1257:-binding protein. Homotrimerisation is a process whereby three of the same subunits, associate to make a complex of three identical YadA proteins. Furthermore, just like other TAAs, it has a head-stalk-anchor protein architecture. The majority of TAAs share strong similarity in the C-terminal membrane anchor region, the only member to differ across TAAs is the head, neck, and stalk regions. The head region of YadA is composed of beta-helices further folded to create a nine-coiled left-handed parallel beta-roll (LPBR).
348:: This particular domain is a trimer of single-stranded, left-handed beta-helices. These associate to form a nine-coiled left-handed beta-roll. It contains sequence motifs, of which there is a strong similarity with other TAA heads. This indicates that there is a lot of similarity when comparing protein structure. The head domain is connected to the stalk by a short, highly conserved sequence, which is often called the neck, or occasionally named the connector.
3350:
706:
43:
1572:
1643:, since they have an outer membrane that makes it hard for them to stick to and infect the host. The outer membrane is useful, as it allows the bacteria to colonize, and adds another layer of protection. However, the outer membrane is a barrier for the secretion of proteins, and it requires energy to transport proteins across the outer membrane. Hence, the T5SS pathway overcomes this problem.
31:
1684:. The signal peptide on the N-terminus acts as a temporary tether to hold it in place. Next, it must move to the outer membrane. The trimerisation aids translocation, and no translocation would occur without its beta-barrel membrane anchor. The type V secretion system is described as non-fimbrious, meaning that the bacterial cells do not use long physical appendages named
329:: There are several roles that the Extended Signal Peptide Region is thought to hold. First, biogenesis of proteins in the Type V Secretion System (T5SS). Second, it is thought to target the protein to the inner membrane to be translocated either by the signal recognition particle pathway (SRP) or by twin
1293:
infections in humans. The structure of UspA1 also has a head domain at N-terminal domain, however it is folded into the beta propeller. Like the other TAAs, it has a coiled-coil stalk region but, in this case it is extended, and it has the TAA typical C-terminal beta barrel membrane anchor domain.
200:
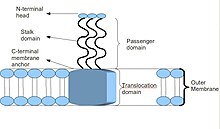
The protein domain arrangement of the
Trimeric Autotransporter Adhesin, BadA This figure shows the head, stalk and anchor domains. It shows the YadA-like head in grey. The stalk contains repeats coloured in green and the membrane anchor in red. The sequence below shows colouring according to domain
398:
YadA-like head is composed of single-stranded, left-handed beta-helices, which associate further to create a nine-coiled left-handed parallel beta-roll (LPBR). It is the tightest beta-roll structure known, and the first to be discovered. The YadA head domain has eight repeat motifs, each fourteen
1330:
arranged in a triangular prism and contains internal symmetry. Additionally, the head domain contains 5 Trp-Ring domains. Furthermore, this protein also contains three neck domains, of which two are IsNeck domains in addition to other domains such as KG, GANG, and TTT domains. It also contains a
658:
that deviate from the standard model due to their unusual properties. From a deeper structural perspective, coiled-coil arranges itself in such a way that the crossing angle between the helices is almost zero. The packing of these helices follows a "knobs-into-holes" arrangement whereby
141:. Once the membrane anchor has been inserted into the outer membrane, the passenger domain passes through it into the host extracellular environment autonomously, hence the description of autotransporter. The head domain, once assembled, then adheres to an element of the host
666:
protrude forming knobs that pack into cavities formed by other residues on another helix. Then, once the knobs are packed into cavities, the three helices are wound in register around each other, so all of the residues in certain positions are at the same height.
389:
There are several types of head domain. Each domain helps the head to bind to a different component of the extracellular matrix. These are as follows: YadA-like head domain, Trp-ring, GIN, FxG, HIN1, and HIN2. This entry focuses on the first three mentioned.
488:
The ISneck domain is a type of neck domain. There are two types of ISneck domain. This first is an ISneck which is interrupted by an insertion. The insertion can take form of either folded (ISneck 1) or much shorter, unfolded (ISneck 2) perturbation.
311:. The function of the ESPR is to aid inner membrane translocation by acting as a temporary tether. This prevents the accumulation of misfolded proteins. The ESPR can be divided into individual regions, they are as follows: N1 (charged), H1 (
476:
and the smaller one of the coiled coil. Furthermore, just like its safety pin structure, it also has a function of pinning all three monomers together and pins it to the head domain. This increases the stability of the neck domain.
136:
Trimeric autotransporter adhesins have a unique structure. The structure they hold is crucial to their function. They all appear to have a head-stalk-anchor structure. Each TAA is made up of three identical proteins, hence the name
454:
are connected by a diagonally running extended beta-sheet. The sheets then further fold to form a beta prism in which each wall is composed of a complete set of five beta-strands. The GIN domain is often followed by a neck domain.
185:, referring to the C-terminal membrane anchor. Although all TAAs carry a membrane anchor in common, they may not all contain both a stalk and a head as well. In addition, all membrane anchor domains are of the left-handed parallel
629:: These domains are fibrous and found in highly repetitive numbers. They contain coiled coils and their length tends to vary among different species. The coiled-coil segments of the stalk domains have two unusual properties:
1252:
adhesin protein A. This protein domain is an example of
Trimeric Autotransporter Adhesins, and it was the first TAA to be discovered. Like other TAAs, YadA also undergoes homotrimerisation to form a stable
875:
or in other words coiled-coil stalk domain translocates through this pore. Additional functions of the membrane anchor is to oligomerise the stalk domain and to anchor the whole protein to the
887:
All
Trimeric Autotransporter Adhesins are crucial virulence factors that cause serious disease in humans. The most-studied and well-known Trimeric Autotransporter Adhesins are listed below:
480:
There are seven different type of neck domains. They are as follows: ISneck1, ISneck2, HANS connector, DALL-1, DALL-2, DALL-3, and the neck domain. This entry focuses on the ISneck domain.
1527:. It is important that these outer-membrane adhesins make physical contact with the receptors found on the host cell. This means that the adhesin must be long enough to extend beyond the
1277:
in humans. Studies have shown that the globular N-terminal head domain of NadA is vital for adhesion. NadA also contains a coiled-coil region and also a C-terminal membrane anchor.
201:
arrangement and protease cleavage sites red (trypsin) and blue (chymotrypsin). (Figure used from open access journal, in the public domain, Public
Library of Science (PLoS) Pathogen
1492:
The process of infection is complicated. The invasive bacterium must overcome many barriers in order to infect its host, including environmental barriers, physical barriers and
385:
Comparison of Head domains in different
Trimeric Autotransporter Adhesins (Figure used from open access journal, in the public domain, Public Library of Science (PLoS) Pathogen)
378:
466:: The neck domain is a homotrimer, where three of the same subunits associate. All three subunits are arranged in such a way that they resemble a "safety pin"-like structure.
193:
1082:
859:, a process known as autotransport activity. The way TAAs translocate across the outer membrane remains to be elucidated, but it is thought that it translocates inside the
1185:
798:
594:
275:
1680:. The mechanism is split into two. First, the protein must move across the inner membrane or, in other words, translocate, in a Sec-dependent manner via the
841:. It also contains a recognition site for signal peptidases, which means the enzyme will recognise the signal peptide and cleave it at a particular point.
