737:
5730:
5729:
5727:Wicked problem
5724:
5719:
5714:
5709:
5704:
5699:
5694:
5689:
5684:
5679:
5674:
5669:
5664:
5659:
5654:
5649:
5644:
5639:
5634:
5629:
5624:
5618:
5615:
5614:
5612:Related topics
5604:
5603:
5600:
5599:
5596:
5595:
5593:
5592:
5587:
5582:
5577:
5572:
5567:
5562:
5557:
5551:
5549:
5545:
5544:
5542:
5541:
5536:
5531:
5529:Design Council
5526:
5521:
5516:
5510:
5508:
5504:
5503:
5501:
5500:
5499:
5498:
5496:European Union
5488:
5481:
5476:
5471:
5466:
5461:
5455:
5453:
5447:
5446:
5444:
5443:
5438:
5433:
5428:
5423:
5418:
5413:
5408:
5403:
5398:
5393:
5388:
5383:
5382:
5381:
5376:
5366:
5361:
5356:
5351:
5345:
5343:
5333:
5332:
5329:
5328:
5325:
5322:
5319:
5315:
5307:
5306:
5303:
5302:
5300:
5299:
5294:
5289:
5284:
5279:
5274:
5269:
5264:
5259:
5254:
5249:
5244:
5239:
5234:
5229:
5224:
5217:
5216:
5215:
5214:
5204:
5199:
5194:
5193:
5192:
5182:
5177:
5175:Usage-centered
5172:
5171:
5170:
5168:Design for All
5160:
5155:
5150:
5148:Transformation
5145:
5140:
5135:
5130:
5129:
5128:
5118:
5113:
5108:
5103:
5098:
5096:Research-based
5093:
5088:
5083:
5078:
5073:
5068:
5063:
5061:Platform-based
5058:
5053:
5048:
5043:
5038:
5033:
5028:
5023:
5018:
5013:
5011:KISS principle
5008:
5003:
4998:
4993:
4988:
4983:
4978:
4973:
4968:
4963:
4958:
4953:
4948:
4943:
4938:
4933:
4928:
4923:
4921:Fault-tolerant
4918:
4916:Error-tolerant
4913:
4912:
4911:
4901:
4899:Energy neutral
4896:
4891:
4886:
4881:
4880:
4879:
4869:
4864:
4859:
4858:
4857:
4855:Design fiction
4847:
4842:
4837:
4832:
4827:
4822:
4817:
4812:
4807:
4802:
4797:
4792:
4787:
4782:
4777:
4772:
4766:
4763:
4762:
4752:
4751:
4748:
4747:
4744:
4743:
4741:
4740:
4738:Systems design
4735:
4730:
4725:
4720:
4715:
4710:
4708:Protein design
4705:
4700:
4698:Process design
4695:
4690:
4685:
4680:
4675:
4674:
4673:
4668:
4663:
4661:Circuit design
4653:
4648:
4643:
4638:
4633:
4628:
4623:
4618:
4613:
4608:
4602:
4600:
4589:
4588:
4586:
4585:
4583:Textile design
4580:
4575:
4570:
4565:
4560:
4555:
4550:
4549:
4548:
4543:
4541:Costume design
4536:Fashion design
4533:
4524:
4518:
4516:
4508:
4507:
4505:
4504:
4499:
4494:
4489:
4484:
4479:
4474:
4469:
4464:
4463:
4462:
4457:
4447:
4446:
4445:
4434:
4432:
4424:
4423:
4421:
4420:
4418:Service design
4415:
4413:Sensory design
4410:
4405:
4403:Product design
4400:
4395:
4390:
4385:
4380:
4379:
4378:
4368:
4363:
4358:
4353:
4348:
4342:
4340:
4332:
4331:
4329:
4328:
4323:
4321:Spatial design
4318:
4313:
4312:
4311:
4301:
4299:Keyline design
4296:
4295:
4294:
4284:
4279:
4274:
4269:
4268:
4267:
4265:Computer-aided
4257:
4252:
4247:
4246:
4245:
4235:
4230:
4224:
4222:
4214:
4213:
4211:
4210:
4205:
4200:
4191:
4182:
4177:
4172:
4167:
4162:
4157:
4152:
4151:
4150:
4145:
4140:
4133:Graphic design
4130:
4125:
4123:Exhibit design
4120:
4115:
4110:
4104:
4102:
4090:
4089:
4079:
4078:
4076:
4075:
4070:
4064:
4061:
4060:
4053:
4052:
4045:
4038:
4030:
4021:
4020:
4018:
4017:
4012:
4007:
4002:
3998:
3993:
3988:
3983:
3978:
3973:
3968:
3963:
3958:
3953:
3948:
3943:
3938:
3933:
3928:
3923:
3918:
3913:
3908:
3903:
3898:
3893:
3888:
3883:
3878:
3873:
3868:
3863:
3858:
3856:Cephalosporins
3853:
3848:
3844:
3839:
3834:
3829:
3827:ACE inhibitors
3824:
3818:
3816:
3810:
3809:
3807:
3806:
3805:
3804:
3803:
3802:
3792:
3782:
3777:
3775:Drug discovery
3771:
3769:
3765:
3764:
3757:
3756:
3749:
3742:
3734:
3728:
3727:
3720:
3708:
3707:External links
3705:
3703:
3702:
3667:
3656:(4): 160–178.
