87:
560:. MAMs contain tandem repeats of mammalian cell entry (MCE) domains which specifically bind to extracellular matrix proteins and anionic lipids on host tissues. Since they are abundant in many pathogens of clinical importance, Multivalent Adhesion Molecules are a potential target for prophylactic or therapeutic anti-infectives. The use of a MAM targeting adhesion inhibitor was shown to significantly decrease the colonization of burn wounds by multidrug resistant
79:. In the crudest sense, bacterial adhesins serve as anchors allowing bacteria to overcome these environmental shear forces, thus remaining in their desired environment. However, bacterial adhesins do not serve as a sort of universal bacterial Velcro. Rather, they act as specific surface recognition molecules, allowing the targeting of a particular bacterium to a particular surface such as root tissue in plants,
357:
171:. The bacterial adhesin consists primarily of an intramembranous structural protein which provides a scaffold upon which several extracellular adhesins may be attached. However, as in the case of the CFA1 fimbriae, the structural protein itself can sometimes act as an adhesin if a portion of the protein extends into the
297:
A number of problems create challenges for the researcher exploring the anti-adhesin immunity concept. First, a large number of different bacterial adhesins target the same human tissues. Further, an individual bacterium can produce multiple different types of adhesin, at different times, in
203:
links the two domains. Additionally, a carbohydrate-binding pocket has been identified at the tip of the N-terminal adhesive domain. This basic structure is conserved across type 1 fimbrial adhesins though recent studies have shown that in vitro induced mutations can lead to the addition of
1086:
Langermann S, Möllby R, Burlein J, Palaszynski S, Auguste C, DeFusco A, Strouse R, Schenerman M, Hultgren S, Pinkner J, Winberg J, Guldevall L, Söderhäll M, Ishikawa K, Normark S, Koenig S (2000). "Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by
159:
Through the mechanisms of evolution, different species of bacteria have developed different solutions to the problem of attaching receptor specific proteins to the bacteria surface. Today many different types and subclasses of bacterial adhesins may be observed in the literature.
641:
FimH provides an example of conformation specific immune response which enhances impact on the protein. By studying this particular adhesion, researchers hope to develop adhesion-specific vaccines which may serve as a model for antibody-mediation of pathogen adhesion.
216:. "A large number of bacterial adhesins with individual receptor specificities have been identified." Many bacterial pathogens are able to express an array of different adhesins. Expression of these adhesins at different phases during
510:
to the urinary tract. They do so by inducing the development of long cellular extensions that wrap around the bacteria. They also confer the mannose-resistant hemaglutination phenotype, which can be inhibited by
616:
strains most known for causing diarrhea can be found in the intestinal tissue of pigs and humans where they express the K88 and CFA1. to attach to the intestinal lining. Additionally, UPEC causes about 90% of
191:
then processes the protein by removing several signal peptides ultimately leaving a 279 amino acid protein. Mature FimH is displayed on the bacterial surface as a component of the type 1 fimbrial organelle.
323:
adhesins FHA and pertactin are components of three of the four acellular pertussis vaccines currently licensed for use in the U.S. Additionally, anti-adhesion vaccines are being explored as a solution to
236:
comes from early studies which indicated that an important component of protective immunity against certain bacteria came from an ability to prevent adhesin binding. Additionally, adhesins are attractive
220:
play the most important role in adhesion based virulence. Numerous studies have shown that inhibiting a single adhesin in this coordinated effort can often be enough to make a pathogenic bacterium
298:
different places, and in response to different environmental triggers. Finally, many adhesins present as different immunologically distinct antigenic varieties, even within the same
199:. FimH is folded into two domains. The N terminal adhesive domain plays the main role in surface recognition while the C-terminal domain is responsible for organelle integration. A
271:
during the first three years of life. If the child survives this initial period of susceptibility, infection rates typically drop substantially. Field studies show that this
1345:
Huebinger, Ryan M.; Stones, Daniel H.; de Souza Santos, Marcela; Carlson, Deborah L.; Song, Juquan; Vaz, Diana
Pereira; Keen, Emma; Wolf, Steven E.; Orth, Kim (2016-12-20).
