372:
describe the hydrophobic features of the protein using turns in the main-chain atoms. Yet another approach is to use a
Fourier shape descriptor technique. Whereas the shape complementarity based approaches are typically fast and robust, they cannot usually model the movements or dynamic changes in the ligand/protein conformations accurately, although recent developments allow these methods to investigate ligand flexibility. Shape complementarity methods can quickly scan through several thousand ligands in a matter of seconds and actually figure out whether they can bind at the protein's active site, and are usually scalable to even protein-protein interactions. They are also much more amenable to
193:
309:(where on the surface of the lock is the key hole, which direction to turn the key after it is inserted, etc.). Here, the protein can be thought of as the “lock” and the ligand can be thought of as a “key”. Molecular docking may be defined as an optimization problem, which would describe the “best-fit” orientation of a ligand that binds to a particular protein of interest. However, since both the ligand and the protein are flexible, a
402:
201:
753:
385:
body transformations such as translations and rotations, as well as internal changes to the ligand's structure including torsion angle rotations. Each of these moves in the conformation space of the ligand induces a total energetic cost of the system. Hence, the system's total energy is calculated after every move.
557:
Docking programs generate a large number of potential ligand poses, of which some can be immediately rejected due to clashes with the protein. The remainder are evaluated using some scoring function, which takes a pose as input and returns a number indicating the likelihood that the pose represents a
773:
for complexes between proteins and high affinity ligands, but comparatively fewer for low affinity ligands as the latter complexes tend to be less stable and therefore more difficult to crystallize. Scoring functions trained with this data can dock high affinity ligands correctly, but they will also
384:
Simulating the docking process is much more complicated. In this approach, the protein and the ligand are separated by some physical distance, and the ligand finds its position into the protein's active site after a certain number of “moves” in its conformational space. The moves incorporate rigid
757:
The components consist of solvent effects, conformational changes in the protein and ligand, free energy due to protein-ligand interactions, internal rotations, association energy of ligand and receptor to form a single complex and free energy due to changes in vibrational modes. A low (negative)
535:
Computational capacity has increased dramatically over the last decade making possible the use of more sophisticated and computationally intensive methods in computer-assisted drug design. However, dealing with receptor flexibility in docking methodologies is still a thorny issue. The main reason
466:
of the protein paired with the ligand. However, in practice with current computational resources, it is impossible to exhaustively explore the search space — this would involve enumerating all possible distortions of each molecule (molecules are dynamic and exist in an ensemble of conformational
388:
The obvious advantage of docking simulation is that ligand flexibility is easily incorporated, whereas shape complementarity techniques must use ingenious methods to incorporate flexibility in ligands. Also, it more accurately models reality, whereas shape complementary techniques are more of an
371:
and the ligand's molecular surface is described in terms of its matching surface description. The complementarity between the two surfaces amounts to the shape matching description that may help finding the complementary pose of docking the target and the ligand molecules. Another approach is to
803:
The interdependence between sampling and scoring function affects the docking capability in predicting plausible poses or binding affinities for novel compounds. Thus, an assessment of a docking protocol is generally required (when experimental data is available) to determine its predictive
518:
Conformations of the ligand may be generated in the absence of the receptor and subsequently docked or conformations may be generated on-the-fly in the presence of the receptor binding cavity, or with full rotational flexibility of every dihedral angle using fragment based docking.
204:
208:
207:
203:
202:
1677:
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (Mar 2004). "Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy".
209:
838:” molecules. In this way, the success of a docking screen is evaluated by its capacity to enrich the small number of known active compounds in the top ranks of a screen from among a much greater number of decoy molecules in the database. The area under the
526:
Peptides are both highly flexible and relatively large-sized molecules, which makes modeling their flexibility a challenging task. A number of methods were developed to allow for efficient modeling of flexibility of peptides during protein-peptide docking.
573:
761:
Alternative approaches use modified scoring functions to include constraints based on known key protein-ligand interactions, or knowledge-based potentials derived from interactions observed in large databases of protein-ligand structures (e.g. the
960:
lead optimization – docking can be used to predict in where and in which relative orientation a ligand binds to a protein (also referred to as the binding mode or pose). This information may in turn be used to design more potent and selective
536:
behind this difficulty is the large number of degrees of freedom that have to be considered in this kind of calculations. Neglecting it, however, in some of the cases may lead to poor docking results in terms of binding pose prediction.