2634:"Mapping of the Neisseria meningitidis NadA cell-binding site: relevance of predicted {alpha}-helices in the NH2-terminal and dimeric coiled-coil regions"
863:, leading to transportation of the passenger domain from the C terminus to the N terminus through the beta-barrel lumen. In essence, the beta-barrel is a
1402:
or, in other words, a combination of YadA and Hia head domains. This combination gives insight into how the pathogenicity of Gram-negative bacteria has
17:
2802:
2155:
Koretke KK, Szczesny P, Gruber M, Lupas AN (2006). "Model structure of the prototypical non-fimbrial adhesin YadA of
Yersinia enterocolitica".
1322:. Hia has a slightly unusual N-terminal head made of beta-prisms. The beta-prism is an unusual type of protein architecture first described by
2686:"Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding"
1635:
bacteria, mainly due to the difference in cell wall structure. Trimeric
Autotransporter Adhesins use a particular secretion pathway, named
186:
2586:
Białas N, Kasperkiewicz K, Radziejewska-Lebrecht J, Skurnik M (2012). "Bacterial cell surface structures in
Yersinia enterocolitica".
2537:
Cotter, S. E.; Surana, N. K.; St. Geme, J. W. (2005). "Trimeric autotransporters: A distinct subfamily of autotransporter proteins".
1809:
Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA (2006). "Trimeric autotransporter adhesins: variable structure, common function".
1851:"The translocation domain in trimeric autotransporter adhesins is necessary and sufficient for trimerization and autotransportation"
1654:. Such Sec-dependent systems do not need to use energy, unlike Sec-independent machinery, which uses other forms of energy such as
1665:. Since it can transport things across the outer membrane without the need to generate a new form of energy, it earned the name
1696:
The TAAs can help prevent the bacteria from being destroyed by the host's immune system. In particular in the case of certain
2426:
1228:
1223:
1133:
891:
542:
1205:
818:
614:
295:
423:
found in the C-terminal part of the Head domain. These work by stabilising the transition between the coiled-coil and the
307:
The
Extended Signal Peptide Region (ESPR) is found in the N-terminus of the signal peptides of proteins belonging to the
3184:
Gerlach RG, Hensel M (2007). "Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens".
2478:
2464:
2450:
2268:
1406:
over time. BadA also contains a neck domain, an extended coil-coil stalk, and beta-barrel C terminal membrane anchor.
1265:
Another example of a TAA is the NadA protein. The NadA protein is found in a species of Gram-negative bacteria called
2513:
2388:
848:
1604:
in comparison to Gram-positive bacteria. Gram-negative bacteria have three layers: The innermost layer is named the
50:
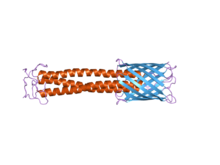
The C-terminal membrane anchor domain can clearly be seen on the right in blue. The stalk domain can be seen in red.
1949:"A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism"
3385:
1716:
resistance. All of these methods prevent the bacteria from being eliminated by the host and ensure its survival.
1507:. Then the Trimeric Autotransporter Adhesin must adhere to the layer of cells found on the internal surface, the
673:: Their role is to act as spacers by moving the head domains away from the bacterial cell surface and toward the
2970:"Structural classification of proteins and structural genomics: new insights into protein folding and evolution"
2867:"Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter"
1676:
The Sec-dependent system is divided into three pathways. TAAs are one of those pathways and also go by the name
333:
translocated (TAT). Third, it has been observed and believed to regulate the rate of protein migration into the
1193:
806:
602:
283:
109:
TAAs are just one of many methods bacteria use to infect their hosts, infection resulting in diseases such as
2047:"The unusual extended signal peptide region of the type V secretion system is phylogenetically restricted"
837:. It is thought that, once trimerisation has occurred, these beta strands further fold into a 12-stranded
3062:"The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain"
2284:"UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli"
1535:
layer of the host cell. Once it has done so, it may bind to the ECM of the host cell. TAAs are a type of
448:
427:
where the head meets the neck or stalk. In many cases, the Trp ring is often followed by the GIN domain.
2571:
3395:
3380:
3340:
2103:"The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll"
1189:
802:
681:. They also have a role in protecting the bacterial cell against host defences. They do this by aiding
598:
279:
2743:
Casutt-Meyer S, Renzi F, Schmaler M, Jann NJ, Amstutz M, Cornelis GR (2010). Bereswill, Stefan (ed.).
2190:
Edwards TE, Phan I, Abendroth J, Dieterich SH, Masoudi A, Guo W, et al. (2010). Kursula P (ed.).
1995:"From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis"
651:
Furthermore, the stalk is made up entirely of pentadecads. Hence, the stalk domains can be considered
3370:
2409:Łyskowski A, Leo JC, Goldman A (2011). "Structure and Biology of Trimeric Autotransporter Adhesins".
1146:
555:
2742:
1636:
1588:
308:
130:
3375:
1617:
1597:
1395:
876:
868:
833:: The structure of this protein domain is a left-handed coiled-coil followed by four transmembrane
67:
896:
685:
resistance. The stalk protein domain is also alternatively named the internal passenger domain.
2331:
Caserta R, Takita MA, Targon ML, Rosselli-Murai LK, de Souza AP, Peroni L, et al. (2010).
1743:
Szczesny P, Linke D, Ursinus A, Bär K, Schwarz H, Riess TM, et al. (2008). Ghosh P (ed.).
1655:
1651:
1605:
1364:
1307:
920:
904:
643:
often interrupted by small globular domains, which owes to their appearance of segmented ropes.
710:
The beta barrel structure found in the C-terminus of the bacterial adhesin anchor domain, YadA
2796:
1601:
912:
320:
2981:
2816:
Comanducci M, Bambini S, Brunelli B, Adu-Bobie J, Aricò B, Capecchi B, et al. (2002).
2756:
2697:
2344:
2333:"Expression of Xylella fastidiosa fimbrial and afimbrial proteins during biofilm formation"
2203:
1512:
1172:
785:
674:
581:
355:
262:
142:
2632:
Tavano R, Capecchi B, Montanari P, Franzoso S, Marin O, Sztukowska M, et al. (2011).
2045:
Desvaux M, Cooper LM, Filenko NA, Scott-Tucker A, Turner SM, Cole JA, et al. (2006).
472:: The function of the neck domain is to be the adaptor between the larger diameter of the
8:
1947:
Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, et al. (2007).
1621:
1386:, restricting the flow of blood. This may be due to BadA's inducing the transcription of
1348:
928:
162:
2985:
2760:
2701:
2348:
2282:
Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, et al. (2008).
2207:
3390:
3294:
3269:
3245:
3220:
3127:
3102:
3002:
2969:
2945:
2920:
2891:
2866:
2842:
2817:
2779:
2744:
2720:
2685:
2658:
2633:
2611:
2365:
2332:
2308:
2283:
2226:
2191:
2024:
1924:
1899:
1875:
1850:
1771:
1744:
1563:
olecules (MSCRAMMs). In other words, they are a complex that aids adhesion to the ECM.
1528:
1399:
1379:
688:
There are two types of stalk domain: the FGG domain and the right-handed stalk domain.