449:
633:
overcomes the antibody based immune response by natural conversion from the high to the low affinity state. Through this conversion, FimH adhesion may shed the
75:
Bacteria are typically found attached to and living in close association with surfaces. During the bacterial lifespan, a bacterium is subjected to frequent
128:
surface layer serves as the specific adhesin. To effectively achieve adherence to host surfaces, many bacteria produce multiple adherence factors called
1182:"Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coli isolates and identification of new subtypes"
515:. The N-terminal portion of the mature protein is thought to be responsible for chloramphenicol sensitivity. Also, they induce activation of several
879:
Choudhury D, Thompson A, Stojanoff V, et al. (August 1999). "X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic
1280:"Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens"
294:
consumption. This research has opened the door to further exploration of orally administered vaccines which exploit bacterial adhesins.
1167:
1231:"Molecular structure of the Dr adhesin: nucleotide sequence and mapping of receptor-binding domain by use of fusion constructs"
585:
virulence factors." These studies have shown that only strains capable of expressing fimbriae are pathogenic. High survival of
1576:
Adhesins are also used in cell communication, and bind to surface communicators. Can also be used to bind to other bacteria.
469:
263:
stems from observations of human acquired immunity. Children in third world countries may suffer from several episodes of
204:
C-terminal domain specificity resulting in a bacterial adhesion with dual bending sites and related binding phenotypes.
34:
or adherence to other cells or to surfaces, usually in the host they are infecting or living in. Adhesins are a type of
1070:
988:
544:
Multivalent
Adhesion Molecules (MAMs) are a widespread family of adhesins found in Gram negative bacteria, including
241:
candidates because they are often essential to infection and are surface-located, making them readily accessible to
1347:"Targeting bacterial adherence inhibits multidrug-resistant Pseudomonas aeruginosa infection following burn injury"
656:
279:
224:. This has led to the exploration of adhesin activity interruption as a method of bacterial infection treatment.
504:
253:
601:
phagocytic activity. This action facilitates the spread of the pathogen throughout the epithelial cell layer.
579:
is host restricted almost entirely to humans. "Extensive studies have established type 4 fimbrial adhesins of
457:
311:
Despite these challenges, progress is being made in the creation of anti-adhesion vaccines. In animal models,
183:
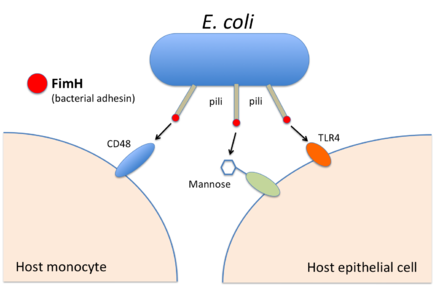
The best characterized bacterial adhesin is the type 1 fimbrial FimH adhesin. This adhesin is responsible for
835:
Kline, Kimberly A.; Fälker, Stefan; Dahlberg, Sofia; Normark, Staffan; Henriques-Normark, Birgitta (2009).
1006:"Oral consumption of cranberry juice cocktail inhibits molecular-scale adhesion of clinical uropathogenic
328:(UTIs). The use of synthetic FimH adhesion peptides was shown to prevent urogenital mucosal infection by
1610:
453:
56:
496:
489:
1170:, State Key Laboratory for Moleclular Virology and Genetic Engineering, Beijing. Retrieved July 2011
1589:
151:
in addition to a bacterium's ability to produce toxins and resist the immune defenses of the host.
17:
626:
618:
325:
287:
598:
272:
164:
124:
at the tip of the fimbriae that is the actual adhesin. In gram-positive bacteria, a protein or
113:
979:
Levine, M. M.; Giron, J. A.; Noriega, E. R. (1994). "Fimbrial vaccines". In Klemm, Per (ed.).
1605:
1585:
581:
575:
304:
196:
172:
64:
675:
Coutte L, Alonso S, Reveneau N, Willery E, Quatannens B, Locht C, Jacob-Dubuisson F (2003).