206:
317:. During the course of the docking process, the ligand and the protein adjust their conformation to achieve an overall "best-fit" and this kind of conformational adjustment resulting in the overall binding is referred to as
479:. Most docking programs in use account for the whole conformational space of the ligand (flexible ligand), and several attempt to model a flexible protein receptor. Each "snapshot" of the pair is referred to as a
392:
Clearly, simulation is computationally expensive, having to explore a large energy landscape. Grid-based techniques, optimization methods, and increased computer speed have made docking simulation more realistic.
1385:
Feig M, Onufriev A, Lee MS, Im W, Case DA, Brooks CL (Jan 2004). "Performance comparison of generalized born and
Poisson methods in the calculation of electrostatic solvation energies for protein structures".
826:
Docking accuracy represents one measure to quantify the fitness of a docking program by rationalizing the ability to predict the right pose of a ligand with respect to that experimentally observed.
781:
One way to reduce the number of false positives is to recalculate the energy of the top scoring poses using (potentially) more accurate but computationally more intensive techniques such as
2547:
Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW (Feb 2007). "Diverse, high-quality test set for the validation of protein-ligand docking performance".
1464:
Cai W, Shao X, Maigret B (Jan 2002). "Protein-ligand recognition using spherical harmonic molecular surfaces: towards a fast and efficient filter for large virtual throughput screening".
1345:
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998). "Automated docking using a
Lamarckian genetic algorithm and an empirical binding free energy function".
205:
748:{\displaystyle \bigtriangleup G_{bind}=\bigtriangleup G_{solvent}+\bigtriangleup G_{conf}+\bigtriangleup G_{int}+\bigtriangleup G_{rot}+\bigtriangleup G_{t/t}+\bigtriangleup G_{vib}}
1156:
Mostashari-Rad T, Arian R, Mehridehnavi A, Fassihi A, Ghasemi F (June 13, 2019). "Study of CXCR4 chemokine receptor inhibitors using QSPR andmolecular docking methodologies".
436:
construction. This protein structure and a database of potential ligands serve as inputs to a docking program. The success of a docking program depends on two components: the
409:
To perform a docking screen, the first requirement is a structure of the protein of interest. Usually the structure has been determined using a biophysical technique such as
2781:
Project of
Conformational Sampling and Docking on Grids : one aim is to deploy some intrinsic distributed docking algorithms on computational Grids, download
539:
Multiple static structures experimentally determined for the same protein in different conformations are often used to emulate receptor flexibility. Alternatively
364:
2796:
359:
Geometric matching/shape complementarity methods describe the protein and ligand as a set of features that make them dockable. These features may include
509:
to "evolve" new low energy conformations and where the score of each pose acts as the fitness function used to select individuals for the next iteration.
328:
process. It aims to achieve an optimized conformation for both the protein and ligand and relative orientation between protein and ligand such that the
2753:
2094:
Taylor RD, Jewsbury PJ, Essex JW (Oct 2003). "FDS: flexible ligand and receptor docking with a continuum solvent model and soft-core energy function".
1013:
798:
543:
of amino acid side chains that surround the binding cavity may be searched to generate alternate but energetically reasonable protein conformations.
834:
Docking screens can also be evaluated by the enrichment of annotated ligands of known binders from among a large database of presumed non-binding, “
2320:
Ballante F, Marshall GR (January 2016). "An
Automated Strategy for Binding-Pose Selection and Docking Assessment in Structure-Based Drug Design".
1113:
Kitchen DB, Decornez H, Furr JR, Bajorath J (Nov 2004). "Docking and scoring in virtual screening for drug discovery: methods and applications".
453:
2754:"Step by step installation of MGLTools 1.5.2 (AutoDockTools, Python Molecular Viewer and Visual Programming Environment) on Ubuntu Linux 8.04"
867:
measurements). Only prospective studies constitute conclusive proof of the suitability of a technique for a particular target. In the case of
1959:
Cerqueira NM, Bras NF, Fernandes PA, Ramos MJ (January 2009). "MADAMM: a multistaged docking with an automated molecular modeling protocol".
1234:
Wei BQ, Weaver LH, Ferrari AM, Matthews BW, Shoichet BK (Apr 2004). "Testing a flexible-receptor docking algorithm in a model binding site".
568:
that estimate the energy of the pose within the binding site. The various contributions to binding can be written as an additive equation:
1368:
523:
energy evaluation are most often used to select energetically reasonable conformations, but knowledge-based methods have also been used.
2688:
1287:
1270:
2355:
Bursulaya BD, Totrov M, Abagyan R, Brooks CL (November 2003). "Comparative study of several algorithms for flexible ligand docking".
871:(GPCRs), which are targets of more than 30% of marketed drugs, molecular docking led to the discovery of more than 500 GPCR ligands.
