371:
Cartesian space may not be a straight line trajectory due to the prohibitions of the interconnected bonds. Thus, it is very common for computational optimizing programs to flip back and forth between representations during their iterations. This can dominate the calculation time of the potential itself and in long chain molecules introduce cumulative numerical inaccuracy. While all conversion algorithms produce mathematically identical results, they differ in speed and numerical accuracy. Currently, the fastest and most accurate torsion to
Cartesian conversion is the Natural Extension Reference Frame (NERF) method.
85:
333:. Integration of Newton's laws of motion, using different integration algorithms, leads to atomic trajectories in space and time. The force on an atom is defined as the negative gradient of the potential energy function. The energy minimization method is useful to obtain a static picture for comparing between states of similar systems, while molecular dynamics provides information about the dynamic processes with the intrinsic inclusion of temperature effects.
1827:
1851:
288:, computes the molecular potential energy as a sum of energy terms that describe the deviation of bond lengths, bond angles and torsion angles away from equilibrium values, plus terms for non-bonded pairs of atoms describing van der Waals and electrostatic interactions. The set of parameters consisting of equilibrium bond lengths, bond angles, partial charge values, force constants and van der Waals parameters are collectively termed a
296:. The common force fields in use today have been developed by using chemical theory, experimental reference data, and high level quantum calculations. The method, termed energy minimization, is used to find positions of zero gradient for all atoms, in other words, a local energy minimum. Lower energy states are more stable and are commonly investigated because of their role in chemical and biological processes. A
1863:
1839:
370:
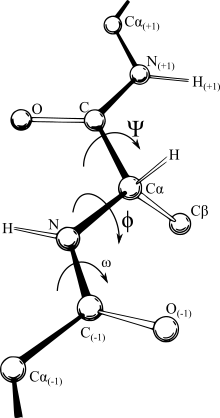
or torsion angle representation. Unfortunately, continuous motions in
Cartesian space often require discontinuous angular branches in internal coordinates, making it relatively hard to work with force fields in the internal coordinate representation, and conversely a simple displacement of an atom in
379:
Molecular modelling methods are used routinely to investigate the structure, dynamics, surface properties, and thermodynamics of inorganic, biological, and polymeric systems. A large number of molecular models of force field are today readily available in databases. The types of biological activity
67:
to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of
111:) to describe the physical basis behind the models. Molecular models typically describe atoms (nucleus and electrons collectively) as point charges with an associated mass. The interactions between neighbouring atoms are described by spring-like interactions (representing
361:
Most force fields are distance-dependent, making the most convenient expression for these
Cartesian coordinates. Yet the comparatively rigid nature of bonds which occur between specific atoms, and in essence, defines what is meant by the designation
222:
279:
135:
and is related to the system internal energy (U), a thermodynamic quantity equal to the sum of potential and kinetic energies. Methods which minimize the potential energy are termed energy minimization methods (e.g.,
366:, make an internal coordinate system the most logical representation. In some fields the IC representation (bond length, angle between bonds, and twist angle of the bond as shown in the figure) is termed the
131:, and can also be assigned velocities in dynamical simulations. The atomic velocities are related to the temperature of the system, a macroscopic quantity. The collective mathematical expression is termed a
30:
331:
154:
300:
simulation, on the other hand, computes the behaviour of a system as a function of time. It involves solving Newton's laws of motion, principally the second law,
420:
230:
602:
Parsons J, Holmes JB, Rojas JM, Tsai J, Strauss CE (July 2005). "Practical conversion from torsion space to
Cartesian space for in silico protein synthesis".