86:
1423:
1358:
1291:
935:
436:
312:
299:
1122:
Langermann S, Palaszynski S, Barnhart M, et al. (April 1997). "Prevention of mucosal
212:
The majority of bacterial pathogens exploit specific adhesion to host cells as their main
8:
516:
200:
1529:"Type 1 fimbrial adhesin FimH elicits an immune response that enhances cell adhesion of
1427:
1362:
1295:
939:
1557:
1528:
1446:
1411:
1387:
1346:
1322:
1279:
1034:
1005:
754:
729:
701:
676:
1499:
1470:
1255:
1230:
1206:
1181:
956:
919:
813:
503:
contain both fimbriated and afimbriated adherence structures and mediate adherence of
1562:
1504:
1451:
1392:
1374:
1327:
1309:
1260:
1211:
1143:
1104:
1066:
1039:
984:
961:
900:
858:
817:
759:
706:
485:
444:
1197:
947:
385:
187:
sensitive adhesion. The bacterium synthesizes a precursor protein consisting of 300
1552:
1544:
1494:
1486:
1441:
1431:
1382:
1366:
1317:
1299:
1250:
1242:
1201:
1193:
1135:
1096:
1029:
1021:
951:
943:
892:
848:
809:
749:
741:
696:
688:
612:
432:
330:
213:
148:
92:
35:
1246:
896:
398:
1490:
1436:
1139:
800:
Klemm P, Schembri MA (March 2000). "Bacterial adhesins: function and structure".
512:
410:
291:
283:
1284:
Proceedings of the
National Academy of Sciences of the United States of America
1180:
Zhang L, Foxman B, Tallman P, Cladera E, Le
Bouguenec C, Marrs CF (June 1997).
853:
836:
651:
531:
125:
80:
60:
1599:
1378:
1313:
117:
1304:
730:"Targeting the bacteria-host interface: strategies in anti-adhesion therapy"
1566:
1455:
1396:
1331:
1108:
1043:
904:
862:
821:
763:
710:
76:
48:
1508:
1264:
1215:
1147:
1025:
965:
677:"Role of adhesin release for mucosal colonization by a bacterial pathogen"
593:
infections. Additionally, recent studies out of
Stockholm have shown that
1548:
1057:
Davies, J. K.; Koomey, J. M.; Seifert, H. S. (1994). "Pili (fimbriae) of
692:
586:
520:
290:
researchers have shown that adhesion forces decrease with time following
144:
63:. Adhesion and bacterial adhesins are also a potential target either for
1056:
978:
745:
188:
140:
120:
bacteria function as adhesins, but in many cases it is a minor subunit
1370:
30:
are cell-surface components or appendages of bacteria that facilitate
1344:
597:
can hitchhike on PMNs using their adhesin pili thus hiding them from
221:
217:
52:
96:
to bind to host cells and their receptors (here: the human proteins
1412:"Pathogenic Neisseria hitchhike on the uropod of human neutrophils"
1100:
725:
634:
527:
405:
268:
249:
242:
45:
31:
1527:
Tchesnokova V, Aprikian P, Kisiela D, et al. (October 2011).
1085:
500:
492:
316:
238:
233:
184:
136:
121:
105:
1471:"Host-specific fimbrial adhesins of noninvasive enterotoxigenic
464:
356:
1003:
1121:
168:
1409:
1278:
Krachler, Anne Marie; Ham, Hyeilin; Orth, Kim (2011-07-12).
1229:
Swanson TN, Bilge SS, Nowicki B, Moseley SL (January 1991).
1063:
Fimbriae : adhesion, genetics, biogenesis, and vaccines
981:
Fimbriae : adhesion, genetics, biogenesis, and vaccines
674:
526:
The Dr family of adhesins are particularly associated with
426:
392:
380:
101:
97:
1526:
1228:
878:
834:
319:
significantly reduced colonization by UPEC. Moreover, the
163:
The typical structure of a bacterial adhesin is that of a
1179:
90:
FimH is a bacterial adhesin that helps bacteria such as
1004:
Tao Y, Pinzón-Arango PA, Howell AB, Camesano TA (2011).
1126:
infection by FimH-adhesin-based systemic vaccination".
1168:
Identified
Virulence Factors of UPEC : Adherence
232:
The study of adhesins as a point of exploitation for
252:
is illustrated by studies with FimH, the adhesin of
1410:Söderholm N, Vielfort K, Hultenby K, Aro H (2011).