424:
89:
3221:"Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane"
3038:
2505:
2127:
2102:
1285:
UspA1 is another example of a
Trimeric Autotransporter Adhesin found in the bacterium
3299:
3285:
3250:
3201:
3163:
3132:
3083:
3042:
3007:
2950:
2896:
2847:
2784:
2725:
2684:
Agnew C, Borodina E, Zaccai NR, Conners R, Burton NM, Vicary JA, et al. (2011).
2663:
2603:
2554:
2519:
2509:
2432:
2422:
2370:
2313:
2231:
2172:
2132:
2068:
2063:
2046:
2016:
1970:
1929:
1880:
1826:
1776:
1709:
1701:
1609:
1180:
1138:
793:
682:
589:
547:
371:
270:
99:
55:
2921:"Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter"
2615:
2028:
1915:
1114:
734:
523:
236:
3289:
3281:
3240:
3232:
3193:
3155:
3122:
3114:
3073:
3034:
2997:
2989:
2940:
2932:
2886:
2878:
2837:
2829:
2774:
2764:
2715:
2705:
2653:
2645:
2595:
2546:
2501:
2414:
2360:
2352:
2303:
2295:
2221:
2211:
2164:
2122:
2114:
2058:
2006:
1960:
1919:
1911:
1870:
1862:
1818:
1766:
1756:
1508:
1168:
781:
577:
258:
1398:
1. The head domain of BadA is more complex than other TAAs. It is thought to be a
747:
3322:
3159:
2769:
2413:. Advances in Experimental Medicine and Biology. Vol. 715. pp. 143–58.
2216:
1761:
1579:
A schematic diagram illustrating the Trimeric Autotransporter Adhesins in Type V
1126:
864:
759:
535:
2418:
2192:"Structure of a Burkholderia pseudomallei trimeric autotransporter adhesin head"
1627:
In Gram-negative bacteria, the secretary pathway is very different from that of
3354:
3197:
3150:
Harris LG, Richards RG (2006). "Staphylococci and implant surfaces: a review".
2585:
2254:
978:
678:
138:
81:
78:
2993:
2936:
2599:
2550:
2168:
1822:
1331:
coiled-coil stalk and the typically conserved TAA C terminal membrane anchor.
3364:
2882:
2118:
1705:
1632:
1613:
1493:
1375:
1352:
1323:
856:
316:
85:
71:
2710:
1849:
Mikula KM, Leo JC, Łyskowski A, Kedracka-Krok S, Pirog A, Goldman A (2012).
370:. The head domain is very important for attachment to the host cell and for
3303:
3254:
3236:
3205:
3167:
3136:
3087:
3078:
3061:
3011:
2954:
2900:
2851:
2788:
2729:
2667:
2607:
2558:
2523:
2436:
2374:
2317:
2235:
2176:
2136:
2072:
2020:
1974:
1933:
1884:
1830:
1780:
1713:
1387:
1356:
638:
3046:
2492:
Lupas AN, Gruber M (2005). "The structure of alpha-helical coiled coils".
1965:
1948:
1142:
551:
165:, the structure has been described as a "lollipop" shape consisting of an
3118:
2833:
2356:
2101:
Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M, Goldman A (2004).
1524:
1383:
1327:
1034:
860:
852:
838:
834:
660:
655:
652:
359:
312:
178:
150:
92:
38:
Schematic diagram of the basic Trimeric Autotransporter Adhesin structure
3333:
2649:
2299:
2011:
1994:
1866:
1628:
1511:, in the intestine by using its head to bind to proteins found in the
1496:
barriers. The bacterium must enter the host's body and, in the case of
1315:
1290:
1274:
991:
970:
663:
473:
451:
444:
440:
420:
416:
412:
400:
170:
166:
118:
1302:
The Hia protein is a TAA found on the outer membrane of the bacterium
3103:"Intruders below the radar: molecular pathogenesis of Bartonella spp"
3025:
Shimizu T, Morikawa K (1996). "The beta-prism: a new folding motif".
2572:
http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl
1681:
1593:
1580:
1501:
1403:
1311:
334:
126:
122:
110:
3267:
435:
The GIN domain is a head domain named after its sequence motif GIN (
3349:
3318:
1662:
1640:
1531:
layer in the outer membrane of the bacterium and interact with the
1516:
1254:
1233:
1121:
754:
530:
363:
330:
146:
103:
102:, factors that make the bacteria harmful and infective to the host
95:
74:
3270:"Mechanisms of protein export across the bacterial outer membrane"
2815:
1391:
2330:
1520:
1371:
436:
367:
173:
membrane anchor domain. Often, the literature refers to these as
2044:
447:) motif. It has an all-beta structure, whereby the two pairs of
2631:
1946:
1900:"Domain annotation of trimeric autotransporter adhesins--daTAA"
1848:
1659:
1647:
1532:
1504:
1368:
1319:
1270:
1200:
813:
705:
609:
290:
161:
Most TAAs have a similar protein structure. When observed with
114:
42:
1382:, causing knots to form in the smaller blood vessels, such as
1351:, a normally harmless disease, but, in people with a weakened
377:
27:
Proteins found on the outer membrane of Gram-negative bacteria
3268:
Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C (2005).
2745:"Oligomeric coiled-coil adhesin YadA is a double-edged sword"
2281:
2189:
1685:
121:. Most bacteria infect their host through a method named the
3059:
1231:
is a protein domain found in Gram-negative bacteria such as
192:
30:
3314:
2818:"NadA, a novel vaccine candidate of Neisseria meningitidis"
1360:
1162:
1109:
1081:
775:
741:
729:
571:
518:
411:
The Trp ring is the second-most-common TAA head. Trp is an
252:
231:
2683:
2154:
2100:
1808:
2864:
419:. The Trp ring obtains its name from the high levels of
354:: The function of this protein domain is to bind to the
205:
1992:
1742:
1571:
1470:
pneumonia and some strains cause meningitis and sepsis
319:
site) domains. N1 and H1 form the ESPR and have strong
3313:
This article incorporates text from the public domain
3060:
Surana NK, Cutter D, Barenkamp SJ, St Geme JW (2004).
1745:"Structure of the head of the Bartonella adhesin BadA"
1339:
The BadA protein is another example of a TAA found in
1027:
Coiled-coil alpha helices followed by a linker region
3338:
2536:
2249:
2247:
2245:
1596:
is one method of transferring substances across the
1086:
Crystal structure of the collagen-binding domain of
691:
3334:
The TAA database, based at the Max Planck Institute
2408:
1691:
1415:Clinical effect of infection with virulence factor
98:, which is shortened to OCAs. In essence, they are
2918:
2865:Meng G, Surana NK, St Geme JW, Waksman G (2006).
2242:
1566:
1326:and Murzin. As the name suggests, it holds three
3362:
637:they alternate from right-handed to left-handed
3179:
3177:
3024:
2914:
2912:
2910:
2679:
2677:
2627:
2625:
2581:
2579:
2404:
2402:
2400:
2398:
3149:
2967:
2150:
2148:
2146:
2096:
2094:
2092:
2090:
2088:
2086:
2084:
2082:
2040:
2038:
1988:
1986:
1984:
1844:
1842:
1840:
1804:
1802:
1800:
1798:
1796:
1794:
1792:
1790:
1738:
1736:
1734:
1732:
1730:
1728:
1650:Sec translocase is found to be present on the
3183:
1993:Leyton DL, Rossiter AE, Henderson IR (2012).