1626:
Kearsley SK, Underwood DJ, Sheridan RP, Miller MD (Oct 1994). "Flexibases: a way to enhance the use of molecular docking methods".
347:
The second approach simulates the actual docking process in which the ligand-protein pairwise interaction energies are calculated.
1018:
472:
128:
The process of classifying which ligands are most likely to interact favorably to a particular receptor based on the predicted
1713:
Zsoldos Z, Reid D, Simon A, Sadjad SB, Johnson AP (Jul 2007). "eHiTS: a new fast, exhaustive flexible ligand docking system".
2423:
2199:
1540:
Kahraman A, Morris RJ, Laskowski RA, Thornton JM (Apr 2007). "Shape variation in protein binding pockets and their ligands".
540:
2782:
2582:
Chachulski L, Windshügel B (Dec 2020). "LEADS-FRAG: A Benchmark Data Set for
Assessment of Fragment Docking Performance".
1501:"Real spherical harmonic expansion coefficients as 3D shape descriptors for protein binding pocket and ligand comparisons"
2231:
Gohlke H, Hendlich M, Klebe G (January 2000). "Knowledge-based scoring function to predict protein-ligand interactions".
196:
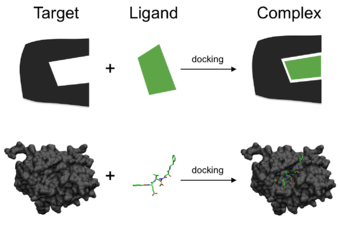
Schematic illustration of docking a small molecule ligand (green) to a protein target (black) producing a stable complex.
2706:
2652:
Suresh PS, Kumar A, Kumar R, Singh VP (Jan 2008). "An in silico approach to bioremediation: laccase as a case study".
988:
2500:"Structure-Based Virtual Screening for Ligands of G Protein-Coupled Receptors: What Can Molecular Docking Do for You?"
262:. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g.,
839:
811:
the correlation between a docking score and the experimental response or determination of the enrichment factor (EF);
182:
2821:
1807:
Klebe G, Mietzner T (October 1994). "A fast and efficient method to generate biologically relevant conformations".
486:
A variety of conformational search strategies have been applied to the ligand and to the receptor. These include:
2793:
2774:
368:
2406:
Ballante F (2018). "Protein-Ligand
Docking in Drug Design: Performance Assessment and Binding-Pose Selection".
221:
782:
778:
hits, i.e., ligands predicted to bind to the protein that actually don't when placed together in a test tube.
2051:
Hartmann C, Antes I, Lengauer T (Feb 2009). "Docking and scoring with alternative side-chain conformations".
1058:
942:
552:
425:
186:
105:
2757:
897:
An evaluation of docking programs for their potential to reproduce peptide binding modes can be assessed by
2831:
2816:
2746:
Internet service that calculates the site, geometry and energy of small molecules interacting with proteins
2147:
Murcko MA (Dec 1995). "Computational
Methods to Predict Binding Free Energy in Ligand-Receptor Complexes".
344:
One approach uses a matching technique that describes the protein and the ligand as complementary surfaces.
2617:
Hauser AS, Windshügel B (Dec 2015). "A Benchmark Data Set for
Assessment of Peptide Docking Performance".
1900:
983:
786:
2826:
868:
218:
181:. Knowledge of the preferred orientation in turn may be used to predict the strength of association or
1023:
267:
129:
2108:
1359:
2687:
Basharat Z, Yasmin A, Bibi M (2020). "Implications of
Molecular Docking Assay for Bioremediation".
351:
Both approaches have significant advantages as well as some limitations. These are outlined below.
178:
146:
93:
1271:"QSD quadratic shape descriptors. 2. Molecular docking using quadratic shape descriptors (QSDock)"
2841:
2836:
894:(DUD) for evaluation of virtual screening performance., or the LEADS-FRAG data set for fragments
565:
29:
2693:. Advances in Environmental Engineering and Green Technologies. IGI Global. pp. 1556–1577.
967:– protein ligand docking can also be used to predict pollutants that can be degraded by enzymes.
376:
based approaches, since they use geometric descriptions of the ligands to find optimal binding.
2103:
1354:
993:
926:
864:
774:
give plausible docked conformations for ligands that do not bind. This gives a large number of
156:
1310:
Meng EC, Shoichet BK, Kuntz ID (1992). "Automated docking with grid-based energy evaluation".
886:
For small molecules, several benchmark data sets for docking and virtual screening exist e.g.