315:with anti FimH-antibodies and vaccination with the
275:is directed primarily against bacterial adhesins.
837:"Bacterial Adhesins in Host-Microbe Interactions"
1597:
795:
793:
195:In 1999, the structure of FimH was resolved via
1468:
1277:
1079:
791:
789:
787:
785:
783:
781:
779:
777:
775:
773:
286:may inhibit the action of UPEC adhesins. Using
1522:
1520:
1518:
147:. This prevalence marks them as key microbial
135:Bacterial adhesins provide species and tissue
67:or for the treatment of bacterial infections.
917:
799:
227:
770:
723:
1515:
1222:
1173:
1065:. Boca Raton: CRC Press. pp. 147–155.
983:. Boca Raton: CRC Press. pp. 255–270.
874:
872:
1115:
668:
355:
83:tissues in mammals, or even tooth enamel.
1588:at the U.S. National Library of Medicine
1556:
1498:
1445:
1435:
1403:
1386:
1321:
1303:
1254:
1205:
1033:
955:
852:
753:
700:
178:
869:
85:
1462:
997:
911:
657:Trimeric autotransporter adhesins (TAA)
629:, 95% express type 1 fimbriae. FimH in
207:
14:
1598:
1163:
1161:
1159:
1157:
1050:
972:
1469:Gaastra W, de Graaf FK (June 1982).
920:"Heterobinary adhesins based on the
337:
1154:
24:
519:cascades, including activation of
361:drae adhesin from escherichia coli
248:The effectiveness of anti-adhesin
25:
1622:
1579:
1087:uropathogenic Escherichia coli".
918:Schembri MA, Klemm P (May 1998).
567:
139:. Adhesins are expressed by both
1338:
1271:
1198:10.1128/iai.65.6.2011-2018.1997
948:10.1128/AEM.64.5.1628-1633.1998
280:Worcester Polytechnic Institute
828:
717:
539:Multivalent Adhesion Molecules
13:
1:
1247:10.1128/iai.59.1.261-268.1991
897:10.1126/science.285.5430.1061
814:10.1016/S1438-4221(00)80102-2
662:
587:polymorphonuclear neutrophils
421:Available protein structures:
282:show that the consumption of
154:
70:
1491:10.1128/mr.46.2.129-161.1982
1437:10.1371/journal.pone.0024353
1140:10.1126/science.276.5312.607
342:
7:
645:
10:
1627:
854:10.1016/j.chom.2009.05.011
604:
228:Vaccines based on adhesins
530:and pregnancy-associated
497:decay-accelerating factor
463:
443:
425:
420:
416:
404:
391:
379:
371:
366:
354:
349:
1590:Medical Subject Headings
1061:". In Klemm, Per (ed.).
928:Appl. Environ. Microbiol
619:urinary tract infections
44:is an essential step in
1305:10.1073/pnas.1102360108
841:Cell Host & Microbe
326:urinary tract infection
288:atomic force microscopy
1235:Infection and Immunity
1186:Infection and Immunity
924:FimH fimbrial protein"
802:Int. J. Med. Microbiol
562:Pseudomonas aeruginosa
558:Pseudomonas aeruginosa
179:FimH adhesin—structure
109:
1059:Neisseria gonorrhoeae
1026:10.1089/jmf.2010.0154
591:Neisseria gonorrhoeae
589:(PMNs) characterizes
483:Dr family of adhesins
305:Neisseria gonorrhoeae
197:x-ray crystallography
89:
1549:10.1128/IAI.05169-11
693:10.1084/jem.20021153
321:Bordetella pertussis
313:passive immunization
278:Recent studies from
208:As virulence factors
145:saprophytic bacteria
1428:2011PLoSO...624353S
1363:2016NatSR...639341H
1296:2011PNAS..10811614K
1290:(28): 11614–11619.
940:1998ApEnM..64.1628S
517:signal transduction
302:(as is the case in
141:pathogenic bacteria
1351:Scientific Reports
746:10.4161/viru.24606
259:(UPEC). Work with
201:tetra-peptide loop
110:
28:Bacterial adhesins
1611:Virulence factors
1586:Bacterial+Adhesin
1371:10.1038/srep39341
479:
478:
475:
474:
470:structure summary
338:Specific examples
273:acquired immunity
149:virulence factors
16:(Redirected from
1618:
1571:
1570:
1560:
1543:(10): 3895–904.