68:
molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit (a
441:
922:
415:
341:
Molecules can be modelled either in vacuum, or in the presence of a solvent such as water. Simulations of systems in vacuum are referred to as
562:"Simulations of inorganic-bioorganic interfaces to discover new materials: insights, comparisons to experiment, challenges, and opportunities"
436:
349:
simulations. In another type of simulation, the effect of solvent is estimated using an empirical mathematical expression; these are termed
1023:
446:
1028:
410:
751:"CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field"
72:
approach), or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons (a
1006:
491:
1050:
890:
536:
425:
1062:
1001:
292:. Different implementations of molecular mechanics use different mathematical expressions and different parameters for the
391:, protein stability, conformational changes associated with biomolecular function, and molecular recognition of proteins,
915:
486:
293:
285:
132:
431:
17:
869:
850:
831:
812:
1904:
880:
1867:
303:
1746:
908:
702:
Eggimann, Becky L.; Sunnarborg, Amara J.; Stern, Hudson D.; Bliss, Andrew P.; Siepmann, J. Ilja (2014-01-02).
1189:
943:
1889:
1466:
953:
1899:
1843:
1392:
1363:
1343:
1296:
981:
646:
501:
471:
451:
703:
1736:
1652:
1291:
1894:
1674:
1585:
1548:
1432:
1358:
1179:
1162:
1105:
289:
120:
52:
1592:
1580:
1471:
1336:
1110:
976:
506:
217:{\displaystyle E=E_{\text{bonds}}+E_{\text{angle}}+E_{\text{dihedral}}+E_{\text{non-bonded}}\,}
123:
is commonly used to describe the latter. The electrostatic interactions are computed based on
1741:
1638:
1623:
1553:
1476:
1308:
1258:
1167:
1092:
991:
456:
60:
144:), while methods that model the behaviour of the system with propagation of time are termed
1731:
1686:
1461:
1281:
1211:
968:
948:
367:
345:
simulations, while those that include the presence of solvent molecules are referred to as
128:
29:
8:
1754:
1708:
1633:
1606:
1504:
1486:
1439:
1377:
1273:
1253:
1122:
1018:
274:{\displaystyle E_{\text{non-bonded}}=E_{\text{electrostatic}}+E_{\text{van der Waals}}\,}
116:
108:
104:
100:
69:
1831:
1797:
1659:
1628:
1509:
1451:
1149:
1132:
1127:
1082:
1045:
1035:
996:
775:
750:
749:
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, et al. (January 2016).
731:
684:
658:
627:
481:
461:
297:
145:
141:
84:
647:"MolMod – an open access database of force fields for molecular simulations of fluids"
1850:
1812:
1777:
1760:
1698:
1616:
1611:
1539:
1524:
1494:
1415:
1382:
1353:
1348:
1323:
1313:
1233:
1221:
1100:
1013:
886:
865:
846:
827:
808:
780:
723:
688:
676:
619:
581:
542:
532:
496:
73:
64:
735:
1855:
1772:
1427:
1286:
1263:
1216:
1157:
770:
762:
715:
668:
631:
611:
573:
137:
124:
44:
672:
1713:
1669:
1664:
1558:
1534:
1368:
1331:
1184:
1174:
1057:
719:
476:
466:
405:
381:
1597:
1575:
1570:
1565:
1520:
1516:
1499:
1456:
1387:
1248:
1243:
1228:
1040:
958:
89:
1883:
1802:
1691:
1647:
1372:
1206:
1201:
1194:
1072:
766:
727:
680:
645:
Stephan, Simon; Horsch, Martin T.; Vrabec, Jadran; Hasse, Hans (2019-07-03).
546:
112:
1679:
1529:
1444:
1420:
1410:
1402:
1303:
1238:
1137:
986:
784:
623:
585:
882:
Computational
Chemistry and Molecular Modeling Principles and Applications
1077:
862:
Molecular
Simulation of Fluids: Theory, Algorithms and Object-Orientation
56:
900:
1703:
577:
561:
704:"An online parameter and property database for the TraPPE force field"
615:
1765:
1067:
932:
388:
1787:
824:
Understanding
Molecular Simulation: From Algorithms to Applications
663:
48:
878:
1807:
93:
103:
is one aspect of molecular modelling, as it involves the use of
43:
encompasses all methods, theoretical and computational, used to
385:
380:
that have been investigated using molecular modelling include
701:
1782:
127:. Atoms are assigned coordinates in Cartesian space or in
1792:
392:
644:
879:
Ramachandran KI, Deepa G, Krishnan
Namboori PK (2008).