3640:
3621:(4): 445–451.
3605:
3556:
3527:(7): 503–513.
3507:
3489:
3468:10.1038/nrd839
3462:(7): 493–502.
3446:
3439:
3421:
3386:
3380:978-1849731669
3379:
3349:
3314:
3295:(3): 475–482.
3279:
3244:(3): 243–256.
3228:
3222:978-1849731669
3221:
3195:
3157:
3106:
3079:(8): 649–663.
3063:
3028:
3001:(5): 269–281.
2982:
2971:(3): 365–374.
2955:
2948:
2930:
2924:978-0521887236
2923:
2901:
2894:
2876:
2857:(1): 221–230.
2840:
2821:(2): 337–344.
2805:
2786:(1): 380–391.
2770:
2741:(2): 397–403.
2721:
2685:
2649:
2614:
2603:(3): 308–315.
2587:
2581:978-1849731669
2580:
2554:
2499:
2470:(3): 335–344.
2450:
2415:
2408:
2390:
2383:
2361:
2326:
2286:
2279:
2257:
2228:(8): 531–543.
2208:
2157:
2108:
2057:
2022:
2015:
1997:
1978:(3): 209–215.
1962:
1941:(9): 787–797.
1921:
1878:
1829:
1794:
1767:(7): 475–486.
1751:
1708:
1673:
1629:
1608:(1): 122–128.
1585:
1579:978-0521887236
1578:
1555:
1548:
1529:
1527:
1524:
1522:
1521:
1516:
1511:
1506:
1501:
1496:
1494:Drug discovery
1491:
1486:
1481:
1479:Bioinformatics
1476:
1470:
1468:
1465:
1460:
1457:
1455:
1454:
1449:
1444:
1439:
1434:
1429:
1424:
1419:
1414:
1409:
1404:
1399:
1394:
1389:
1384:
1379:
1374:
1369:
1363:
1361:
1358:
1341:
1340:Drug screening
1338:
1336:
1335:
1333:antiviral drug
1326:
1317:
1307:
1294:
1288:
1278:
1272:
1264:
1257:
1201:
1198:
1181:
1177:
1176:
1173:
1170:
1163:
1158:desolvation –
1131:
1127:
1124:
1115:
1111:
1108:
1099:
1095:
1092:
1083:
1079:
1076:
1067:
1063:
1060:
1059:
1052:
1044:
1039:
1031:
1028:
1020:
1013:
1004:
1000:
999:
990:
986:
983:
980:
977:
974:
971:
962:
958:
955:
954:
941:
940:
936:
933:
929:
925:
922:
918:
915:
911:
908:
904:
884:
881:
878:
875:
864:
860:
857:
853:
850:
847:
837:
833:
830:
825:
822:
819:
816:
813:
810:
807:
804:
801:
797:
787:
783:
780:
775:
772:
769:
766:
763:
760:
757:
753:
743:
739:
736:
727:
723:
720:
711:
707:
666:Main article:
663:
660:
628:
625:
593:homology model
572:
569:
547:
544:
527:
524:
523:
522:
521:
520:
517:
511:
510:
509:
506:
487:
486:
480:
470:
432:polarizability
420:Semi-empirical
412:small molecule
395:
392:
357:virtual screen
325:
322:
313:
310:
308:
307:Drug discovery
305:
300:gene silencing
190:
187:
175:drug discovery
134:
131:
79:small molecule
43:
42:
32:
31:
30:
29:
15:
9:
6:
4:
3:
2:
6108:
6097:
6094:
6092:
6089:
6087:
6084:
6083:
6081:
6066:
6063:
6061:
6060:Pharmacophore
6058:
6056:
6053:
6051:
6048:
6046:
6043:
6041:
6038:
6036:
6033:
6031:
6028:
6026:
6023:
6021:
6018:
6016:
6013:
6011:
6008:
6006:
6003:
6001:
5998:
5996:
5993:
5991:
5988:
5986:
5985:Drug delivery
5983:
5981:
5978:
5976:
5975:Chemogenomics
5973:
5971:
5968:
5966:
5963:
5962:
5959:
5955:
5947:
5942:
5940:
5935:
5933:
5928:
5927:
5924:
5912:
5907:
5905:
5900:
5898:
5893:
5891:
5886:
5884:
5879:
5877:
5872:
5871:
5868:
5858:
5855:
5853:
5850:
5848:
5845:
5843:
5842:specification
5840:
5838:
5835:
5833:
5830:
5828:
5825:
5823:
5820:
5818:
5815:
5813:
5810:
5808:
5805:
5803:
5800:
5798:
5795:
5793:
5790:
5788:
5785:
5783:
5780:
5778:
5775:
5773:
5770:
5768:
5765:
5761:
5758:
5756:
5755:architectural
5753:
5752:
5751:
5748:
5746:
5743:
5741:
5738:
5736:
5732:
5731:
5728:
5725:
5723:
5722:Visualization
5720:
5718:
5715:
5713:
5710:
5708:
5705:
5703:
5700:
5698:
5695:
5693:
5690:
5688:
5685:
5683:
5680:
5678:
5675:
5673:
5670:
5668:
5665:
5663:
5660:
5658:
5655:
5653:
5650:
5648:
5645:
5643:
5642:Cultural icon
5640:
5638:
5635:
5633:
5630:
5628:
5625:
5623:
5620:
5619:
5616:
5609:
5605:
5591:
5588:
5586:
5583:
5581:
5578:
5576:
5573:
5571:
5568:
5566:
5563:
5561:
5558:
5556:
5553:
5552:
5550:
5546:
5540:
5537:
5535:
5532:
5530:
5527:
5525:
5522:
5520:
5517:
5515:
5512:
5511:
5509:
5507:Organizations
5505:
5497:
5494:
5493:
5492:
5489:
5487:
5486:
5482:
5480:
5477:
5475:
5474:Design patent
5472:
5470:
5467:
5465:
5464:Design around
5462:
5460:
5457:
5456:
5454:
5448:
5442:
5439:
5437:
5434:
5432:
5429:
5427:
5424:
5422:
5419:
5417:
5414:
5412:
5409:
5407:
5404:
5402:
5399:
5397:
5394:
5392:
5389:
5387:
5384:
5380:
5377:
5375:
5372:
5371:
5370:
5367:
5365:
5362:
5360:
5357:
5355:
5352:
5350:
5347:
5346:
5344:
5342:
5338:
5334:
5326:
5324:Organizations
5323:
5320:
5317:
5316:
5312:
5308:
5298:
5295:
5293:
5290:
5288:
5285:
5283:
5280:
5278:
5275:
5273:
5270:
5268:
5265:
5263:
5260:
5258:
5255:
5253:
5250:
5248:
5245:
5243:
5240:
5238:
5235:
5233:
5230:
5228:
5225:
5223:
5219:
5218:
5213:
5210:
5209:
5208:
5205:
5203:
5200:
5198:
5195:
5191:
5188:
5187:
5186:
5185:User-centered
5183:
5181:
5178:
5176:
5173:
5169:
5166:
5165:
5164:
5161:
5159:
5156:
5154:
5151:
5149:
5146:
5144:
5141:
5139:
5136:
5134:
5133:Tableless web
5131:
5127:
5124:
5123:
5122:
5119:
5117:
5114:
5112:
5109:
5107:
5104:
5102:
5099:
5097:
5094:
5092:
5089:
5087:
5084:
5082:
5079:
5077:
5074:
5072:
5069:
5067:
5064:
5062:
5059:
5057:
5056:Participatory
5054:
5052:
5049:
5047:
5044:
5042:
5039:
5037:
5034:
5032:
5029:
5027:
5024:
5022:
5019:
5017:
5014:
5012:
5009:
5007:
5004:
5002:
4999:
4997:
4994:
4992:
4989:
4987:
4984:
4982:
4979:
4977:
4974:
4972:
4969:
4967:
4964:
4962:
4959:
4957:
4954:
4952:
4949:
4947:
4946:For Six Sigma
4944:
4942:
4939:
4937:
4934:
4932:
4929:
4927:
4924:
4922:
4919:
4917:
4914:
4910:
4907:
4906:
4905:
4902:
4900:
4897:
4895:
4892:
4890:
4889:Domain-driven
4887:
4885:
4882:
4878:
4877:architect-led
4875:
4874:
4873:
4870:
4868:
4865:
4863:
4860:
4856:
4853:
4852:
4851:
4848:
4846:
4843:
4841:
4838:
4836:
4833:
4831:
4828:
4826:
4823:
4821:
4820:Configuration
4818:
4816:
4813:
4811:
4808:
4806:
4803:
4801:
4798:
4796:
4793:
4791:
4788:
4786:
4785:Brainstorming
4783:
4781:
4778:
4776:
4773:
4771:
4768:
4767:
4764:
4757:
4753:
4739:
4736:
4734:
4731:
4729:
4726:
4724:
4721:
4719:
4718:Social design
4716:
4714:
4711:
4709:
4706:
4704:
4701:
4699:
4696:
4694:
4691:
4689:
4686:
4684:
4681:
4679:
4676:
4672:
4669:
4667:
4664:
4662:
4659:
4658:
4657:
4654:
4652:
4649:
4647:
4644:
4642:
4641:Filter design
4639:
4637:
4634:
4632:
4629:
4627:
4624:
4622:
4619:
4617:
4616:Boiler design
4614:
4612:
4609:
4607:
4604:
4603:
4601:
4599:
4590:
4584:
4581:
4579:
4576:
4574:
4571:
4569:
4568:Scenic design
4566:
4564:
4561:
4559:
4556:
4554:
4553:Floral design
4551:
4547:
4544:
4542:
4539:
4538:
4537:
4534:
4532:
4528:
4525:
4523:
4520:
4519:
4517:
4515:
4509:
4503:
4500:
4498:
4495:
4493:
4490:
4488:
4485:
4483:
4480:
4478:
4475:
4473:
4470:
4468:
4465:
4461:
4458:
4456:
4453:
4452:
4451:
4448:
4444:
4441:
4440:
4439:
4436:
4435:
4433:
4431:
4425:
4419:
4416:
4414:
4411:
4409:
4406:
4404:
4401:
4399:
4396:
4394:
4391:
4389:
4386:
4384:
4381:
4377:
4374:
4373:
4372:
4369:
4367:
4364:
4362:
4359:
4357:
4354:
4352:
4349:
4347:
4344:
4343:
4341:
4339:
4333:
4327:
4324:
4322:
4319:
4317:
4314:
4310:
4307:
4306:
4305:
4302:
4300:
4297:
4293:
4290:
4289:
4288:
4285:
4283:
4280:
4278:
4275:
4273:
4270:
4266:
4263:
4262:
4261:
4260:Garden design
4258:
4256:
4253:
4251:
4248:
4244:
4243:Passive solar
4241:
4240:
4239:
4236:
4234:
4231:
4229:
4226:
4225:
4223:
4221:
4218:Environmental
4215:
4209:
4206:
4204:
4201:
4199:
4195:
4192:
4190:
4186:
4183:
4181:
4180:Retail design
4178:
4176:
4173:
4171:
4168:
4166:
4163:
4161:
4158:
4156:
4153:
4149:
4146:
4144:
4141:
4139:
4136:
4135:
4134:
4131:
4129:
4126:
4124:
4121:
4119:
4116:
4114:
4111:
4109:
4106:
4105:
4103:
4101:
4098:Communication
4095:
4091:
4084:
4080:
4074:
4071:
4069:
4066:
4065:
4062:
4058:
4051:
4046:
4044:
4039:
4037:
4032:
4031:
4028:
4016:
4013:
4011:
4008:
4006:
3999:
3997:
3994:
3992:
3989:
3987:
3984:
3982:
3979:
3977:
3974:
3972:
3969:
3967:
3964:
3962:
3959:
3957:
3954:
3952:
3949:
3947:
3944:
3942:
3939:
3937:
3934:
3932:
3929:
3927:
3924:
3922:
3919:
3917:
3914:
3912:
3909:
3907:
3904:
3902:
3899:
3897:
3894:
3892:
3889:
3887:
3884:
3882:
3879:
3877:
3874:
3872:
3869:
3867:
3864:
3862:
3859:
3857:
3854:
3852:
3845:
3843:
3842:Beta-blockers
3840:
3838:
3837:Antiandrogens
3835:
3833:
3830:
3828:
3825:
3823:
3820:
3819:
3817:
3811:
3801:
3798:
3797:
3796:
3793:
3791:
3788:
3787:
3786:
3783:
3781:
3778:
3776:
3773:
3772:
3770:
3766:
3762:
3755:
3750:
3748:
3743:
3741:
3736:
3735:
3732:
3725:
3721:
3718:
3714:
3711:
3710:
3698:
3694:
3690:
3686:
3682:
3678:
3671:
3663:
3659:
3655:
3651:
3644:
3636:
3632:
3628:
3624:
3620:
3616:
3609:
3601:
3597:
3593:
3589:
3584:
3579:
3575:
3571:
3567:
3560:
3552:
3548:
3543:
3538:
3534:
3530:
3526:
3522:
3518:
3511:
3503:
3502:Press Release
3499:
3493:
3485:
3481:
3477:
3473:
3469:
3465:
3461:
3457:
3450:
3442:
3436:
3432:
3425:
3417:
3413:
3409:
3405:
3401:
3397:
3390:
3382:
3376:
3372:
3368:
3364:
3360:
3353:
3345:
3341:
3337:
3333:
3329:
3325:
3318:
3310:
3306:
3302:
3298:
3294:
3290:
3283:
3275:
3271:
3267:
3263:
3259:
3255:
3251:
3247:
3243:
3239:
3232:
3224:
3218:
3214:
3210:
3206:
3199:
3191:
3187:
3183:
3179:
3175:
3171:
3164:
3162:
3153:
3149:
3145:
3141:
3137:
3133:
3129:
3125:
3121:
3117:
3110:
3102:
3098:
3094:
3090:
3086:
3082:
3078:
3074:
3067:
3059:
3055:
3051:
3047:
3043:
3039:
3032:
3024:
3020:
3016:
3012:
3008:
3004:
3000:
2996:
2989:
2987:
2978:
2974:
2970:
2966:
2959:
2951:
2945:
2941:
2934:
2926:
2920:
2916:
2912:
2905:
2897:
2891:
2887:
2880:
2872:
2868:
2864:
2860:
2856:
2852:
2844:
2836:
2832:
2828:
2824:
2820:
2816:
2809:
2801:
2797:
2793:
2789:
2785:
2781:
2774:
2766:
2762:
2757:
2752:
2748:
2744:
2740:
2736:
2732:
2725:
2717:
2713:
2709:
2705:
2701:
2697:
2689:
2681:
2677:
2673:
2669:
2665:
2661:
2653:
2645:
2641:
2637:
2633:
2629:
2625:
2618:
2610:
2606:
2602:
2598:
2591:
2583:
2577:
2573:
2569:
2565:
2558:
2550:
2546:
2542:
2538:
2534:
2530:
2526:
2522:
2518:
2514:
2510:
2503:
2495:
2491:
2486:
2481:
2477:
2473:
2469:
2465:
2461:
2454:
2446:
2442:
2438:
2434:
2430:
2426:
2419:
2411:
2405:
2401:
2394:
2386:
2380:
2376:
2372:
2365:
2357:
2353:
2349:
2345:
2341:
2337:
2330:
2322:
2318:
2314:
2310:
2306:
2302:
2295:
2293:
2291:
2282:
2280:9780123971760
2276:
2272:
2268:
2261:
2253:
2249:
2245:
2241:
2236:
2231:
2227:
2223:
2219:
2212:
2204:
2200:
2195:
2190:
2185:
2180:
2176:
2172:
2171:F1000Research
2168:
2161:
2153:
2149:
2145:
2141:
2136:
2131:
2127:
2123:
2119:
2112:
2104:
2100:
2095:
2090:
2085:
2080:
2076:
2072:
2068:
2061:
2053:
2049:
2045:
2041:
2037:
2033:
2026:
2018:
2012:
2008:
2001:
1993:
1989:
1985:
1981:
1977:
1973:
1966:
1958:
1954:
1949:
1944:
1940:
1936:
1932:
1925:
1917:
1913:
1909:
1905:
1901:
1897:
1893:
1889:
1882:
1874:
1870:
1865:
1860:
1856:
1852:
1848:
1844:
1840:
1833:
1825:
1821:
1817:
1813:
1809:
1805:
1798:
1790:
1786:
1782:
1778:
1774:
1770:
1766:
1762:
1755:
1747:
1743:
1739:
1735:
1731:
1727:
1723:
1719:
1712:
1704:
1700:
1696:
1692:
1688:
1684:
1677:
1669:
1665:
1660:
1655:
1651:
1647:
1643:
1636:
1634:
1625:
1621:
1616:
1611:
1607:
1603:
1599:
1592:
1590:
1581:
1575:
1571:
1564:
1562:
1560:
1551:
1545:
1541:
1534:
1530:
1520:
1517:
1515:
1512:
1510:
1507:
1505:
1502:
1500:
1497:
1495:
1492:
1490:
1487:
1485:
1482:
1480:
1477:
1475:
1472:
1471:
1464:
1453:
1450:
1448:
1445:
1443:
1440:
1438:
1435:
1433:
1430:
1428:
1425:
1423:
1420:
1418:
1415:
1413:
1410:
1408:
1405:
1403:
1400:
1398:
1395:
1393:
1390:
1388:
1385:
1383:
1380:
1378:
1375:
1373:
1370:
1368:
1365:
1364:
1357:
1355:
1351:
1347:
1334:
1330:
1327:
1325:
1321:
1318:
1315:
1314:HIV integrase
1311:
1308:
1306:
1302:
1298:
1295:
1292:
1289:
1287:
1283:
1279:
1276:
1268:
1265:
1263:
1259:
1258:
1256:
1253:
1251:
1247:
1243:
1239:
1235:
1231:
1227:
1223:
1219:
1214:
1212:
1208:
1197:
1195:
1191:
1185:
1174:
1171:
1168:
1164:
1161:
1157:
1156:
1155:
1152:
1129:
1122:
1118:configuration
1113:
1106:
1097:
1090:
1081:
1074:
1065:
1011:
1002:
988:
984:
981:
978:
975:
972:
969:
960:
944:
934:
923:
916:
909:
902:
901:
900:
897:
862:
855:
851:
848:
845:
835:
828:
823:
820:
817:
814:
811:
808:
805:
802:
799:
785:
778:
773:
770:
767:
764:
761:
758:
755:
741:
734:
725:
718:
709:
697:
694:
692:
687:
683:
679:
675:
669:
659:
657:
653:
649:
645:
641:
637:
633:
624:
620:
618:
614:
608:
606:
602:
598:
594:
590:
586:
582:
578:
568:
566:
562:
557:
556:pharmacophore
553:
543:
541:
532:
518:
515:
514:
512:
507:
504:
503:
501:
500:
499:
491:
484:
481:
478:
474:
471:
468:
464:
463:
462:
459:
455:
452:
448:
444:
440:
435:
433:
429:
425:
421:
417:
413:
409:
405:
401:
391:
389:
385:
381:
376:
374:
370:
366:
362:
358:
352:
350:
346:
342:
338:
333:
331:
321:
318:
304:
301:
297:
293:
289:
286:
281:
279:
275:
271:
267:
263:
259:
255:
251:
247:
243:
239:
235:
231:
227:
224:
220:
216:
212:
208:
204:
200:
196:
186:
184:
183:toxicological
180:
176:
172:
168:
164:
160:
155:
153:
149:
145:
140:
130:
128:
124:
120:
116:
112:
108:
104:
100:
96:
92:
88:
84:
80:
77:
73:
69:
65:
61:
57:
53:
49:
36:
28:
27:
22:
21:Designer drug
6055:Pharmacology
5989:
5712:STEAM fields
5682:Lean startup
5667:Indie design
5483:
5450:Intellectual
5202:Value-driven
5180:Use-centered
5086:Regenerative
5066:Policy-based
5026:Mind mapping
4931:For assembly
4872:Design–build
4790:By committee
4775:Adaptive web
4625:
4573:Sound design
4531:glass design
4529: /
4514:applied arts
4455:Level design
4326:Urban design
4277:Hotel design
4228:Architecture
4203:Video design
4196: /
4187: /
4155:Illustration
4148:Print design
4118:Brand design
3760:
3680:
3676:
3670:
3653:
3649:
3643:
3618:
3614:
3608:
3573:
3569:
3559:
3524:
3520:
3510:
3501:
3492:
3459:
3455:
3449:
3430:
3424:
3399:
3395:
3389:
3362:
3352:
3327:
3323:
3317:
3292:
3288:
3282:
3241:
3237:
3231:
3204:
3198:
3173:
3169:
3119:
3115:
3109:
3076:
3072:
3066:
3041:
3037:
3031:
2998:
2994:
2968:
2964:
2958:
2939:
2933:
2914:
2904:
2885:
2879:
2854:
2850:
2843:
2818:
2814:
2808:
2783:
2779:
2773:
2738:
2734:
2724:
2699:
2695:
2688:
2663:
2659:
2652:
2627:
2623:
2617:
2600:
2596:
2590:
2563:
2557:
2516:
2512:
2502:
2467:
2463:
2453:
2428:
2424:
2418:
2399:
2393:
2374:
2364:
2342:(2): 86–96.