1531:Escherichia coli
1524:
1513:
1512:
1502:
1473:Escherichia coli
1466:
1460:
1459:
1449:
1439:
1407:
1401:
1400:
1390:
1342:
1336:
1335:
1325:
1307:
1275:
1269:
1268:
1258:
1226:
1220:
1219:
1209:
1177:
1171:
1165:
1152:
1151:
1134:(5312): 607–11.
1124:Escherichia coli
1119:
1113:
1112:
1083:
1077:
1076:
1054:
1048:
1047:
1037:
1008:Escherichia coli
1001:
995:
994:
976:
970:
969:
959:
922:Escherichia coli
915:
909:
908:
891:(5430): 1061–6.
881:Escherichia coli
876:
867:
866:
856:
832:
826:
825:
797:
768:
767:
757:
721:
715:
714:
704:
672:
639:Escherichia coli
613:Escherichia coli
507:Escherichia coli
418:
417:
359:
347:
346:
256:Escherichia coli
214:virulence factor
93:Escherichia coli
36:virulence factor
21:
1626:
1625:
1621:
1620:
1619:
1617:
1616:
1615:
1596:
1595:
1582:
1574:
1525:
1516:
1467:
1463:
1408:
1404:
1343:
1339:
1276:
1272:
1227:
1223:
1178:
1174:
1166:
1155:
1120:
1116:
1084:
1080:
1073:
1055:
1051:
1020:(7–8): 739–45.
1002:
998:
991:
977:
973:
916:
912:
877:
870:
833:
829:
798:
771:
722:
718:
673:
669:
665:
648:
609:
572:
540:
513:chloramphenicol
362:
345:
340:
292:cranberry juice
284:cranberry juice
230:
210:
181:
157:
73:
55:, required for
23:
22:
15:
12:
11:
5:
1624:
1614:
1613:
1608:
1594:
1593:
1581:
1580:External links
1578:
1573:
1572:
1514:
1479:Microbiol. Rev
1461:
1402:
1337:
1270:
1221:
1172:
1153:
1114:
1101:10.1086/315258
1078:
1072:978-0849348945
1071:
1049:
996:
990:978-0849348945
989:
971:
934:(5): 1628–33.
910:
868:
847:(6): 580–592.
827:
769:
724:Krachler, AM;
716:
666:
664:
661:
660:
659:
654:
652:Fungal adhesin
647:
644:
608:
603:
582:N. gonorrhoeae
576:N. gonorrhoeae
571:
569:N. gonorroheae
566:
538:
532:pyelonephritis
505:uropathogenic
477:
476:
473:
472:
467:
461:
460:
447:
441:
440:
430:
423:
422:
414:
413:
408:
402:
401:
396:
389:
388:
383:
377:
376:
373:
369:
368:
364:
363:
360:
352:
351:
344:
341:
339:
336:
254:uropathogenic
229:
226:
209:
206:
180:
177:
156:
153:
126:polysaccharide
72:
69:
9:
6:
4:
3:
2:
1623:
1612:
1609:
1607:
1604:
1603:
1601:
1591:
1587:
1584:
1583:
1577:
1568:
1564:
1559:
1554:
1550:
1546:
1542:
1538:
1537:Infect. Immun
1534:
1532:
1523:
1521:
1519:
1510:
1506:
1501:
1496:
1492:
1488:
1485:(2): 129–61.
1484:
1480:
1476:
1474:
1465:
1457:
1453:
1448:
1443:
1438:
1433:
1429:
1425:
1422:(9): e24353.
1421:
1417:
1413:
1406:
1398:
1394:
1389:
1384:
1380:
1376:
1372:
1368:
1364:
1360:
1356:
1352:
1348:
1341:
1333:
1329:
1324:
1319:
1315:
1311:
1306:
1301:
1297:
1293:
1289:
1285:
1281:
1274:
1266:
1262:
1257:
1252:
1248:
1244:
1240:
1236:
1232:
1225:
1217:
1213:
1208:
1203:
1199:
1195:
1192:(6): 2011–8.