270:). Therefore, docking is useful for predicting both the strength and type of signal produced.
1858:
Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, Kmiecik S (May 2018).
880:
860:
770:
520:
463:
413:
360:
325:
170:
57:
2456:
2364:
2186:. Methods in Molecular Biology. Vol. 2266. New York, NY: Springer US. pp. 39–72.
1816:
1761:
1635:
1200:
954:
286:
111:
8:
2376:
1191:
Jorgensen WL (Nov 1991). "Rusting of the lock and key model for protein-ligand binding".
998:
562:
441:
367:
descriptors. In this case, the receptor's molecular surface is described in terms of its
259:
174:
71:
2460:
2368:
1820:
1765:
1750:"Preference of small molecules for local minimum conformations when binding to proteins"
1639:
1204:
169:
is a method which predicts the preferred orientation of one molecule to a second when a
2712:
2529:
2480:
2388:
2297:
2272:
2213:
2129:
2076:
2028:
2003:
1984:
1941:
1840:
1784:
1749:
1659:
1603:
1576:
1446:
1411:
1327:
1173:
1138:
500:
459:
162:
119:
1477:
1429:
Shoichet BK, Kuntz ID, Bodian DL (2004). "Molecular docking using shape descriptors".
1091:
850:
Resulting hits from docking screens are subjected to pharmacological validation (e.g.
2702:
2669:
2634:
2599:
2564:
2533:
2521:
2472:
2429:
2419:
2380:
2337:
2302:
2248:
2217:
2205:
2195:
2164:
2121:
2068:
2033:
2004:"Flexible ligand docking to multiple receptor conformations: a practical alternative"
1976:
1933:
1881:
1844:
1832:
1789:
1730:
1695:
1651:
1608:
1557:
1522:
1481:
1403:
1292:
1251:
1216:
1177:
1130:
1095:
1053:
1003:
950:
804:
capability. Docking assessment can be performed using different strategies, such as:
763:
506:
433:
329:
226:
214:
2716:
2698:
2484:
2392:
2133:
2080:
1988:
1517:
1500:
1450:
1331:
2694:
2661:
2626:
2591:
2556:
2511:
2464:
2411:
2372:
2329:
2292:
2284:
2240:
2187:
2156:
2113:
2060:
2023:
2015:
1968:
1945:
1923:
1915:
1871:
1824:
1779:
1769:
1722:
1687:
1663:
1643:
1598:
1588:
1549:
1512:
1473:
1438:
1415:
1395:
1364:
1319:
1282:
1243:
1208:
1165:
1142:
1122:
1087:
934:
918:
437:
1919:
1078:
Lengauer T, Rarey M (Jun 1996). "Computational methods for biomolecular docking".
2800:
2778:
1876:
1859:
1774:
949:
to identify molecules that are likely to bind to protein target of interest (see
468:
340:
Two approaches are particularly popular within the molecular docking community.
2516:
2499:
2415:
2191:
110:
The process of evaluating a particular pose by counting the number of favorable
2665:
1726:
1008:
964:
910:
775:
494:
278:
75:
2468:
2019:
1553:
1247:
1169:
879:
The potential of docking programs to reproduce binding modes as determined by
2810:
2630:
2595:
2333:
1369:
10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
1043:
1033:
373:
251:
247:
115:
2735:"Molecular Docking Server - Ligand Protein Docking & Molecular Modeling"
2690:
Data Analytics in Medicine: Concepts, Methodologies, Tools, and Applications
1212:
2673:
2638:
2603:
2568:
2525:
2476:
2433:
2384:
2341:
2306:
2252:
2244:
2209:
2125:
2072:
2037:
1980:
1937:
1885:
1793:
1734:
1699:
1612:
1561:
1526:
1485:
1407:
1296:
1255:
1134:
1028:
282:
192:
2498:
Ballante F, Kooistra AJ, Kampen S, de Graaf C, Carlsson J (October 2021).
2168:
1836:
1655:
1442:
1323:
1288:
10.1002/(SICI)1097-0134(20000101)38:1<79::AID-PROT9>3.0.CO;2-U
1220:
1099:
921:
of the enzyme. If the protein is a receptor, ligand binding may result in
814:
the distance between an ion-binding moiety and the ion in the active site;
1593:
1048:
978:
930:
476:
419:
274:
2160:
890:
consisting of high quality protein−ligand X-ray crystal structures, the
758:
energy indicates a stable system and thus a likely binding interaction.
558:
favorable binding interaction and ranks one ligand relative to another.