421:
Comparison of software for molecular mechanics modeling
37:
Discovering chemical properties by physical simulations
601:
529:
Molecular modelling : principles and applications
306:
233:
157:
559:
442:
List of software for Monte Carlo molecular modeling
748:
325:
273:
216:
1881:
802:
597:
595:
416:Comparison of nucleic acid simulation software
916:
437:List of protein structure prediction software
356:
447:List of software for nanostructures modeling
821:
592:
560:Heinz H, Ramezani-Dakhel H (January 2016).
923:
909:
755:Journal of Chemical Theory and Computation
326:{\displaystyle \mathbf {F} =m\mathbf {a} }
930:
774:
662:
411:Comparison of force field implementations
270:
213:
92:are included in the molecular model of a
843:The Art of Molecular Dynamics Simulation
840:
83:
51:. The methods are used in the fields of
28:
492:Semi-empirical quantum chemistry method
14:
1882:
79:
904:
859:
526:
1838:
1862:
487:Quantum chemistry computer programs
24:
795:
604:Journal of Computational Chemistry
432:List of molecular graphics systems
25:
1916:
1861:
1849:
1837:
1826:
1825:
319:
308:
284:This function, referred to as a
803:Allen MP, Tildesley DJ (1989).
374:
845:. Cambridge University Press.
805:Computer simulation of liquids
742:
695:
638:
553:
520:
13:
1:
1190:Interface and colloid science
944:Glossary of chemical formulae
673:10.1080/08927022.2019.1601191
513:
720:10.1080/08927022.2013.842994
336:
7:
1467:Bioorganometallic chemistry
954:List of inorganic compounds
807:. Oxford University Press.
398:
10:
1921:
1393:Dynamic covalent chemistry
1364:Enantioselective synthesis
1344:Physical organic chemistry
1297:Organolanthanide chemistry
822:Frenkel D, Smit B (1996).
395:, and membrane complexes.
357:Coordinate representations
47:or mimic the behaviour of
1821:
1724:
1485:
1401:
1322:
1272:
1148:
1091:
982:Electroanalytical methods
967:
939:
531:. Pearson Prentice Hall.
502:Structural bioinformatics
472:Molecular modeling on GPU
452:Molecular design software
426:Density functional theory
1737:Nobel Prize in Chemistry
1653:Supramolecular chemistry
1292:Organometallic chemistry
885:. Springer-Verlag GmbH.
767:10.1021/acs.jctc.5b00935
566:Chemical Society Reviews
33:Modeling of ionic liquid
1905:Computational chemistry
1675:Combinatorial chemistry
1586:Food physical chemistry
1549:Environmental chemistry
1433:Bioorthogonal chemistry
1359:Retrosynthetic analysis
1180:Chemical thermodynamics
1163:Spectroelectrochemistry
1106:Computational chemistry
121:Lennard-Jones potential
53:computational chemistry
1747:of element discoveries
1593:Agricultural chemistry
1581:Carbohydrate chemistry
1472:Bioinorganic chemistry
1337:Alkane stereochemistry
1282:Coordination chemistry
1111:Mathematical chemistry
977:Instrumental chemistry
507:Z-matrix (mathematics)
327:
275:
218:
97:
34:
1742:Timeline of chemistry
1639:Post-mortem chemistry
1624:Clandestine chemistry
1554:Atmospheric chemistry
1477:Biophysical chemistry
1309:Solid-state chemistry
1259:Equilibrium chemistry
1168:Photoelectrochemistry
457:Molecular engineering
328:
276:
219:
87:
61:computational biology
32:
1732:History of chemistry
1687:Chemical engineering
1462:Bioorganic chemistry
1212:Structural chemistry
949:List of biomolecules
841:Rapaport DC (2004).