1191:
1187:
1183:
1176:
1169:
1164:
1162:
1160:
1158:
1149:
1145:
1141:
1137:
1133:
1129:
1125:
1118:
1110:
1106:
1102:
1098:
1094:
1090:
1082:
1074:
1068:
1064:
1060:
1053:
1045:
1041:
1036:
1031:
1027:
1023:
1019:
1015:
1011:
1009:
1000:
992:
986:
982:
975:
967:
963:
958:
953:
949:
945:
941:
937:
933:
929:
925:
923:
914:
906:
902:
898:
894:
890:
886:
882:
875:
873:
864:
860:
855:
850:
846:
842:
838:
831:
823:
819:
815:
811:
807:
803:
796:
794:
792:
790:
788:
786:
784:
782:
780:
778:
776:
774:
765:
761:
756:
751:
747:
743:
740:(4): 284–94.
739:
735:
731:
727:
720:
712:
708:
703:
698:
694:
690:
687:(6): 735–42.
686:
682:
678:
671:
667:
658:
655:
653:
650:
649:
643:
640:
637:bound to it.
636:
632:
628:
624:
620:
615:
614:
607:
602:
600:
596:
592:
588:
584:
583:
578:
577:
570:
565:
563:
559:
555:
551:
547:
542:
541:
535:
533:
529:
524:
522:
518:
514:
509:
508:
502:
499:(DAF). These
498:
495:component of
494:
491:
487:
484:
471:
468:
466:
462:
459:
455:
451:
448:
446:
442:
438:
434:
431:
428:
424:
419:
415:
412:
409:
407:
403:
400:
397:
394:
390:
387:
384:
382:
378:
374:
370:
365:
358:
353:
348:
335:
333:
332:
327:
322:
318:
314:
309:
307:
306:
301:
295:
293:
289:
285:
281:
276:
274:
270:
266:
262:
258:
257:
251:
246:
244:
240:
235:
225:
223:
219:
215:
205:
202:
198:
193:
190:
186:
176:
174:
170:
166:
161:
152:
150:
146:
142:
138:
133:
131:
127:
123:
119:
118:gram-negative
115:
107:
103:
99:
95:
94:
88:
84:
82:
81:lacrimal duct
78:
68:
66:
62:
58:
54:
50:
47:
43:
39:
37:
33:
29:
19:
1606:Bacteriology
1575:
1540:
1536:
1530:
1482:
1478:
1472:
1464:
1419:
1415:
1405:
1354:
1350:
1340:
1287:
1283:
1273:
1241:(1): 261–8.
1238:
1234:
1224:
1189:
1185:
1175:
1131:
1127:
1123:
1117:
1095:(2): 774–8.
1092:
1089:J Infect Dis
1088:
1081:
1062:
1058:
1052:
1017:
1013:
1007:
999:
980:
974:
931:
927:
921:
913:
888:
884:
880:
844:
840:
830:
808:(1): 27–35.
805:
801:
737:
733:
719:
684:
680:
670:
638:
630:
625:which cause
622:
611:
610:
605:
594:
590:
580:
574:
573:
568:
561:
557:
553:
549:
545:
543:
537:
536:
525:
506:
482:
480:
329:
320:
310:
303:
296:
277:
264:
260:
255:
247:
231:
222:non-virulent
211:
194:
182:
162:
158:
134:
129:
111:
91:
77:shear-forces
74:
49:pathogenesis
41:
40:
27:
26:
621:. Of those
521:PI-3 kinase
490:blood group
367:Identifiers
267:associated
189:amino acids
65:prophylaxis
1600:Categories
1014:J Med Food
663:References
635:antibodies
599:neutrophil
488:to the Dr
433:structures
375:Adhesin_Dr
350:Adhesin_Dr
250:antibodies
243:antibodies
155:Structures
108:residues).
71:Background
57:colonizing
1379:2045-2322
1357:: 39341.
1314:1091-6490
734:Virulence
681:J Exp Med
595:Neisseria
564:in rats.
411:IPR006713
343:Dr family
334:in mice.