1897:
1600:. Gram-negative bacteria have very different
177:, containing the N-terminal, head, neck, and
77:. Bacteria use TAAs in order to infect their
3261:
3218:
3212:
3174:
3143:
3094:
3053:
2961:
2907:
2858:
2809:
2801:: CS1 maint: multiple names: authors list (
2736:
2674:
2622:
2576:
2530:
2485:
2395:
2324:
2275:
2183:
1646:T5SS uses Sec-machinery system to work. The
1639:(T5SS). Gram-negative bacteria must secrete
3018:
2491:
2143:
2079:
2035:
1981:
1940:
1891:
1837:
1787:
1725:
990:Similar to YadA head, contains left handed
3100:
1080:
704:
3293:
3244:
3126:
3077:
3001:
2944:
2890:
2841:
2778:
2768:
2719:
2709:
2657:
2364:
2307:
2225:
2215:
2126:
2062:
2010:
1964:
1923:
1874:
1770:
1760:
340:
1612:, is a space containing a thin layer of
1570:
376:
191:
41:
29:
1363:, it is more serious as it can lead to
14:
3363:
2919:Meng G, St Geme JW, Waksman G (2008).
156:
1224:YadA bacterial adhesin protein domain
940:
937:
206:Extended Signal Peptide Region domain
1487:
699:YadA bacterial adhesin anchor domain
1616:; and the third layer is named the
1409:
169:head domain, a stalk domain, and a
24:
3225:Philos Trans R Soc Lond B Biol Sci
1310:in humans. This protein can cause
25:
3407:
3327:
2496:. Advances in Protein Chemistry.
882:
692:C-terminal membrane anchor domain
393:
129:pathway, to be more specific the
60:trimeric autotransporter adhesins
18:Trimeric Autotransporter Adhesins
3348:
3286:10.1128/JB.187.13.4306-4314.2005
3219:Leo JC, Grin I, Linke D (2012).
2064:10.1111/j.1574-6968.2006.00425.x
1692:Evading the host's immune system
1280:
483:
88:. TAAs also go by another name,
2565:
2471:
2457:
2443:
2381:
2261:
1669:, since it transports proteins
1334:
1260:
1217:
492:
125:pathway. TAAs are part of the
2968:Andreeva A, Murzin AG (2010).
1608:; the middle layer, named the
1567:Type V secretion system (T5SS)
1297:
1040:Three-alpha helix coiled-coil
851:is to aid the movement of the
458:
13:
1:
3039:10.1016/s0968-0004(06)80018-6
2588:Arch Immunol Ther Exp (Warsz)
2506:10.1016/S0065-3233(05)70003-6
1916:10.1093/bioinformatics/btn118
1719:
1700:, the TAA YadA has a role in
1673:, in other words, by itself.
1289:, found as a common cause of
1157:Available protein structures:
969:Single-stranded, left-handed
770:Available protein structures:
566:Available protein structures:
247:Available protein structures:
66:), are proteins found on the
3160:10.1016/j.injury.2006.04.003
2770:10.1371/journal.pone.0015159
2217:10.1371/journal.pone.0012803
1898:Szczesny P, Lupas A (2008).
1762:10.1371/journal.ppat.1000119
1390:factors, as it activates of
7:
2419:10.1007/978-94-007-0940-9_9
1413:
1355:, such as those undergoing
1246:Yersinia pseudotuberculosis
1048:C terminal Membrane anchor
406:
10:
3412:
3312:
3198:10.1016/j.ijmm.2007.03.017
1688:to attach to one another.
1586:
1347:is the causative agent of
1221:
358:of the host, most notably
3154:. 37 Suppl 2 (2): S3–14.
3101:Harms A, Dehio C (2012).
2994:10.1107/S1744309110007177
2937:10.1016/j.jmb.2008.09.085
2600:10.1007/s00005-012-0168-z
2551:10.1016/j.tim.2005.03.004
2169:10.1016/j.jsb.2006.03.012
1823:10.1016/j.tim.2006.04.005
1678:type Vc secretion pathway
1367:. This a condition where
1199:
1179:
1161:
1156:
1152:
1132:
1120:
1108:
1100:
1095:
1079:
1074:
1024:Right-handed coiled-coil
960:
957:
954:
951:
948:
945:
812:
792:
774:
769:
765:
753:
740:
728:
720:
715:
703:
698:
608:
588:
570:
565:
561:
541:
529:
517:
509:
504:
499:
289:
269:
251:
246:
242:
230:
222:
217:
212:
2883:10.1038/sj.emboj.7601132
2690:Proc Natl Acad Sci U S A
2479:"Bioinformatics Toolkit"
2465:"Bioinformatics Toolkit"
2451:"Bioinformatics Toolkit"
2389:"Bioinformatics Toolkit"
2269:"Bioinformatics Toolkit"
2255:"Bioinformatics Toolkit"
2119:10.1038/sj.emboj.7600100
1598:bacterial outer membrane
1434:Yersinia enterocolitica
1396:hypoxia-inducible factor
877:bacterial outer membrane
869:bacterial outer membrane
309:Type V secretion systems
131:type Vc secretion system
2711:10.1073/pnas.1106341108
1710:complement inactivation
1637:type V secretion system
1589:Type V secretion system
1467:Haemophilus influenzae
1445:Neisseria meningitidis
1314:and some strains cause
897:Yersinia enterocolitica
3386:Gram-negative bacteria
3237:10.1098/rstb.2011.0208
3079:10.1074/jbc.M311496200
2539:Trends in Microbiology
2337:Appl Environ Microbiol
1656:adenosine triphosphate
1584:
1456:Moraxella catarrhalis
1448:sepsis and meningitis
1365:bacillary angiomatosis
1308:respiratory epithelium
1304:Haemophilus influenzae
1267:Neisseria meningitidis
1066:Beta barrel structure
1063:Beta barrel structure
1060:Beta barrel structure
1057:Beta barrel structure
1054:Beta barrel structure
1051:Beta barrel structure
921:Haemophilus influenzae
905:Neisseria meningitidis
849:membrane anchor domain
847:: The function of the
430:
386:
374:, sticking to itself.
341:N-terminal head domain
202:
51:
39:
1966:10.1099/mic.0.29091-0
1574:
1459:middle ear infection
1378:undergo uncontrolled
1287:Moraxella catarrhalis
1043:Extended coiled-coil
1030:Extended coiled-coil
913:Moraxella catarrhalis
867:that sits within the
380:
195:
84:via a process called
45:
33:
3119:10.1128/CMR.05009-11
2834:10.1084/jem.20020407
2357:10.1128/AEM.02114-09
1602:cell wall structures
1513:extracellular matrix
1481:cat scratch disease
1478:Bartonella henselae
1306:. It adheres to the
675:extracellular matrix
356:extracellular matrix
183:Translocation domain
143:extracellular matrix
3186:Int J Med Microbiol
2986:2010AcCrF..66.1190A
2761:2010PLoSO...515159C
2702:2011PNAS..10815174A
2650:10.1128/JB.00430-10
2349:2010ApEnM..76.4250C
2300:10.1128/JB.00122-08
2208:2010PLoSO...512803E
2051:FEMS Microbiol Lett
2012:10.1038/nrmicro2733
1867:10.1128/JB.05322-11
1622:lipopolysaccharides
1416:
1349:cat scratch disease
1345:Bartonella henselae
1341:Bartonella henselae
929:Bartonella henselae
163:electron microscopy
157:Molecular structure
3231:(1592): 1088–101.
3107:Clin Microbiol Rev
3027:Trends Biochem Sci
2974:Acta Crystallogr F
2411:Bacterial Adhesion
1585:
1529:lipopolysaccharide
1500:, invade the host
1423:Bacterial species
1414:
1248:. YadA stands for
855:chain through the
387:
203:
52:
40:
3396:Membrane proteins
3381:Virulence factors
2980:(Pt 10): 1190–7.
2428:978-94-007-0939-3
1999:Nat Rev Microbiol
1702:autoagglutination
1620:, which contains
1610:periplasmic space
1488:Infection process
1485:
1484:
1215:
1214:
1211:
1210:
1206:structure summary
1070:
1069:
828:
827:
824:
823:
819:structure summary
624:
623:
620:
619:
615:structure summary
372:autoagglutination
315:), N2, H2 and C (
305:
304:
301:
300:
296:structure summary
100:virulence factors
56:molecular biology
16:(Redirected from
3403:
3371:Protein families
3353:
3352:
3344:
3308:
3307:
3297:
3265:
3259:
3258:
3248:
3216:
3210:
3209:
3181:
3172:
3171:
3147:
3141:
3140:
3130:
3098:
3092:
3091:
3081:
3072:(15): 14679–85.
3057:
3051:
3050:
3022:
3016:
3015:
3005:
2965:
2959:
2958:
2948:
2916:
2905:
2904:
2894:
2877:(11): 2297–304.
2862:
2856:
2855:
2845:
2813:
2807:
2806:
2800:
2792:
2782:
2772:
2740:
2734:
2733:
2723:
2713:
2681:
2672:
2671:
2661:
2629:
2620:
2619:
2583:
2574:
2569:
2563:
2562:
2534:
2528:
2527:
2494:Adv Protein Chem
2489:
2483:
2482:
2475:
2469:
2468:
2461:
2455:
2454:
2447:
2441:
2440:
2406:
2393:
2392:
2385:
2379:
2378:
2368:
2328:
2322:
2321:
2311:
2279:
2273:
2272:
2265:
2259:
2258:
2251:
2240:
2239:
2229:
2219:
2187:
2181:
2180:
2152:
2141:
2140:
2130:
2098:
2077:
2076:
2066:
2042:
2033:
2032:
2014:
1990:
1979:
1978:
1968:
1944:
1938:
1937:
1927:
1895:
1889:
1888:
1878:
1846:
1835:
1834:
1811:Trends Microbiol
1806:
1785:
1784:
1774:
1764:
1740:
1509:epithelial cells
1426:Diseases caused
1417:
1410:Clinical effects
1154:
1153:
1088:Yersinia adhesin
1084:
1075:YadA head domain
1072:
1071:
966:N terminal Head
935:
934:
910:UspA1 and A2 of
873:passenger domain
767:
766:
708:
696:
695:
563:
562:
497:
496:
244:
243:
210:
209:
175:Passenger domain
21:
3411:
3410:
3406:
3405:
3404:
3402:
3401:
3400:
3376:Protein domains
3361:
3360:
3359:
3347:
3339:
3330:
3325:
3311:
3280:(13): 4306–14.
3266:
3262:
3217:
3213:
3182:
3175:
3148:
3144:
3099:
3095:
3058:
3054:
3023:
3019:
2966:
2962:
2917:
2908:
2863:
2859:
2828:(11): 1445–54.
2814:
2810:
2794:
2793:
2741:
2737:
2696:(37): 15174–8.
2682:
2675:
2630:
2623:
2584:
2577:
2570:
2566:
2535:
2531:
2516:
2490:
2486:
2477:
2476:
2472:
2463:
2462:
2458:
2449:
2448:
2444:
2429:
2407:
2396:
2387:
2386:
2382:
2329:
2325:
2294:(12): 4147–61.
2280:
2276:
2267:
2266:
2262:
2253:
2252:
2243:
2188:
2184:
2153:
2144:
2099:
2080:
2043:
2036:
1991:
1982:
1959:(Pt 1): 59–70.
1945:
1941:
1896:
1892:
1847:
1838:
1807:
1788:
1755:(8): e1000119.
1741:
1726:
1722:
1694:
1667:autotransporter
1591:
1578:
1577:
1569:
1490:
1420:Protein Domain
1412:
1337:
1300:
1283:
1269:, which causes
1263:
1242:Yersinia pestis
1226:
1220:
1091:
918:Hia and Hsf of
885:
711:
694:
495:
486:
461:
433:
409:
396:
384:
383:
343:
208:
199:
198:
181:stalk, and the
159:
145:, for example,
49:
48:
37:
36:
28:
23:
22:
15:
12:
11:
5:
3409:
3399:
3398:
3393:
3388:
3383:
3378:
3373:
3358:
3357:
3337:
3336:
3329:
3328:External links
3326:
3310:
3309:
3260:
3211:
3173:
3142:
3093:
3052:
3017:
2960:
2906:
2857:
2808:
2755:(12): e15159.
2735:
2673:
2621:
2594:(3): 199–209.
2575:
2564:
2545:(5): 199–205.
2529:
2514:
2484:
2470:
2456:
2442:
2427:
2394:
2380:
2343:(13): 4250–9.
2323:
2274:
2260:
2241:
2182:
2142:
2078:
2034:
1980:
1939:
1910:(10): 1251–6.