708:Molecular Simulation
651:Molecular Simulation
304:
231:
155:
129:internal coordinates
117:Van der Waals forces
1890:Molecular modelling
1755:The central science
1709:Ceramic engineering
1634:Forensic toxicology
1607:Chemistry education
1505:Radiation chemistry
1487:Interdisciplinarity
1440:Medicinal chemistry
1378:Fullerene chemistry
1254:Microwave chemistry
1123:Molecular mechanics
1118:Molecular modelling
109:Newtonian mechanics
105:classical mechanics
101:Molecular mechanics
80:Molecular mechanics
70:molecular mechanics
41:Molecular modelling
1798:Chemical substance
1660:Chemical synthesis
1629:Forensic chemistry
1510:Actinide chemistry
1452:Clinical chemistry
1133:Molecular geometry
1128:Molecular dynamics
1083:Elemental analysis
1036:Separation process
826:. Academic Press.
578:10.1039/C5CS00890E
482:Monte Carlo method
462:Molecular graphics
351:implicit solvation
323:
298:molecular dynamics
294:potential function
286:potential function
271:
214:
146:molecular dynamics
142:conjugate gradient
133:potential function
98:
35:
18:Molecular modeling
1900:Molecular biology
1877:
1876:
1813:Quantum mechanics
1778:Chemical compound
1761:Chemical reaction
1699:Materials science
1617:General chemistry
1612:Amateur chemistry
1540:Photogeochemistry
1525:Stellar chemistry
1495:Nuclear chemistry
1416:Molecular biology
1383:Polymer chemistry
1354:Organic synthesis
1349:Organic reactions
1314:Ceramic chemistry
1304:Cluster chemistry
1234:Chemical kinetics
1222:Molecular physics
1101:Quantum chemistry
1014:Mass spectrometry
892:978-3-540-77302-3
860:Sadus RJ (2002).
616:10.1002/jcc.20237
538:978-0-582-38210-7
527:Leach AR (2009).
497:Simulated reality
267:
254:
241:
210:
197:
184:
171:
74:quantum chemistry
65:materials science
16:(Redirected from
1912:
1865:
1864:
1853:
1841:
1840:
1829:
1828:
1773:Chemical element
1428:Chemical biology
1287:Magnetochemistry
1264:Mechanochemistry
1217:Chemical physics
1158:Electrochemistry
1063:Characterization
925:
918:
911:
902:
901:
896:
875:
856:
837:
818:
789:
788:
778:
746:
740:
739:
714:(1–3): 101–105.
699:
693:
692:
666:
642:
636:
635:
599:
590:
589:
557:
551:
550:
524:
347:explicit solvent
332:
330:
329:
324:
322:
311:
280:
278:
277:
272:
269:
268:
265:
256:
255:
252:
243:
242:
239:
223:
221:
220:
215:
212:
211:
208:
199:
198:
195:
186:
185:
182:
173:
172:
169:
138:steepest descent
21:
1920:
1919:
1915:
1914:
1913:
1911:
1910:
1909:
1880:
1879:
1878:
1873:
1817:
1720:
1714:Polymer science
1670:Click chemistry
1665:Green chemistry
1559:Ocean chemistry
1535:Biogeochemistry
1481:
1397:
1369:Total synthesis
1332:Stereochemistry
1318:
1268:
1185:Surface science
1175:Thermochemistry
1144:
1087:
1058:Crystallography
963:
935:
929:
899:
893:
872:
853:
834:
815:
798:
796:Further reading
793:
792:
747:
743:
700:
696:
657:(10): 806–814.