2064:
1972:
1828:
1647:
491:
475:
orientations of the ligand relative to the protein at a given level of
285:. Characterisation of the binding behaviour plays an important role in
51:
18:
2560:
2288:
2117:
1928:
1691:
1399:
1155:
230:
138:
Procedure to quantify the predictive capability of a docking protocol.
92:
The orientation of the ligand relative to the receptor as well as the
2771:
2447:
Irwin JJ (2008-02-14). "Community benchmarks for virtual screening".
1038:
946:
324:
Molecular docking research focuses on computationally simulating the
301:, in which one wants to find the correct relative orientation of the
86:
Computational simulation of a candidate ligand binding to a receptor.
2182:
Arcon JP, Turjanski AG, Martí MA, Forli S (2021). Ballante F (ed.).
1901:"Understanding the challenges of protein flexibility in drug design"
1126:
401:
239:
43:
2546:
1498:
945:
can be used to quickly screen large databases of potential drugs
922:
263:
243:
238:
The associations between biologically relevant molecules such as
47:
2410:. Methods in Molecular Biology. Vol. 1824. pp. 67–88.
273:
Molecular docking is one of the most frequently used methods in
2734:
1625:
1539:
1499:
Morris RJ, Najmanovich RJ, Kahraman A, Thornton JM (May 2005).
914:
255:
2497:
1575:
Torres PH, Sodero AC, Jofily P, Silva-Jr FP (September 2019).
835:
2787:
2354:
1857:
277:, due to its ability to predict the binding-conformation of
1958:
1112:
851:
289:
as well as to elucidate fundamental biochemical processes.
1676:
1574:
1344:
2181:
1233:
899:
Lessons for Efficiency Assessment of Docking and Scoring
1860:"Protein-peptide docking: opportunities and challenges"
1712:
883:
can be assessed by a range of docking benchmark sets.
2651:
2050:
1898:
1748:
Wang Q, Pang YP (September 2007). Romesberg F (ed.).
576:
2581:
462:
in theory consists of all possible orientations and
96:
of the ligand and receptor when bound to each other.
2230:
2093:
1899:Antunes DA, Devaurs D, Kavraki LE (December 2015).
1428:
297:One can think of molecular docking as a problem of
2732:
2686:
2270:
1158:Journal of Theoretical and Computational Chemistry
842:curve is widely used to evaluate its performance.
747:
2184:Biased Docking for Protein-Ligand Pose Prediction
1577:"Key Topics in Molecular Docking for Drug Design"
1384:
1309:
957:routinely uses docking for target identification.
799:Critical Assessment of Prediction of Interactions
2808:
2616:
2319:
929:. Docking is most commonly used in the field of
2790:- Directory of computational drug design tools.
1463:
1380:
1378:
454:Searching the conformational space for docking
2654:Journal of Molecular Graphics & Modelling
2266:
2264:
2262:
2001:
1806:
1715:Journal of Molecular Graphics & Modelling
1466:Journal of Molecular Graphics & Modelling
1422:
1077:
941:hit identification – docking combined with a
213:Docking of a small molecule (green) into the
2645:
2619:Journal of Chemical Information and Modeling
2584:Journal of Chemical Information and Modeling
2322:Journal of Chemical Information and Modeling
2313:
2087:
2044:
1995:
1952:
1741:
1706:
1670:
1533:
1492:
1457:
1268:
769:There are a large number of structures from
2271:Huang N, Shoichet BK, Irwin JJ (Nov 2006).
1800:
1619:
1581:International Journal of Molecular Sciences
1375:
1227:
1106:
2803:with MOE (Molecular Operating Environment)
2449:Journal of Computer-Aided Molecular Design
2357:Journal of Computer-Aided Molecular Design
2259:
1809:Journal of Computer-Aided Molecular Design
1628:Journal of Computer-Aided Molecular Design
1338:
1184:
1071:
937:molecules, and docking may be applied to:
185:between two molecules using, for example,
2515:
2296:
2273:"Benchmarking sets for molecular docking"
2107:
2027:
1927:
1875:
1783:
1773:
1602:
1592:
1516:
1358:
1286:
1262:
1190:
1014:Software for molecular mechanics modeling
561:Most scoring functions are physics-based
74:to the receptor. Ligands are most often
70:The complementary partner molecule which
2405:
1747:
1303:
400:
354:
292:
199:
191:
2751:
1019:List of protein-ligand docking software
840:receiver operating characteristic (ROC)
530:
396:
16:Prediction method in molecular modeling
2809:
2783:Docking@GRID open-source Linux version
2733:Bikadi Z, Kovacs S, