362:
522:
354:
108:
421:
is the second most common haplogroup in the
Khoisan groups closer to the Sanid side with (following L0d) being more Khoid but is largely restricted to the Khoisan as a whole. Although the Khoisan associated L0d haplogroup were found in high frequencies in sections of the Coloured population of South
1172:
Age estimates (ka, 95% CI in angular brackets): ML whole-mtDNA age estimate: 128.2 , ρ whole-mtDNA age estimate: 121.3 , ρ synonymous age estimate (ka): 131.0 : Rito T, Richards MB, Fernandes V, Alshamali F, Cerny V, Pereira L, Soares P., "The first modern human dispersals across Africa",
1778:
Based on the previous knowledge of
African complete sequences paraphyletic clade L1 is split into two monophyletic units L0, capturing previously defined L1a and L1d lineages, and L1 clade that includes L1b and L1c
111:
The region in Africa where
Tishkoff found the greatest level of mitochondrial diversity (green) and the region Behar et al. postulated the most ancient division in the human population began to occur (light
458:
group showing increased frequency of about 11%. Haplogroup L0a has a
Paleolithic time depth of about 33,000 years and likely reached Guinea between 10,000 and 4,000 years ago. It also is often seen in the
120:(MRCA) for the shared human maternal lineage. The haplogroup consists of five main branches (L0a, L0b, L0d, L0f, L0k). Four of them were originally classified into L1 subclades, L1a, L1d, L1f and L1k.
415:
population of South Africa and frequencies range from 60% to 71%. This illustrates the massive maternal contribution of
Khoisan people to sections of the Coloured population of South Africa.
1641:
Rídl, Jakub; Edens, Christopher M.; Černý, Viktor (2009). "Mitochondrial DNA Structure of Yemeni
Population: Regional Differences and the Implications for Different Migratory Contributions".
1734:
Rosa, Alexandra; Brehm, Antonio; Kivisild, Toomas; Metspalu, Ene; Villems, Richard (2004). "MtDNA Profile of West Africa
Guineans: Towards a Better Understanding of the Senegambia Region".
1336:
Rosa, Alexandra; Brehm, Antonio; Kivisild, Toomas; Metspalu, Ene; Villems, Richard (2004). "MtDNA Profile of West Africa
Guineans: Towards a Better Understanding of the Senegambia Region".
1293:
1592:
Quintana-Murci, Lluis; Harmant, Christine; Quach, Hélène; Balanovsky, Oleg; Zaporozhchenko, Valery; Bormans, Connie; Van Helden, Paul D.; Hoal, Eileen G.; Behar, Doron M. (2010).
1675:
Mitochondrial DNA subhaplogroups L0a2 and L2a modify susceptibility to peripheral neuropathy in malawian adults on stavudine containing highly active antiretroviral therapy.
1869:
Knight, Alec; Underhill, Peter A.; Mortensen, Holly M.; Zhivotovsky, Lev A.; Lin, Alice A.; Henn, Brenna M.; Louis, Dorothy; Ruhlen, Merritt; Mountain, Joanna L. (2003).
1490:
Knight, Alec; Underhill, Peter A.; Mortensen, Holly M.; Zhivotovsky, Lev A.; Lin, Alice A.; Henn, Brenna M.; Louis, Dorothy; Ruhlen, Merritt; Mountain, Joanna L. (2003).
127:
found that the specimen belonged to the L0d2c1c mtDNA subclade. This maternal haplogroup is today most closely associated with the Ju, a subgroup of the indigenous
411:
closer to the Khoid side with (following L0k) being more Sanid but is largely restricted to the
Khoisan as a whole. L0d is also commonly found in sections of the
1686:
1546:
Schlebusch, Carina M.; Naidoo, Thijessen; Soodyall, Himla (2009). "SNaPshot minisequencing to resolve mitochondrial macro-haplogroups found in Africa".
1123:
Soares, Pedro; Ermini, Luca; Thomson, Noel; Mormina, Maru; Rito, Teresa; Röhl, Arne; Salas, Antonio; Oppenheimer, Stephen; MacAulay, Vincent (2009).