643:
639:
600:
593:
558:
554:
539:
525:
521:
516:
511:
477:Molecule editor
467:Molecular model
406:Cheminformatics
401:
382:protein folding
377:
359:
339:
318:
307:
305:
302:
301:
264:
260:
251:
247:
238:
234:
232:
229:
228:
207:
203:
194:
190:
181:
177:
168:
164:
156:
153:
152:
90:dihedral angles
82:
38:
23:
22:
15:
12:
11:
5:
1918:
1908:
1907:
1902:
1897:
1895:Bioinformatics
1892:
1875:
1874:
1872:
1871:
1859:
1847:
1835:
1822:
1819:
1818:
1816:
1815:
1810:
1805:
1800:
1795:
1790:
1785:
1780:
1775:
1770:
1769:
1768:
1758:
1751:
1750:
1749:
1739:
1734:
1728:
1726:
1722:
1721:
1719:
1718:
1717:
1716:
1711:
1706:
1696:
1695:
1694:
1684:
1683:
1682:
1677:
1672:
1667:
1657:
1656:
1655:
1644:
1643:
1642:
1641:
1636:
1626:
1621:
1620:
1619:
1614:
1603:
1602:
1601:
1600:
1598:Soil chemistry
1590:
1589:
1588:
1583:
1576:Food chemistry
1573:
1571:Carbochemistry
1568:
1566:Clay chemistry
1563:
1562:
1561:
1556:
1545:
1544:
1543:
1542:
1537:
1527:
1521:Astrochemistry
1517:Cosmochemistry
1514:
1513:
1512:
1507:
1502:
1500:Radiochemistry
1491:
1489:
1483:
1482:
1480:
1479:
1474:
1469:
1464:
1459:
1457:Neurochemistry
1454:
1449:
1448:
1447:
1437:
1436:
1435:
1425:
1424:
1423:
1418:
1407:
1405:
1399:
1398:
1396:
1395:
1390:
1388:Petrochemistry
1385:
1380:
1375:
1366:
1361:
1356:
1351:
1346:
1341:
1340:
1339:
1328:
1326:
1320:
1319:
1317:
1316:
1311:
1306:
1301:
1300:
1299:
1289:
1284:
1278:
1276:
1270:
1269:
1267:
1266:
1261:
1256:
1251:
1249:Spin chemistry
1246:
1244:Photochemistry
1241:
1236:
1231:
1229:Femtochemistry
1226:
1225:
1224:
1214:
1209:
1204:
1199:
1198:
1197:
1187:
1182:
1177:
1172:
1171:
1170:
1165:
1154:
1152:
1146:
1145:
1143:
1142:
1141:
1140:
1130:
1125:
1120:
1115:
1114:
1113:
1103:
1097:
1095:
1089:
1088:
1086:
1085:
1080:
1075:
1070:
1065:
1060:
1055:
1054:
1053:
1048:
1041:Chromatography
1038:
1033:
1032:
1031:
1026:
1021:
1011:
1010:
1009:
1004:
999:
994:
984:
979:
973:
971:
965:
964:
962:
961:
959:Periodic table
956:
951:
946:
940:
937:
936:
928:
927:
920:
913:
905:
898:
897:
891:
876:
870:
857:
851:
838:
832:
819:
813:
799:
797:
794:
791:
790:
741:
694:
637:
610:(10): 1063–8.