1904:Bioinformatics
1890:
1836:
1786:
1723:
1721:
1718:
1693:
1690:
1652:inner membrane
1618:outer membrane
1606:inner membrane
1587:Main article:
1575:
1568:
1565:
1489:
1486:
1483:
1482:
1479:
1476:
1472:
1471:
1468:
1465:
1461:
1460:
1457:
1454:
1450:
1449:
1446:
1443:
1439:
1438:
1435:
1432:
1428:
1427:
1424:
1421:
1411:
1408:
1336:
1333:
1299:
1296:
1282:
1279:
1262:
1259:
1238:enterocolitica
1222:Main article:
1219:
1216:
1213:
1212:
1209:
1208:
1203:
1197:
1196:
1183:
1177:
1176:
1166:
1159:
1158:
1150:
1149:
1136:
1130:
1129:
1124:
1118:
1117:
1112:
1106:
1105:
1102:
1098:
1097:
1093:
1092:
1085:
1077:
1076:
1068:
1067:
1064:
1061:
1058:
1055:
1052:
1049:
1045:
1044:
1041:
1038:
1031:
1028:
1025:
1022:
1018:
1017:
1014:
1011:
1008:
1005:
1002:
999:
995:
994:
988:
985:
982:
979:Beta propeller
976:
975:Globular head
973:
967:
963:
962:
959:
956:
953:
950:
947:
943:
942:
939:
933:
932:
924:
916:
908:
900:
884:
883:Model proteins
881:
826:
825:
822:
821:
816:
810:
809:
796:
790:
789:
779:
772:
771:
763:
762:
757:
751:
750:
745:
738:
737:
732:
726:
725:
722:
718:
717:
713:
712:
709:
701:
700:
693:
690:
649:
648:
647:
646:
645:
644:
641:
622:
621:
618:
617:
612:
606:
605:
592:
586:
585:
575:
568:
567:
559:
558:
545:
539:
538:
533:
527:
526:
521:
515:
514:
511:
507:
506:
502:
501:
494:
491:
485:
482:
460:
457:
432:
429:
408:
405:
395:
394:YadA-like head
392:
381:
342:
339:
303:
302:
299:
298:
293:
287:
286:
273:
267:
266:
256:
249:
248:
240:
239:
234:
228:
227:
224:
220:
219:
215:
214:
207:
204:
196:
158:
155:
68:outer membrane
46:
34:
26:
9:
6:
4:
3:
2:
3408:
3397:
3394:
3392:
3389:
3387:
3384:
3382:
3379:
3377:
3374:
3372:
3369:
3368:
3366:
3356:
3351:
3346:
3345:
3342:
3335:
3332:
3331:
3324:
3320:
3316:
3305:
3301:
3296:
3291:
3287:
3283:
3279:
3275:
3271:
3264:
3256:
3252:
3247:
3242:
3238:
3234:
3230:
3226:
3222:
3215:
3207:
3203:
3199:
3195:
3192:(6): 401–15.
3191:
3187:
3180:
3178:
3169:
3165:
3161:
3157:
3153:
3146:
3138:
3134:
3129:
3124:
3120:
3116:
3112:
3108:
3104:
3097:
3089:
3085:
3080:
3075:
3071:
3067:
3063:
3056:
3048:
3044:
3040:
3036:
3032:
3028:
3021:
3013:
3009:
3004:
2999:
2995:
2991:
2987:
2983:
2979:
2975:
2971:
2964:
2956:
2952:
2947:
2942:
2938:
2934:
2931:(4): 824–36.
2930:
2926:
2922:
2915:
2913:
2911:
2902:
2898:
2893:
2888:
2884:
2880:
2876:
2872:
2868:
2861:
2853:
2849:
2844:
2839:
2835:
2831:
2827:
2823:
2819:
2812:
2804:
2798:
2790:
2786:
2781:
2776:
2771:
2766:
2762:
2758:
2754:
2750:
2746:
2739:
2731:
2727:
2722:
2717:
2712:
2707:
2703:
2699:
2695:
2691:
2687:
2680:
2678:
2669:
2665:
2660:
2655:
2651:
2647:
2644:(1): 107–15.
2643:
2639:
2635:
2628:
2626:
2617:
2613:
2609:
2605:
2601:
2597:
2593:
2589:
2582:
2580:
2573:
2568:
2560:
2556:
2552:
2548:
2544:
2540:
2533:
2525:
2521:
2517:
2515:9780120342709
2511:
2507:
2503:
2499:
2495:
2488:
2480:
2474:
2466:
2460:
2452:
2446:
2438:
2434:
2430:
2424:
2420:
2416:
2412:
2405:
2403:
2401:
2399:
2390:
2384:
2376:
2372:
2367:
2362:
2358:
2354:
2350:
2346:
2342:
2338:
2334:
2327:
2319:
2315:
2310:
2305:
2301:
2297:
2293:
2289:
2285:
2278:
2270:
2264:
2256:
2250:
2248:
2246:
2237:
2233:
2228:
2223:
2218:
2213:
2209:
2205:
2202:(9): e12803.
2201:
2197:
2193:
2186:
2178:
2174:
2170:
2166:
2163:(2): 154–61.
2162:
2158:
2157:J Struct Biol
2151:
2149:
2147:
2138:
2134:
2129:
2124:
2120:
2116:
2113:(4): 701–11.
2112:
2108:
2104:
2097:
2095:
2093:
2091:
2089:
2087:
2085:
2083:
2074:
2070:
2065:
2060:
2056:
2052:
2048:
2041:
2039:
2030:
2026:
2022:
2018:
2013:
2008:
2005:(3): 213–25.
2004:
2000:
1996:
1989:
1987:
1985:
1976:
1972:
1967:
1962:
1958:
1954:
1950:
1943:
1935:
1931:
1926:
1921:
1917:
1913:
1909:
1905:
1901:
1894:
1886:
1882:
1877:
1872:
1868:
1864:
1861:(4): 827–38.
1860:
1856:
1852:
1845:
1843:
1841:
1832:
1828:
1824:
1820:
1817:(6): 264–70.
1816:
1812:
1805:
1803:
1801:
1799:
1797:
1795:
1793:
1791:
1782:
1778:
1773:
1768:
1763:
1758:
1754:
1750:
1746:
1739:
1737:
1735:
1733:
1731:
1729:
1724:
1717:
1715:
1711:
1707:
1703:
1699:
1698:Yersinia spp.
1689:
1687:
1683:
1679:
1674:
1672:
1668:
1664:
1661:
1657:
1653:
1649:
1644:
1642:
1638:
1634:
1633:Gram-positive
1630:
1625:
1623:
1619:
1615:
1614:peptidoglycan
1611:
1607:
1603:
1599:
1595:
1590:
1582:
1573:
1564:
1562:
1558:
1554:
1550:
1546:
1542:
1538:
1534:
1530:
1526:
1522:
1518:
1514:
1510:
1506:
1503:
1499:
1495:
1494:immune system
1480:
1477:
1474:
1473:
1469:
1466:
1463:
1462:
1458:
1455:
1452:
1451:
1447:
1444:
1441:
1440:
1436:
1433:
1430:
1429:
1425:
1422:
1419:
1418:
1407:
1405:
1401:
1397:
1393:
1389:
1388:proangiogenic
1385:
1381:
1380:proliferation
1377:
1376:blood vessels
1373:
1370:
1366:
1362:
1358:
1354:
1353:immune system
1350:
1346:
1342:
1332:
1329:
1325:
1321:
1317:
1313:
1309:
1305:
1295:
1292:
1288:
1281:UspA1 protein
1278:
1276:
1272:
1268:
1258:
1256:
1251:
1247:
1243:
1239:
1236:
1235:
1230:
1225:
1207:
1204:
1202:
1198:
1195:
1191:
1187:
1184:
1182:
1178:
1174:
1170:
1167:
1164:
1160:
1155:
1151:
1148:
1144:
1140:
1137:
1135:
1131:
1128:
1125:
1123:
1119:
1116:
1113:
1111:
1107:
1103:
1099:
1094:
1089:
1083:
1078:
1073:
1065:
1062:
1059:
1056:
1053:
1050:
1047:
1046:
1042:
1039:
1036:
1032:
1029:
1026:
1023:
1020:
1019:
1015:
1012:
1009:
1006:
1003:
1000:
997:
996:
993:
989:
986:
983:
980:
977:
974:
972:
968:
965:
964:
944:
936:
931:
930:
925:
923:
922:
917:
915:
914:
909:
907:
906:
901:
899:
898:
893:
890:
889:
888:
880:
878:
874:
870:
866:
862:
858:
857:cell membrane
854:
850:
846:
842:
840:
836:
832:
820:
817:
815:
811:
808:
804:
800:
797:
795:
791:
787:
783:
780:
777:
773:
768:
764:
761:
758:
756:
752:
749:
746:
743:
739:
736:
733:
731:
727:
723:
719:
714:
707:
702:
697:
689:
686:
684:
680:
676:
672:
668:
665:
662:
657:
654:
653:alpha helical
642:
640:
636:
635:
634:
633:
632:
631:
630:
628:
616:
613:
611:
607:
604:
600:
596:
593:
591:
587:
583:
579:
576:
573:
569:
564:
560:
557:
553:
549:
546:
544:
540:
537:
534:
532:
528:
525:
522:
520:
516:
512:
508:
503:
498:
490:
484:ISneck domain
481:
478:
475:
471:
467:
465:
456:
453:
450:
446:
442:
438:
428:
426:
422:
418:
414:
404:
402:
391:
379:
375:
373:
369:
365:
361:
357:
353:
349:
347:
338:
336:
332:
328:
324:
322:
318:
314:
310:
297:
294:
292:
288:
285:
281:
277:
274:
272:
268:
264:
260:
257:
254:
250:
245:
241:
238:
235:
233:
229:
225:
221:
216:
211:
194:
190:
188:
184:
180:
176:
172:
168:
164:
154:
152:
148:
144:
140:
134:
132:
128:
124:
120:
116:
112:
107:
105:
101:
97:
94:
91:
87:
86:cell adhesion
83:
80:
76:
73:
72:Gram-negative
69:
65:
61:
57:
44:
32:
19:
3277:
3273:
3263:
3228:
3224:
3214:
3189:
3185:
3151:
3145:
3113:(1): 42–78.