2339:
2335:
2329:
2304:
2300:
2273:. Elsevier.
2270:
2260:
2225:
2221:
2211:
2174:
2170:
2160:
2125:
2121:
2111:
2077:(10): 1497.
2074:
2071:Biomolecules
2070:
2060:
2035:
2031:
2025:
2006:
2000:
1975:
1971:
1965:
1938:
1934:
1924:
1891:
1887:
1881:
1846:
1842:
1832:
1807:
1803:
1797:
1764:
1760:
1754:
1724:(2): 56–62.
1721:
1717:
1711:
1686:
1682:
1676:
1649:
1645:
1605:
1601:
1569:
1539:
1533:
1462:
1360:Case studies
1343:
1260:Many of the
1254:
1250:chemotherapy
1225:
1215:
1203:
1186:
1178:
1153:
945:
942:
898:
698:
695:
671:
632:Binding site
630:
621:
609:
576:
574:
551:
549:
546:Ligand-based
537:
496:
460:
456:
436:
416:conformation
410:between the
397:
377:
353:
334:
327:
315:
282:
274:side effects
266:binding site
203:nucleic acid
192:
189:Drug targets
156:
152:side effects
136:
114:
110:
51:
47:
46:
5990:Drug design
5750:competition
5707:Slow design
5657:Form factor
5627:Concept art
5436:HTML editor
5116:Sustainable
4951:For testing
4795:By contract
4651:Work design
4626:Drug design
4598:engineering
4472:Icon design
4450:Game design
4428:Interaction
4376:Sustainable
4309:Sustainable
4198:Type design
4175:Photography
4170:News design
4113:Book design
4108:Advertising
4087:Disciplines
4005:antagonists
3790:Preclinical
3780:Hit to lead
3761:Drug design
3713:Drug+Design
3576:(15): 380.
1474:Bioisostere
1310:Raltegravir
1291:Enfuvirtide
1211:dorzolamide
1134:interaction
1086:desolvation
691:antitargets
686:efficacious
652:hydrophobic
601:selectivity
473:hit-to-lead
451:neural nets
384:serendipity
270:antitargets
238:antagonists
219:infectivity
91:therapeutic
83:biomolecule
64:medications
48:Drug design
6080:Categories
5980:Drug class
5952:Topics in
5910:Wiktionary
5903:Wikisource
5857:technology
5827:principles
5426:Storyboard
5252:management
5247:leadership
5212:Privacy by
5051:Parametric
5021:Metadesign
4991:Integrated
4981:High-level
4966:Generative
4961:Functional
4830:Continuous
4825:Contextual
4800:C-K theory
4760:Approaches
4502:Web design
4356:CMF design
4336:Industrial
4194:Typography
3896:Gliflozins
1526:References
1284:inhibitor
1280:Selective
1267:Cimetidine
1232:-positive
840:lipophilic
654:surfaces,
644:apoprotein
254:inhibitors
250:activators
246:modulators
217:or to the
185:profiles.