591:
552:
537:
518:
517:
515:
512:
510:
509:
504:
499:
494:
489:
484:
479:
474:
469:
464:
459:
454:
449:
444:
439:
434:
429:
423:
418:
413:
408:
402:
400:
397:
376:
373:
358:
355:
338:
335:
321:
317:
314:
310:
282:
281:
263:
259:
250:
246:
237:
225:
224:
206:
202:
193:
189:
180:
176:
167:
163:
160:
113:chemical bonds
81:
78:
36:
9:
6:
4:
3:
2:
1917:
1906:
1903:
1901:
1898:
1896:
1893:
1891:
1888:
1887:
1885:
1870:
1869:
1860:
1858:
1857:
1852:
1848:
1846:
1845:
1836:
1834:
1833:
1824:
1823:
1820:
1814:
1811:
1809:
1806:
1804:
1803:Chemical bond
1801:
1799:
1796:
1794:
1791:
1789:
1786:
1784:
1781:
1779:
1776:
1774:
1771:
1767:
1764:
1763:
1762:
1759:
1756:
1752:
1748:
1745:
1744:
1743:
1740:
1738:
1735:
1733:
1730:
1729:
1727:
1723:
1715:
1712:
1710:
1707:
1705:
1702:
1701:
1700:
1697:
1693:
1692:Stoichiometry
1690:
1689:
1688:
1685:
1681:
1678:
1676:
1673:
1671:
1668:
1666:
1663:
1662:
1661:
1658:
1654:
1651:
1650:
1649:
1648:Nanochemistry
1646:
1645:
1640:
1637:
1635:
1632:
1631:
1630:
1627:
1625:
1622:
1618:
1615:
1613:
1610:
1609:
1608:
1605:
1604:
1599:
1596:
1595:
1594:
1591:
1587:
1584:
1582:
1579:
1578:
1577:
1574:
1572:
1569:
1567:
1564:
1560:
1557:
1555:
1552:
1551:
1550:
1547:
1546:
1541:
1538:
1536:
1533:
1532:
1531:
1528:
1526:
1522:
1518:
1515:
1511:
1508:
1506:
1503:
1501:
1498:
1497:
1496:
1493:
1492:
1490:
1488:
1484:
1478:
1475:
1473:
1470:
1468:
1465:
1463:
1460:
1458:
1455:
1453:
1450:
1446:
1443:
1442:
1441:
1438:
1434:
1431:
1430:
1429:
1426:
1422:
1419:
1417:
1414:
1413:
1412:
1409:
1408:
1406:
1404:
1400:
1394:
1391:
1389:
1386:
1384:
1381:
1379:
1376:
1374:
1373:Semisynthesis
1370:
1367:
1365:
1362:
1360:
1357:
1355:
1352:
1350:
1347:
1345:
1342:
1338:
1335:
1334:
1333:
1330:
1329:
1327:
1325:
1321:
1315:
1312:
1310:
1307:
1305:
1302:
1298:
1295:
1294:
1293:
1290:
1288:
1285:
1283:
1280:
1279:
1277:
1275:
1271:
1265:
1262:
1260:
1257:
1255:
1252:
1250:
1247:
1245:
1242:
1240:
1237:
1235:
1232:
1230:
1227:
1223:
1220:
1219:
1218:
1215:
1213:
1210:
1208:
1207:Sonochemistry
1205:
1203:
1202:Cryochemistry
1200:
1196:
1195:Micromeritics
1193:
1192:
1191:
1188:
1186:
1183:
1181:
1178:
1176:
1173:
1169:
1166:
1164:
1161:
1160:
1159:
1156:
1155:
1153:
1151:
1147:
1139:
1136:
1135:
1134:
1131:
1129:
1126:
1124:
1121:
1119:
1116:
1112:
1109:
1108:
1107:
1104:
1102:
1099:
1098:
1096:
1094:
1090:
1084:
1081:
1079:
1076:
1074:
1073:Wet chemistry
1071:
1069:
1066:
1064:
1061:
1059:
1056:
1052:
1049:
1047:
1044:
1043:
1042:
1039:
1037:
1034:
1030:
1027:
1025:
1022:
1020:
1017:
1016:
1015:
1012:
1008:
1005:
1003:
1000:
998:
995:
993:
990:
989:
988:
985:
983:
980:
978:
975:
974:
972:
970:
966:
960:
957:
955:
952:
950:
947:
945:
942:
941:
938:
934:
926:
921:
919:
914:
912:
907:
906:
903:
894:
888:
884:
883:
877:
873:
871:0-444-51082-6
867:
863:
858:
854:
852:0-521-82568-7
848:
844:
839:
835:
833:0-12-267370-0
829:
825:
820:
816:
814:0-19-855645-4
810:
806:
801:
800:
786:
782:
777:
772:
768:
764:
761:(1): 405–13.
760:
756:
752:
745:
737:
733:
729:
725:
721:
717:
713:
709:
705:
698:
690:
686:
682:
678:
674:
670:
665:
660:
656:
652:
648:
641:
633:
629:
625:
621:
617:
613:
609:
605:
598:
596:
587:
583:
579:
575:
572:(2): 412–48.