1863:
haplogroup L0 mtDNAs, the haplogroup that we had previously concluded lies at the base of the human mtDNA. tree based on phylogenetic analysis…
1817:
The first axis of mtDNA, on the other hand, places the ancestral haplogroup L0 at the extreme left, since it gave rise to one sole lineage…
1825:"Mitochondrial DNA-like sequences in the nucleus (NUMTs): Insights into our African origins and the mechanism of foreign DNA integration"
1308:
1394:
Tishkoff, S. A.; Gonder, M. K.; Henn, B. M.; Mortensen, H.; Knight, A.; Gignoux, C.; Fernandopulle, N.; Lema, G.; Nyambo, T. B. (2007).
1095:
Heinz, Tanja; et al. (2017). "Updating the
African human mitochondrial DNA tree: Relevance to forensic and population genetics".
1292:
Frank J. Rühli; Maryna Steyn; Morongwa N. Mosothwane; Lena Öhrström; Molebogeng K. Bodiba; Abigail Bouwman (January–February 2016).
361:
1658:
17:
521:
1438:
Chen, Yu-Sheng; Olckers, Antonel; Schurr, Theodore G.; Kogelnik, Andreas M.; Huoponen, Kirsi; Wallace, Douglas C. (2000).
799:
811:
96:
1923:
1594:"Strong Maternal Khoisan Contribution to the South African Coloured Population: A Case of Gender-Biased Admixture"
1440:"MtDNA Variation in the South African Kung and Khwe—and Their Genetic Relationships to Other African Populations"
537:
This phylogenetic tree of haplogroup L0 subclades is based on the paper by Mannis van Oven and Manfred Kayser
1178:
789:
1396:"History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation"
827:
63:
135:
desiccated mummy from the Tuli region in northern Botswana was also found to belong to haplogroup L0.
1705:
1787:"Utility of JC polyomavirus in tracing the pattern of human migrations dating to prehistoric times"
288:
1871:"African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages"
1492:"African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages"
1291:
1041:
1036:
1003:
910:
888:
883:
878:
867:
859:
854:
779:
505:
422:
Africa, L0k were not observed in two studies involving large groups of Coloured individuals.
353:
303:
1677:, J Acquir Immune Defic Syndr. 2013 Aug 15; 63(5):647-52. doi: 10.1097/QAI.0b013e3182968ea5
1068:
1063:
1058:
1053:
1046:
1029:
1021:
998:
993:
983:
976:
971:
966:
961:
953:
948:
943:
938:
930:
925:
920:
915:
900:
895:
1294:"Radiological and genetic analysis of a Late Iron Age mummy from the Tuli Block, Botswana"
8:
794:
53:
1900:
1854:
1769:
1728:
1618:
1593:
1571:
1521:
1464:
1439:
1371:
1269:
1244:
1225:
1149:
1124:
370:
1887:
1870:
1508:
1491:
1243:
Alan G. Morris; Anja Heinze; Eva K.F. Chan; Andrew B. Smith; Vanessa M. Hayes (2014).
1892:
1846:
1808:
1761:
1747:
1654:
1623:
1563:
1513:
1469:
1417:
1363:
1349:
1274:
1242:
1217:
1196:"Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation"
1154:
1125:"Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock"
1112:
1012:
823:
784:
142:
117:
67:
1904:
1858:
1773:
1575:
1525:
1375:
1229:
1882:
1836:
1798:
1751:
1743:
1646:
1613:
1605:
1555:
1503:
1459:
1451:
1407:
1353:
1345:
1264:
1256:
1207:
1144:
1136:
1104:
539:
Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation
526:
1245:"First Ancient Mitochondrial Human Genome from a Pre-Pastoralist Southern African"
1823:
Mishmar, Dan; Ruiz-Pesini, Eduardo; Brandon, Martin; Wallace, Douglas C. (2004).
1108:
530:
467:
408:
124:
49:
1687:
First Ancient Mitochondrial Human Genome from a Pre-Pastoralist Southern African
1650:
1674:
1609:
1140:
455:
437:
1917:
132:
123:
In 2014, ancient DNA analysis of a 2,330 year old male forager's skeleton in
107:
1412:
1395:
1896:
1850:
1812:
1765:
1627:
1567:
1559:
1517:
1473:
1421:
1367:
1278:
1221:
1158:
1116:
988:
386:
1803:
1786:
1756:
1358:
1260:
433:
1841:
1824:
1212:
1195:
464:
447:
128:
497:
471:
1591:
1868:
1489:
1455:
485:
429:
412:
394:
1722:
131:, which points to population continuity in the region. In 2016, a
1645:. Vertebrate Paleobiology and Paleoanthropology. pp. 69–78.
1090:
378:
374:
1822:
451:
377:
people at 73% on average. Some of the highest frequencies are:
86:
263!, 1048, 3516A, 5442, 6185, 9042, 9347, 10589, 12007, 12720
475:
460:
398:
804:
504:, Haplogroup L0a2 is associated with a higher likelihood of
446:
is most prevalent in South-East African populations (25% in
1711:
1393:
390:
382:
27:
Human mitochondrial DNA grouping indicating common ancestry
1541:
1539:
1537:
1535:
1122:
357:
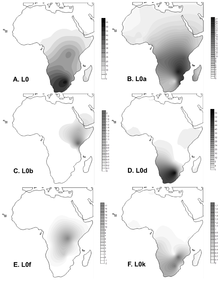
Projected spatial distribution of haplogroup L0 in Africa.
1733:
1437:
1335:
501:
365:
Frequency maps for L0 (total), L0a, L0b, L0d, L0f and L0k
1587:
1585:
1545:
1532:
1673:
Kampira E, Kumwenda J, van Oosterhout JJ, Dandara C.
1582:
1485:
1483:
454:, it has a frequency between 1% and 5%, with the
1915:
1480:
1020:
1017:
863:
491:
470:. L0a is found at a frequency of almost 25% in
1640:
1193:
1089:point estimate 168.5 ka (136.3–201.1 ka
428:is present in relatively small frequencies in
1643:The Evolution of Human Populations in Arabia
1094:
812:human mitochondrial DNA (mtDNA) haplogroups
1433:
1431:
1389:
1387:
1385:
1194:Van Oven, Mannis; Kayser, Manfred (2009).
373:. It reaches its highest frequency in the
1886:
1840:
1802:
1755:
1667:
1617:
1507:
1463:
1411:
1357:
1268:
1211:
1189:
1187:
1148:
1097:Forensic Science International: Genetics
520:
360:
352:
106:
1428:
1382:
153: L0
14:
1916:
1784:
1598:The American Journal of Human Genetics
1444:The American Journal of Human Genetics
1331:
1329:
1184:
1129:The American Journal of Human Genetics
952:
904:
871:
866:
848:
845:
832:
822:
525:Schematic tree of haplogroup L0. MSA:
899:
894:
1045:
1040:
1035:
1025:
1010:
1007:
1002:
997:
992:
987:
982:
975:
970:
965:
960:
957:
947:
942:
937:
934:
929:
924:
919:
914:
909:
887:
882:
877:
874:
858:
853:
840:
835:
819:
1326:
800:Human mitochondrial DNA haplogroups
541:and subsequent published research.
496:In patients who are given the drug
116:L0 is one of two branches from the
24:
546:Most Recent Common Ancestor (MRCA)
25:
1935:
1695:
1179:doi: 10.1371/journal.pone.0080031
407:is found among Khoisan groups of
1748:10.1046/j.1529-8817.2004.00100.x
1350:10.1046/j.1529-8817.2004.00100.x
1301:South African Journal of Science
1680:
1634:
1400:Molecular Biology and Evolution
1121:(table 2). 150 ka suggested in:
348:
1285:
1236:
1166:
1083:
13:
1:
1888:10.1016/S0960-9822(03)00130-1
1509:10.1016/S0960-9822(03)00130-1
1076:
492:Drug and disease interactions
369:L0 is found most commonly in
1249:Genome Biology and Evolution
1109:10.1016/j.fsigen.2016.12.016
790:Human mitochondrial genetics
511:
7:
1791:Journal of General Virology
1651:10.1007/978-90-481-2719-1_5
1177:2013 Nov 13; 8(11):e80031.
773:
118:most recent common ancestor
10:
1940:
1610:10.1016/j.ajhg.2010.02.014
1141:10.1016/j.ajhg.2009.05.001
807:
533:, ka: thousand years ago.
316:
301:
293:
266:
244:
222:
200:
187:
179:
171:
163:
155:
147:
102:
82:
74:
59:
45:
37:
32:
1736:Annals of Human Genetics
1338:Annals of Human Genetics
1067:
1062:
1057:
1052:
1028:
46:Possible place of origin
1924:Human mtDNA haplogroups
516:
97:human mitochondrial DNA
38:Possible time of origin
1706:Mitochondrial DNA Site
1560:10.1002/elps.200900197
534:
366:
358:
113:
1804:10.1099/vir.0.80650-0
1413:10.1093/molbev/msm155
1307:(1/2). Archived from
810:Phylogenetic tree of
780:Genealogical DNA test
524:
506:peripheral neuropathy
364:
356:
110:
18:Haplogroup L0 (mtDNA)
99:(mtDNA) haplogroup.
54:Southern East Africa
1785:Pavesi, A. (2005).
795:Population genetics
1842:10.1002/humu.10304
1710:Mannis van Oven's
1261:10.1093/gbe/evu202
1213:10.1002/humu.20921
535:
508:as a side effect.
371:Sub-Saharan Africa
367:
359:
114:
83:Defining mutations
1660:978-90-481-2718-4
1074:
1073:
1015:
824:Mitochondrial Eve
785:Genetic genealogy
345:
344:
336:
335:
327:
326:
277:
276:
255:
254:
233:
232:
211:
210:
143:MRCA (mtDNA)
90:
89:
68:Mitochondrial Eve
16:(Redirected from
1931:
1908:
1890:
1865:
1844:
1819:
1806:
1781:
1759:
1689:
1684:
1678:
1671:
1665:
1664:
1638:
1632:
1631:
1621:
1589:
1580:
1579:
1543:
1530:
1529:
1511:
1487:
1478:
1477:
1467:
1435:
1426:
1425:
1415:
1391:
1380:
1379:
1361:
1333:
1324:
1323:
1321:
1319:
1313:
1298:
1289:
1283:
1282:
1272:
1240:
1234:
1233:
1215:
1191:
1182:
1170:
1164:
1162:
1152:
1120:
1087:
1011:
805:
527:Middle Stone Age
393:/!Xun) 83%, and
296:
295:
182:
181:
174:
173:
166:
165:
158:
157:
150:
149:
139:
138:
30:
29:
21:
1939:
1938:
1934:
1933:
1932:
1930:
1929:
1928:
1914:
1913:
1875:Current Biology
1698:
1693:
1692:
1685:
1681:
1672:
1668:
1661:
1639:
1635:
1590:
1583:
1554:(21): 3657–64.
1548:Electrophoresis
1544:
1533:
1496:Current Biology
1488:
1481:
1436:
1429:
1406:(10): 2180–95.
1392:
1383:
1334:
1327:
1317:
1315:
1314:on 21 June 2016
1311:
1296:
1290:
1286:
1255:(10): 2647–53.
1241:
1237:
1192:
1185:
1171:
1167:
1093:) according to
1088:
1084:
1079:
776:
531:Later Stone Age
519:
514:
494:
419:Haplogroups L0k
409:Southern Africa
351:
346:
337:
328:
278:
256:
234:
212:
125:Southern Africa
105:
50:Southern Africa
28:
23:
22:
15:
12:
11:
5:
1937:
1927:
1926:
1912:
1911:
1910:
1909:
1866:
1829:Human Mutation
1820:
1797:(5): 1315–26.
1782:
1731:
1725:
1721:YFull MTree's
1718:Haplogroup L0
1716:
1715:
1714:
1708:
1697:
1696:External links
1694:
1691:
1690:
1679:
1666:
1659:
1633:
1581:
1531:
1479:
1456:10.1086/302848
1450:(4): 1362–83.
1427:
1381:
1325:
1284:
1235:
1206:(2): E386–94.
1200:Human Mutation
1183:
1165:
1081:
1080:
1078:
1075:
1072:
1071:
1066:
1061:
1056:
1050:
1049:
1044:
1039:
1033:
1032:
1027:
1024:
1019:
1016:
1009:
1006:
1001:
996:
991:
986:
980:
979:
974:
969:
964:
959:
956:
951:
946:
941:
936:
933:
928:
923:
918:
913:
907:
906:
903:
898:
892:
891:
886:
881:
876:
873:
870:
865:
862:
857:
851:
850:
847:
844:
838:
837:
834:
831:
821:
817:
816:
803:
802:
797:
792:
787:
782:
775:
772:
771:
770:
769:
768:
767:
766:
765:
764:
763:
762:
761:
760:
755:
754:
753:
750:
747:
746:
745:
742:
739:
738:
737:
731:
730:
729:
728:
727:
721:
720:
719:
718:
717:
714:
699:
698:
697:
694:
691:
690:
689:
686:
685:
684:
675:
674:
673:
656:
655:
654:
653:
652:
649:
643:
632:
631:
630:
627:
616:
615:
614:
613:
612:
611:
610:
609:
608:
602:
601:
600:
597:
596:
595:
583:
582:
581:
580:
579:
573:
570:
561:
518:
515:
513:
510:
493:
490:
482:Haplogroup L0b
444:Haplogroup L0a
438:Sandawe people
426:Haplogroup L0f
405:Haplogroup L0d
350:
347:
343:
342:
339:
338:
334:
333:
330:
329:
325:
324:
321:
320:
315:
312:
311:
308:
307:
300:
294:
292:
284:
283:
280:
279:
275:
274:
271:
270:
265:
262:
261:
258:
257:
253:
252:
249:
248:
243:
240:
239:
236:
235:
231:
230:
227:
226:
221:
218:
217:
214:
213:
209:
208:
205:
204:
199:
196:
195:
192:
191:
186:
180:
178:
172:
170:
164:
162:
156:
154:
148:
146:
137:
104:
101:
88:
87:
84:
80:
79:
78:L0a'b'f'k, L0d
76:
72:
71:
61:
57:
56:
47:
43:
42:
39:
35:
34:
26:
9:
6:
4:
3:
2:
1936:
1925:
1922:
1921:
1919:
1906:
1902:
1898:
1894:
1889:
1884:
1881:(6): 464–73.
1880:
1876:
1872:
1867:
1864:
1860:
1856:
1852:
1848:
1843:
1838:
1835:(2): 125–33.
1834:
1830:
1826:
1821:
1818:
1814:
1810:
1805:
1800:
1796:
1792:
1788:
1783:
1780:
1775:
1771:
1767:
1763:
1758:
1757:10400.13/3044
1753:
1749:
1745:
1742:(4): 340–52.
1741:
1737:
1732:
1730:
1729:Haplogroup L0
1726:
1724:
1723:Haplogroup L0
1720:
1719:
1717:
1713:
1709:
1707:
1703:
1702:
1700:
1699:
1688:
1683:
1676:
1670:
1662:
1656:
1652:
1648:
1644:
1637:
1629:
1625:
1620:
1615:
1611:
1607:
1604:(4): 611–20.
1603:
1599:
1595:
1588:
1586:
1577:
1573:
1569:
1565:
1561:
1557:
1553:
1549:
1542:
1540:
1538:
1536:
1527:
1523:
1519:
1515:
1510:
1505:
1502:(6): 464–73.
1501:
1497:
1493:
1486:
1484:
1475:
1471:
1466:
1461:
1457:
1453:
1449:
1445:
1441:
1434:
1432:
1423:
1419:
1414:
1409:
1405:
1401:
1397:
1390:
1388:
1386:
1377:
1373:
1369:
1365:
1360:
1359:10400.13/3044
1355:
1351:
1347:
1344:(4): 340–52.
1343:
1339:
1332:
1330:
1310:
1306:
1302:
1295:
1288:
1280:
1276:
1271:
1266:
1262:
1258:
1254:
1250:
1246:
1239:
1231:
1227:
1223:
1219:
1214:
1209:
1205:
1201:
1197:
1190:
1188:
1180:
1176:
1169:
1160:
1156:
1151:
1146:
1142:
1138:
1135:(6): 740–59.
1134:
1130:
1126:
1118:
1114:
1110:
1106:
1102:
1098:
1092:
1086:
1082:
1070:
1065:
1060:
1055:
1051:
1048:
1043:
1038:
1034:
1031:
1023:
1014:
1005:
1000:
995:
990:
985:
981:
978:
973:
968:
963:
955:
950:
945:
940:
932:
927:
922:
917:
912:
908:
902:
897:
893:
890:
885:
880:
869:
861:
856:
852:
843:
839:
829:
825:
818:
815:
814:
813:
806:
801:
798:
796:
793:
791:
788:
786:
783:
781:
778:
777:
759:
756:
751:
748:
743:
740:
735:
734:
732:
725:
724:
722:
715:
712:
711:
709:
708:
706:
705:
703:
702:
700:
695:
692:
687:
682:
681:
679:
678:
676:
671:
670:
668:
667:
665:
664:
663:
660:
659:
657:
650:
647:
646:
644:
641:
640:
639:
636:
635:
633:
628:
625:
624:
623:
620:
619:
617:
606:
605:
603:
598:
593:
592:
590:
589:
587:
586:
584:
577:
576:
574:
571:
568:
567:
565:
564:
562:
559:
558:
557:
554:
553:
552:
549:
548:
547:
544:
543:
542:
540:
532:
528:
523:
509:
507:
503:
499:
489:
487:
483:
479:
477:
473:
469:
466:
462:
457:
453:
449:
445:
441:
440:of Tanzania.
439:
435:
431:
427:
423:
420:
416:
414:
410:
406:
402:
400:
396:
392:
388:
384:
380:
376:
372:
363:
355:
341:
340:
332:
331:
323:
322:
319:
314:
313:
310:
309:
306:
305:
298:
297:
290:
286:
285:
282:
281:
273:
272:
269:
264:
263:
260:
259:
251:
250:
247:
242:
241:
238:
237:
229:
228:
225:
220:
219:
216:
215:
207:
206:
203:
198:
197:
194:
193:
190:
184:
183:
176:
175:
168:
167:
160:
159:
152:
151:
144:
141:
140:
136:
134:
133:Late Iron Age
130:
126:
121:
119:
109:
100:
98:
94:
93:Haplogroup L0
85:
81:
77:
73:
69:
65:
62:
58:
55:
51:
48:
44:
41:130 to 200 ka
40:
36:
33:Haplogroup L0
31:
19:
1878:
1874:
1862:
1832:
1828:
1816:
1794:
1790:
1777:
1739:
1735:
1704:Ian Logan's
1682:
1669:
1642:
1636:
1601:
1597:
1551:
1547:
1499:
1495:
1447:
1443:
1403:
1399:
1341:
1337:
1316:. Retrieved
1309:the original
1304:
1300:
1287:
1252:
1248:
1238:
1203:
1199:
1174:
1168:
1132:
1128:
1100:
1096:
1085:
841:
809:
808:
757:
661:
637:
621:
555:
550:
545:
538:
536:
495:
484:is found in
481:
480:
443:
442:
425:
424:
418:
417:
404:
403:
387:South Africa
368:
349:Distribution
317:
302:
267:
245:
223:
201:
188:
122:
115:
92:
91:
1103:: 156–159.
434:East Africa
318: L2-6
75:Descendants
1727:MITOMAP's
1077:References
618:L0a'b'f'k
448:Mozambique
436:among the
268: L0d
246: L0k
224: L0f
202: L0b
189: L0a
129:San people
1712:Phylotree
512:Subclades
500:to treat
498:stavudine
472:Hadramawt
450:). Among
1918:Category
1905:52862939
1897:12646128
1859:25109836
1851:14722916
1813:15831942
1774:15391342
1766:15225159
1701:General
1628:20346436
1576:19515426
1568:19810027
1526:52862939
1518:12646128
1474:10739760
1422:17656633
1376:15391342
1368:15225159
1318:26 April
1279:25212860
1230:27566749
1222:18853457
1175:PLoS One
1159:19500773
1117:28086175
774:See also
716:L0a2a1a2
713:L0a2a1a1
710:L0a2a1a
634:L0a'b'f
588:L0d2a'b
486:Ethiopia
452:Guineans
430:Tanzania
413:Coloured
401:) 100%.
395:Botswana
60:Ancestor
1779:clades…
1619:2850426
1465:1288201
1270:4224329
1150:2694979
1026:
1018:
1008:
958:
935:
905:
875:
872:
864:
849:
836:
833:
820:
726:L0a2a2a
723:L0a2a2
707:L0a2a1
683:L0a1b1a
680:L0a1b1
607:L0d2c1c
563:L0d1'2
529:, LSA:
468:Pygmies
456:Balanta
385:) 79%,
379:Namibia
375:Khoisan
299:
291:
185:
177:
169:
161:
145:
1903:
1895:
1857:
1849:
1811:
1772:
1764:
1657:
1626:
1616:
1574:
1566:
1524:
1516:
1472:
1462:
1420:
1374:
1366:
1277:
1267:
1228:
1220:
1157:
1147:
1115:
1091:95% CI
1013:pre-JT
736:L0a2ba
733:L0a2b
704:L0a2a
688:L0a1b2
677:L0a1b
672:L0a1a2
669:L0a1a
658:L0a'b
604:L0d2c
594:L0d2a1
591:L0d2a
578:L0d1c1
575:L0d1c
287:
112:brown)
103:Origin
1901:S2CID
1855:S2CID
1770:S2CID
1572:S2CID
1522:S2CID
1372:S2CID
1312:(PDF)
1297:(PDF)
1226:S2CID
846:L1–6
744:L0a2d
741:L0a2c
701:L0a2
696:L0a1d
693:L0a1c
666:L0a1
651:L0f2b
648:L0f2a
645:L0f2
599:L0d2b
585:L0d2
572:L0d1b
569:L0d1a
566:L0d1
476:Yemen
465:Biaka
461:Mbuti
399:!Kung
95:is a
1893:PMID
1847:PMID
1809:PMID
1762:PMID
1655:ISBN
1624:PMID
1564:PMID
1514:PMID
1470:PMID
1418:PMID
1364:PMID
1320:2016
1275:PMID
1218:PMID
1155:PMID
1113:PMID
752:L0a4
749:L0a3
642:L0f1
629:L0k2
626:L0k1
560:L0d3
517:Tree
463:and
391:Khwe
383:!Xun
289:L1-6
1883:doi
1837:doi
1799:doi
1752:hdl
1744:doi
1647:doi
1614:PMC
1606:doi
1556:doi
1504:doi
1460:PMC
1452:doi
1408:doi
1354:hdl
1346:doi
1305:112
1265:PMC
1257:doi
1208:doi
1145:PMC
1137:doi
1105:doi
758:L0b
662:L0a
638:L0f
622:L0k
556:L0d
502:HIV
478:).
52:or
1920::
1899:.
1891:.
1879:13
1877:.
1873:.
1861:.
1853:.
1845:.
1833:23
1831:.
1827:.
1815:.
1807:.
1795:86
1793:.
1789:.
1776:.
1768:.
1760:.
1750:.
1740:68
1738:.
1653:.
1622:.
1612:.
1602:86
1600:.
1596:.
1584:^
1570:.
1562:.
1552:30
1550:.
1534:^
1520:.
1512:.
1500:13
1498:.
1494:.
1482:^
1468:.
1458:.
1448:66
1446:.
1442:.
1430:^
1416:.
1404:24
1402:.
1398:.
1384:^
1370:.
1362:.
1352:.
1342:68
1340:.
1328:^
1303:.
1299:.
1273:.
1263:.
1251:.
1247:.
1224:.
1216:.
1204:30
1202:.
1198:.
1186:^
1153:.
1143:.
1133:84
1131:.
1127:.
1111:.
1101:27
1099:.
1042:JT
1037:HV
1004:R0
911:CZ
889:L6
884:L5
879:L4
868:L3
860:L2
855:L1
842:L0
830:)
551:L0
488:.
432:,
304:L1
1907:.
1885::
1839::
1801::
1754::
1746::
1663:.
1649::
1630:.
1608::
1578:.
1558::
1528:.
1506::
1476:.
1454::
1424:.
1410::
1378:.
1356::
1348::
1322:.
1281:.
1259::
1253:6
1232:.
1210::
1181:.
1163:.
1161:.
1139::
1119:.
1107::
1069:T
1064:J
1059:V
1054:H
1047:K
1030:U
1022:P
999:F
994:B
989:Z
984:C
977:Y
972:X
967:W
962:I
954:R
949:S
944:A
939:O
931:Q
926:G
921:E
916:D
901:N
896:M
828:L
826:(
474:(
397:(
389:(
381:(
70:)
66:(
64:L
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.