133:Definition
121:including
85:such as a
54:or simply
5896:Wikiquote
5882:Wikibooks
5832:rationale
5797:knowledge
5772:education
5692:OODA loop
5416:Prototype
5401:Flowchart
5359:Blueprint
5227:computing
5163:Universal
5111:Safe-life
5016:Low-level
5006:Iterative
4986:Inclusive
4971:Geodesign
4862:Defensive
4810:Co-design
4780:Affective
3592:1945-3477
2541:142910916
1459:Criticism
1329:Zanamivir
1316:inhibitor
1305:zopiclone
1234:leukemias
1165:motion –
1160:enthalpic
1126:Δ
1110:Δ
1094:Δ
1078:Δ
1062:Δ
985:
973:−
957:Δ
859:Δ
832:Δ
815:−
796:Σ
782:Δ
759:−
752:Σ
738:Δ
722:Δ
706:Δ
678:Selective
361:drug-like
248:; enzyme
223:microbial
215:pathology
211:signaling
207:metabolic
60:inventive
58:, is the
5889:Wikinews
5822:paradigm
5802:language
5782:engineer
5777:elements
5767:director
5452:property
5297:thinking
5287:strategy
5272:research
5232:controls
5190:Empathic
5121:Systemic
5081:Rational
5036:New Wave
4850:Critical
4073:Designer
3966:Triptans
3851:agonists
3795:Clinical
3697:18319418
3635:10959774
3600:72519717
3551:24902068
3476:12120256
3309:25647463
3190:20166931
3144:15031495
3093:16056391
3058:59482623
3023:21314020
3015:10954199
2977:18428090
2871:16426058
2835:14711306
2800:16426072
2765:18831053
2735:Proteins
2716:16722631
2680:14643325
2644:19128212
2609:17554857
2494:17117615
2445:24050140
2356:12565011
2321:23082974
2244:28685762
2203:32850117
2152:27454546
2144:25233993
2103:37892179
2094:10604719
2052:25916823
1992:24981487
1957:14522049
1908:17016423
1873:19740696
1824:12963322
1789:25292436
1781:26091267
1746:21550508
1703:25110827
1668:29733895
1624:25450771
1467:See also
1442:Triptans
1301:zolpidem
1218:imatinib
1200:Examples
1167:entropic
1034:Receptor
682:affinity
597:affinity
345:purified
341:produced
262:blockers
234:agonists
226:pathogen
171:in vitro
123:peptides
5875:Commons
5847:studies
5792:history
5760:student
5745:classic
5733:Design
5647:.design
5575:Graphex
5277:science
5267:pattern
5262:methods
5237:culture
5220:Design
5031:Modular
4884:Diffuse
4805:Closure
4527:Ceramic
4185:Signage
4068:Outline
3956:Statins
3542:4102969
3484:2728341
3416:8164249
3344:8544170
3274:2491616
3266:7964925
3246:Bibcode
3152:1307935
3124:Bibcode
3116:Science
3101:2549851
2756:2656599
2533:4131441
2485:3181823
2252:6180139
2194:7431967
1916:8872470
1864:2787993
1738:8739258
1226:bcr-abl
1154:where:
1047:Complex
899:where:
648:concave
636:homolog
613:docking
479:, etc.)
298:-based
285:organic
258:openers
199:protein
95:patient
87:protein
76:organic
5852:studio
5837:review
5817:museum
5740:change
5548:Awards
5421:Sketch
5406:Mockup
5386:CAutoD
5327:Awards
5292:theory
5282:sprint
5257:marker
5222:choice
4596:&
4594:design
4430:design
4338:design
4220:design
4138:Motion
4100:design
4057:Design
3800:Phases
3719:(MeSH)
3695:
3633:
3598:
3590:
3549:
3539:
3482:
3474:
3437:
3414:
3377:
3342:
3307:
3272:
3264:
3219:
3188:
3150:
3142:
3099:
3091:
3056:
3021:
3013:
2975:
2946:
2921:
2892:
2869:
2833:
2798:
2763:
2753:
2714:
2678:
2642:
2607:
2578:
2549:604123
2547:
2539:
2531:
2492:
2482:
2443:
2406:
2381:
2354:
2319:
2277:
2250:
2242:
2201:
2191:
2150:
2142:
2101:
2091:
2050:
2013:
1990:
1955:
1914:
1906:
1871:
1861:
1822:
1787:
1779:
1744:
1736:
1701:
1666:
1622:
1576:
1546:
1352:, and
1286:NSAIDs
1246:cancer
1102:motion
1023:Ligand
540:ligand
337:cloned
150:, and
139:ligand
103:charge
70:. The
5735:brief
5622:Agile
5341:Tools
5318:Tools
4956:For X
4592:Other
4511:Other
3596:S2CID
3480:S2CID
3270:S2CID
3148:S2CID
3097:S2CID
3054:S2CID
3019:S2CID
2537:S2CID
2529:JSTOR
2248:S2CID
2148:S2CID
1912:S2CID
1785:S2CID
1742:S2CID
1331:, an
1320:SSRIs
1312:, an
1299:like
1282:COX-2
919:ionic
790:ionic
680:high
526:Types
426:, or
244:, or
201:or a
99:shape
5965:ADME
5812:load
5807:life
5787:firm
5374:CAID
5242:flow
5158:TRIZ
5046:Open
4001:5-HT
3847:Beta
3693:PMID
3677:JAMA
3631:PMID
3588:ISSN
3547:PMID
3472:PMID
3435:ISBN
3412:PMID
3375:ISBN
3340:PMID
3305:PMID
3262:PMID
3217:ISBN
3186:PMID
3140:PMID
3089:PMID
3011:PMID
2973:PMID
2944:ISBN
2919:ISBN
2890:ISBN
2867:PMID
2831:PMID
2796:PMID
2761:PMID
2712:PMID
2676:PMID
2640:PMID
2605:PMID
2576:ISBN
2545:SSRN
2490:PMID
2441:PMID
2404:ISBN
2379:ISBN
2352:PMID
2317:PMID
2275:ISBN
2240:PMID
2199:PMID
2140:PMID
2099:PMID
2048:PMID
2011:ISBN
1988:PMID
1953:PMID
1904:PMID
1869:PMID
1820:PMID
1777:PMID
1734:PMID
1699:PMID
1687:1844
1664:PMID
1620:PMID
1574:ISBN
1544:ISBN
1303:and
1220:, a
1070:bind
965:bind
930:lipo
714:bind
599:and
477:QSAR
386:and
343:and
339:and
296:mRNA
179:ADME
101:and
72:drug
5369:CAD
5349:AAD
5126:SOD
5106:RWD
4976:HCD
4443:EED
4292:EID
3685:doi
3681:299
3658:doi
3623:doi
3578:doi
3537:PMC
3529:doi
3464:doi
3404:doi
3367:doi
3332:doi
3297:doi
3254:doi
3209:doi
3178:doi
3132:doi
3120:303
3081:doi
3046:doi
3003:doi
2859:doi
2823:doi
2788:doi
2751:PMC
2743:doi
2704:doi
2668:doi
2632:doi
2568:doi
2521:doi
2480:PMC
2472:doi
2433:doi
2344:doi
2309:doi
2230:doi
2189:PMC
2179:doi
2130:doi
2089:PMC
2079:doi
2040:doi
1980:doi
1943:doi
1896:doi
1859:PMC
1851:doi
1812:doi
1769:doi
1726:doi
1691:doi
1654:doi
1610:doi
937:rot
926:lip
867:rot
676:.
587:or
402:or
260:or
252:or
209:or
161:of
6082::
3691:.
3679:.
3652:.
3629:.
3617:.
3594:.
3586:.
3572:.
3568:.
3545:.
3535:.
3523:.
3519:.
3500:.
3478:.
3470:.
3458:.
3410:.
3400:37
3398:.
3373:.
3338:.
3328:38
3326:.
3303:.
3293:55
3291:.
3268:.
3260:.
3252:.
3240:.
3215:.
3184:.
3174:17
3172:.
3160:^
3146:.
3138:.
3130:.
3118:.
3095:.
3087:.
3075:.
3052:.
3040:.
3017:.
3009:.
2999:78
2997:.
2985:^
2969:11
2967:.
2865:.
2855:46
2853:.
2829:.
2819:47
2817:.
2794:.
2784:46
2782:.
2759:.
2749:.
2739:75
2737:.
2733:.
2710:.
2700:49
2698:.
2674:.
2664:13
2662:.
2638:.
2626:.
2601:10
2599:.
2574:.
2543:.
2535:.
2527:.
2517:49
2515:.
2511:.
2488:.
2478:.
2466:.
2462:.
2439:.
2429:10
2427:.
2350:.
2338:.
2315:.
2305:19
2303:.
2289:^
2269:.
2246:.
2238:.
2226:16
2224:.
2220:.
2197:.
2187:.
2173:.
2169:.
2146:.
2138:.
2126:13
2124:.
2120:.
2097:.
2087:.
2075:13
2073:.
2069:.
2046:.
2036:33
2034:.
1986:.
1974:.
1951:.
1939:10
1937:.
1933:.
1910:.
1902:.
1890:.
1867:.
1857:.
1847:13
1845:.
1841:.
1818:.
1806:.
1783:.
1775:.
1765:14
1763:.
1740:.
1732:.
1722:18
1720:.
1697:.
1685:.
1662:.
1650:23
1648:.
1644:.
1632:^
1618:.
1606:20
1604:.
1600:.
1588:^
1558:^
1348:,
982:ln
935:ΔG
924:ΔG
917:ΔG
912:hb
910:ΔG
903:ΔG
746:hb
619:.
449:,
445:,
422:,
390:.
240:,
236:,
193:A
146:,
5945:e
5938:t
5931:v
4049:e
4042:t
4035:v
4003:3
3849:2
3753:e
3746:t
3739:v
3726:)
3722:(
3699:.
3687::
3664:.
3660::
3654:3
3637:.
3625::
3619:4
3602:.
3580::
3574:5
3553:.
3531::
3525:5
3486:.
3466::
3460:1
3443:.
3418:.
3406::
3383:.
3369::
3346:.
3334::
3311:.
3299::
3276:.
3256::
3248::
3242:8
3225:.
3211::
3192:.
3180::
3154:.
3134::
3126::
3103:.
3083::
3077:4
3060:.
3048::
3042:6
3025:.
3005::
2979:.
2952:.
2927:.
2898:.
2873:.
2861::
2837:.
2825::
2802:.
2790::
2767:.
2745::
2718:.
2706::
2682:.
2670::
2646:.
2634::
2628:9
2611:.
2584:.
2570::
2551:.
2523::
2496:.
2474::
2468:8
2447:.
2435::
2412:.
2387:.
2358:.
2346::
2340:8
2323:.
2311::
2283:.
2254:.
2232::
2205:.
2181::
2175:9
2154:.
2132::
2105:.
2081::
2054:.
2042::
2019:.
1994:.
1982::
1976:1
1959:.
1945::
1918:.
1898::
1892:5
1875:.
1853::
1826:.
1814::
1808:8
1791:.
1771::
1748:.
1728::
1705:.
1693::
1670:.
1656::
1626:.
1612::
1582:.
1552:.
1273:2
1271:H
1236:(
1182:d
1130:G
1123:+
1114:G
1107:+
1098:G
1091:+
1082:G
1075:=
1066:G
1051:]
1043:[
1038:]
1030:[
1027:]
1019:[
1012:=
1007:d
1003:K
993:d
989:K
979:T
976:R
970:=
961:G
905:0
883:T
880:O
877:R
874:N
863:G
856:+
852:|
849:A
846:|
836:G
829:+
824:t
821:n
818:i
812:c
809:i
806:n
803:o
800:i
786:G
779:+
774:s
771:d
768:n
765:o
762:b
756:h
742:G
735:+
730:0
726:G
719:=
710:G
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.