218:infection
185:D-mannose
53:infection
46:bacterial
42:Adherence
1567:21768279
1475:strains"
1456:21949708
1416:PLOS ONE
1397:27996032
1332:21709226
1109:10669375
1044:21480803
905:10446051
863:19527885
822:11043979
764:23799663
728:(2014).
711:12629063
646:See also
554:Yersinia
528:cystitis
501:proteins
450:RCSB PDB
406:InterPro
269:diarrhea
234:vaccines
130:adhesins
32:adhesion
18:Adhesins
1558:3187269
1509:6126799
1447:3174955
1424:Bibcode
1388:5171828
1359:Bibcode
1323:3136308
1292:Bibcode
1265:1670929
1216:9169726
1148:9110982
1128:Science
1035:3133681
966:9572927
936:Bibcode
885:Science
755:3710331
726:Orth, K
702:2193847
631:E. coli
623:E. coli
606:E. coli
546:E. coli
493:antigen
386:PF04619
331:E. coli
317:protein
265:E. coli
261:E. coli
239:vaccine
165:fimbria
137:tropism
122:protein
114:fimbria
106:mannose
1592:(MeSH)
1565:
1555:
1507:
1500:281536
1497:
1454:
1444:
1395:
1385:
1377:
1330:
1320:
1312:
1263:
1256:257736
1253:
1214:
1207:175278
1204:
1146:
1107:
1069:
1042:
1032:
987:
964:
957:106206
954:
903:
861:
820:
762:
752:
709:
699:
556:, and
550:Vibrio
465:PDBsum
439:
429:
399:CL0204
372:Symbol
59:a new
300:clone
169:pilus
112:Most
104:, or
1563:PMID
1505:PMID
1452:PMID
1393:PMID
1375:ISSN
1328:PMID
1310:ISSN
1261:PMID
1212:PMID
1144:PMID
1105:PMID
1067:ISBN
1040:PMID
985:ISBN
962:PMID
901:PMID
859:PMID
818:PMID
760:PMID
707:PMID
627:UTIs
486:bind
481:The
458:PDBj
454:PDBe
437:ECOD
427:Pfam
395:clan
393:Pfam
381:Pfam
143:and
102:TLR4
100:and
98:CD48
61:host
1553:PMC
1545:doi
1495:PMC
1487:doi
1442:PMC
1432:doi
1383:PMC
1367:doi
1318:PMC
1300:doi
1288:108
1251:PMC
1243:doi
1202:PMC
1194:doi
1136:doi
1132:276
1097:doi
1093:181
1030:PMC
1022:doi
952:PMC
944:doi
893:doi
889:285
883:".
849:doi
810:doi
806:290
750:PMC
742:doi
697:PMC
689:doi
685:197
445:PDB
308:).
173:ECM
167:or
116:of
51:or
1602::
1561:.
1551:.
1541:79
1539:.
1535:.
1517:^
1503:.
1493:.
1483:46
1481:.
1477:.
1450:.
1440:.
1430:.
1418:.
1414:.
1391:.
1381:.
1373:.
1365:.
1353:.
1349:.
1326:.
1316:.
1308:.
1298:.
1286:.
1282:.
1259:.
1249:.
1239:59
1237:.
1233:.
1210:.
1200:.
1190:65
1188:.
1184:.
1156:^
1142:.
1130:.
1103:.
1091:.
1038:.
1028:.
1018:14
1016:.
1012:.
960:.
950:.
942:.
932:64
930:.
926:.
899:.
887:.
871:^
857:.
843:.
839:.
816:.
804:.
772:^
758:.
748:.
736:.
732:.
705:.
695:.
683:.
679:.
552:,
548:,
534:.
523:.
456:;
452:;
435:/
245:.
175:.
132:.
38:.
1569:.
1547::
1533:"
1511:.
1489::
1458:.
1434::
1426::
1420:6
1399:.
1369::
1361::
1355:6
1334:.
1302::
1294::
1267:.
1245::
1218:.
1196::
1150:.
1138::
1111:.
1099::
1075:.
1046:.
1024::
1010:"
993:.
968:.
946::
938::
907:.
895::
865:.
851::
845:5
824:.
812::
766:.
744::
738:4
713:.
691::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.