3110:
3106:
3096:
3069:
3065:
3055:
3030:
3026:
3020:
2977:
2973:
2963:
2928:
2924:
2874:
2870:
2860:
2825:
2821:
2811:
2797:cite journal
2752:
2748:
2738:
2693:
2689:
2641:
2637:
2591:
2587:
2567:
2542:
2538:
2532:
2497:
2493:
2487:
2473:
2459:
2445:
2410:
2383:
2340:
2336:
2326:
2291:
2287:
2277:
2263:
2199:
2195:
2185:
2160:
2156:
2110:
2106:
2057:(1): 22–30.
2054:
2050:
2002:
1998:
1956:
1953:Microbiology
1952:
1942:
1907:
1903:
1893:
1858:
1854:
1814:
1810:
1752:
1748:
1714:phagocytosis
1708:resistance,
1697:
1695:
1677:
1675:
1671:autonomously
1670:
1666:
1645:
1626:
1592:
1560:
1556:
1552:
1548:
1544:
1540:
1536:
1498:Yersinia sp.
1497:
1491:
1437:yersiniosis
1359:or fighting
1357:chemotherapy
1344:
1340:
1338:
1335:BadA protein
1303:
1301:
1286:
1284:
1266:
1264:
1261:NadA protein
1249:
1245:
1241:
1237:
1232:
1227:
1218:YadA protein
1087:
1037:coiled-coil
1004:Not present
987:Beta prisms
984:Not present
927:
919:
911:
903:
895:
886:
872:
844:
843:
835:beta strands
830:
829:
687:
670:
669:
656:coiled-coils
650:
639:supercoiling
626:
625:
493:Stalk domain
487:
479:
474:beta-helices
469:
468:
463:
462:
449:antiparallel
434:
425:beta-meander
410:
397:
388:
351:
350:
345:
344:
326:
325:
321:conservation
306:
182:
174:
160:
135:
108:
63:
59:
53:
3274:J Bacteriol
3066:J Biol Chem
2638:J Bacteriol
2288:J Bacteriol
1855:J Bacteriol
1749:PLOS Pathog
1658:(ATP) or a
1551:ecognizing
1525:fibronectin
1394:as well as
1384:capillaries
1328:beta sheets
1298:Hia protein
1096:Identifiers
1035:alpha helix
861:beta-barrel
853:polypeptide
839:beta-barrel
724:YadA_anchor
716:Identifiers
661:hydrophobic
505:Identifiers
459:Neck domain
452:beta sheets
403:in length.
360:fibronectin
313:hydrophobic
218:Identifiers
179:coiled-coil
151:fibronectin
93:coiled-coil
3365:Categories
3033:(1): 3–6.
2925:J Mol Biol
1720:References
1629:eukaryotes
1547:omponents
1502:intestinal
1343:bacteria.
1316:meningitis
1291:middle ear
1275:meningitis
1169:structures
992:beta helix
971:beta helix
782:structures
683:complement
578:structures
513:YadA_stalk
500:YadA_stalk
445:Asparagine
441:Isoleucine
421:tryptophan
417:tryptophan
413:amino acid
259:structures
171:C-terminal
167:N-terminal
119:meningitis
90:oligomeric
3391:Secretion
3323:IPR005594
2822:J Exp Med
2500:: 37–78.
1682:periplasm
1594:Secretion
1581:Secretion
1539:icrobial
1312:pneumonia
1127:IPR008640
1104:YadA_head
941:Proteins
831:Structure
760:IPR005594
627:Structure
536:IPR008635
464:Structure
346:Structure
335:periplasm
187:beta-roll
127:secretion
123:secretion
111:pneumonia
3319:InterPro
3304:15968039
3255:22411980
3206:17482513
3168:16651069
3137:22232371
3088:14726537
3012:20944210
2955:18948113
2901:16688217
2852:12045242
2789:21170337
2749:PLOS ONE
2730:21876142
2668:20971901
2616:10237335
2608:22484801
2559:15866036
2524:15837513
2437:21557062
2375:20472735
2318:18424525
2236:20862217
2196:PLOS ONE
2177:16675268
2137:14765110
2073:17020545
2029:19562964
2021:22337167
1975:17185535
1934:18397894
1885:22155776
1831:16678419
1781:18688279
1663:gradient
1641:adhesins
1576:Figure 5
1555:dhesive
1517:collagen
1515:such as
1255:collagen
1250:Yersinia
1234:Yersinia
1186:RCSB PDB
1122:InterPro
1016:Present
1013:Present
1010:Present
1007:Present
1001:Present
926:BadA of
902:NadA of
845:Function
799:RCSB PDB
755:InterPro
677:of the
671:Function
664:residues
595:RCSB PDB
531:InterPro
470:Function
407:Trp ring
401:residues
382:Figure 4
364:collagen
352:Function
331:arginine
327:Function
317:cleavage
276:RCSB PDB
197:Figure 3
147:collagen
139:trimeric
104:organism
96:adhesins
75:bacteria
47:Figure 2
35:Figure 1
3355:Biology
3295:1151778
3246:3297439
3128:3255967
3047:8848836
3003:2954204
2982:Bibcode
2946:2597055
2892:1478200
2843:2193550
2780:2999546
2757:Bibcode
2721:3174611
2698:Bibcode
2659:3019930
2366:2897468
2345:Bibcode
2309:2446758
2227:2942831
2204:Bibcode
1925:2373917
1876:3272944
1772:2483945
1583:System.
1543:urface
1521:laminin
1404:evolved
1400:chimera
1374:of the
1372:tumours
1324:Chothia
1115:PF05658
938:Domain
735:PF03895
524:PF05662
437:Glycine
368:laminin
237:PF13018
153:, etc.
3341:Portal
3302:
3292:
3253:
3243:
3204:
3166:
3152:Injury
3135:
3125:
3086:
3045:
3010:
3000:
2953:
2943:
2899:
2889:
2871:EMBO J
2850:
2840:
2787:
2777:
2728:
2718:
2666:
2656:
2614:
2606:
2557:
2522:
2512:
2435:
2425:
2373:
2363:
2316:
2306:
2234:
2224:
2175:
2135:
2128:381008
2125:
2107:EMBO J
2071:
2027:
2019:
1973:
1932:
1922:
1883:
1873:
1829:
1779:
1769:
1712:, and
1660:proton
1648:enzyme
1559:atrix
1533:glycan
1523:, and
1505:mucosa
1453:UspA1
1369:benign
1320:sepsis
1271:sepsis
1244:, and
1201:PDBsum
1175:
1165:
1147:SUPFAM
1101:Symbol
1033:Three-
1021:Stalk
952:UspA1
871:. The
814:PDBsum
788:
778:
748:CL0327
721:Symbol
610:PDBsum
584:
574:
556:SUPFAM
510:Symbol
415:named
366:, and
291:PDBsum
265:
255:
223:Symbol
189:type.
117:, and
115:sepsis
2612:S2CID
2025:S2CID
1706:serum
1475:BadA
1442:NadA
1431:YadA
1392:NF-κB
1143:SCOPe
1134:SCOP2
998:Neck
981:head
961:BadA
955:HadA
949:NadA
946:YadA
865:porin
552:SCOPe
543:SCOP2
82:cells
3317:and
3315:Pfam
3300:PMID
3251:PMID
3202:PMID
3164:PMID
3133:PMID
3084:PMID
3043:PMID
3008:PMID
2951:PMID
2897:PMID
2848:PMID
2803:link
2785:PMID
2726:PMID
2664:PMID
2604:PMID
2555:PMID
2520:PMID
2510:ISBN
2433:PMID
2423:ISBN
2371:PMID
2314:PMID
2232:PMID
2173:PMID
2133:PMID
2069:PMID
2017:PMID
1971:PMID
1930:PMID
1881:PMID
1827:PMID
1777:PMID
1686:pili
1464:Hia
1361:AIDS
1318:and
1273:and
1229:YadA
1194:PDBj
1190:PDBe
1173:ECOD
1163:Pfam
1139:1p9h
1110:Pfam
1090:YadA
958:Hia
892:YadA
807:PDBj
803:PDBe
786:ECOD
776:Pfam
744:clan
742:Pfam
730:Pfam
679:host
603:PDBj
599:PDBe
582:ECOD
572:Pfam
548:1s7m
519:Pfam
284:PDBj
280:PDBe
263:ECOD
253:Pfam
232:Pfam
226:ESPR
213:ESPR
79:host
64:TAAs
3290:PMC
3282:doi
3278:187
3241:PMC
3233:doi
3229:367
3194:doi
3190:297
3156:doi
3123:PMC
3115:doi
3074:doi
3070:279
3035:doi
2998:PMC
2990:doi
2941:PMC
2933:doi
2929:384
2887:PMC
2879:doi
2838:PMC
2830:doi
2826:195
2775:PMC
2765:doi
2716:PMC
2706:doi
2694:108
2654:PMC
2646:doi
2642:193
2596:doi
2547:doi
2502:doi
2415:doi
2361:PMC
2353:doi
2304:PMC
2296:doi
2292:190
2222:PMC
2212:doi
2165:doi
2161:155
2123:PMC
2115:doi
2059:doi
2055:264
2007:doi
1961:doi
1957:153
1920:PMC
1912:doi
1871:PMC
1863:doi
1859:194
1819:doi
1767:PMC
1757:doi
1631:or
1181:PDB
894:of
794:PDB
590:PDB
431:GIN
271:PDB
70:of
54:In
3367::
3321::
3298:.
3288:.
3276:.
3272:.
3249:.
3239:.
3227:.
3223:.
3200:.
3188:.
3176:^
3162:.
3131:.
3121:.
3111:25
3109:.
3105:.
3082:.
3068:.
3064:.
3041:.
3031:21
3029:.
3006:.
2996:.
2988:.
2978:66
2976:.
2972:.
2949:.
2939:.
2927:.
2923:.
2909:^
2895:.
2885:.
2875:25
2873:.
2869:.
2846:.
2836:.
2824:.
2820:.
2799:}}
2795:{{
2783:.
2773:.
2763:.
2751:.
2747:.
2724:.
2714:.
2704:.
2692:.
2688:.
2676:^
2662:.
2652:.
2640:.
2636:.
2624:^
2610:.
2602:.
2592:60
2590:.
2578:^
2553:.
2543:13
2541:.
2518:.
2508:.
2498:70
2431:.
2421:.
2397:^
2369:.
2359:.
2351:.
2341:76
2339:.
2335:.
2312:.
2302:.
2290:.
2286:.
2244:^
2230:.
2220:.
2210:.
2198:.
2194:.
2171:.
2159:.
2145:^
2131:.
2121:.
2111:23
2109:.
2105:.
2081:^
2067:.
2053:.
2049:.
2037:^
2023:.
2015:.
2003:10
2001:.
1997:.
1983:^
1969:.
1955:.
1951:.
1928:.
1918:.
1908:24
1906:.
1902:.
1879:.
1869:.
1857:.
1853:.
1839:^
1825:.
1815:14
1813:.
1789:^
1775:.
1765:.
1751:.
1747:.
1727:^
1704:,
1624:.
1519:,
1240:,
1192:;
1188:;
1171:/
1145:/
1141:/
879:.
805:;
801:;
784:/
601:;
597:;
580:/
554:/
550:/
362:,
337:.
323:.
282:;
278:;
261:/
149:,
133:.
113:,
106:.
58:,
3343::
3306:.
3284::
3257:.
3235::
3208:.
3196::
3170:.
3158::
3139:.
3117::
3090:.
3076::
3049:.
3037::
3014:.
2992::
2984::
2957:.
2935::
2903:.
2881::
2854:.
2832::
2805:)
2791:.
2767::
2759::
2753:5
2732:.
2708::
2700::
2670:.
2648::
2618:.
2598::
2561:.
2549::
2526:.
2504::
2481:.
2467:.
2453:.
2439:.
2417::
2391:.
2377:.
2355::
2347::
2320:.
2298::
2271:.
2257:.
2238:.
2214::
2206::
2200:5
2179:.
2167::
2139:.
2117::
2075:.
2061::
2031:.
2009::
1977:.
1963::
1936:.
1914::
1887:.
1865::
1833:.
1821::
1783:.
1759::
1753:4
1561:m
1557:m
1553:a
1549:r
1545:c
1541:s
1537:m
443:-
439:-
62:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.