571:
567:
563:
556:
548:
544:
540:
534:
530:
523:
519:
508:
505:
503:
500:
498:
495:
493:
490:
488:
485:
483:
480:
478:
475:
473:
470:
468:
465:
463:
460:
458:
455:
453:
450:
448:
445:
443:
440:
438:
435:
433:
430:
427:
424:
422:
419:
417:
414:
412:
409:
407:
404:
403:
396:
394:
390:
387:
383:
372:
369:
365:
354:
353:simulations.
352:
348:
344:
334:
315:
312:
299:
295:
291:
287:
266:van der Waals
261:
257:
253:electrostatic
248:
244:
235:
227:
226:
204:
200:
191:
187:
178:
174:
165:
161:
158:
151:
150:
149:
147:
143:
139:
134:
130:
126:
125:Coulomb's law
122:
118:
114:
110:
106:
102:
95:
91:
88:The backbone
86:
77:
75:
71:
66:
62:
58:
54:
50:
46:
42:
31:
27:
19:
1866:
1854:
1842:
1830:
1680:Biosynthesis
1530:Geochemistry
1445:Pharmacology
1421:Cell biology
1411:Biochemistry
1239:Spectroscopy
1138:VSEPR theory
1117:
987:Spectroscopy
931:Branches of
881:
864:. Elsevier.
861:
842:
823:
804:
758:
754:
744:
711:
707:
697:
654:
650:
640:
607:
603:
569:
565:
555:
528:
522:
378:
375:Applications
363:
360:
350:
346:
342:
340:
283:
99:
40:
39:
26:
1868:WikiProject
1093:Theoretical
1078:Calorimetry
290:force field
76:approach).
57:drug design
1884:Categories
1704:Metallurgy
1403:Biological
969:Analytical
664:1904.05206
514:References
240:non-bonded
209:non-bonded
1766:Catalysis
1274:Inorganic
1068:Titration
933:chemistry
728:0892-7022
689:119199372
681:0892-7022
547:635267533
389:catalysis
343:gas-phase
337:Variables
49:molecules
1832:Category
1788:Molecule
1725:See also
1150:Physical
785:26631602
736:95716947
624:15898109
586:26750724
428:software
399:See also
368:Z-matrix
364:molecule
196:dihedral
1844:Commons
1808:Alchemy
1324:Organic
776:4712441
632:2279574
94:protein
1856:Portal
1002:UV-Vis
889:
868:
849:
830:
811:
783:
773:
734:
726:
687:
679:
630:
622:
584:
545:
535:
386:enzyme
119:. The
115:) and
1029:MALDI
997:Raman
732:S2CID
685:S2CID
659:arXiv
628:S2CID
183:angle
170:bonds
45:model
1783:Atom
1051:HPLC
887:ISBN
866:ISBN
847:ISBN
828:ISBN
809:ISBN
781:PMID
724:ISSN
677:ISSN
620:PMID
582:PMID
543:OCLC
533:ISBN
140:and
63:and
1793:Ion
1024:ICP
1007:NMR
771:PMC
763:doi
716:doi
669:doi
612:doi
574:doi
393:DNA
1886::
1523:/
1519:/
1371:/
1046:GC
1019:EI
992:IR
779:.
769:.
759:12
757:.
753:.
730:.
722:.
712:40
710:.
706:.
683:.
675:.
667:.
655:45
653:.
649:.
626:.
618:.
608:26
606:.
594:^
580:.
570:45
568:.
564:.
541:.
384:,
148:.
59:,
55:,
1757:"
1753:"
924:e
917:t
910:v
895:.
874:.
855:.
836:.
817:.
787:.
765::
738:.
718::
691:.
671::
661::
634:.
614::
588:.
576::
549:.
320:a
316:m
313:=
309:F
262:E
258:+
249:E
245:=
236:E
205:E
201:+
192:E
188:+
179:E
175:+
166:E
162:=
159:E
107:(
96:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.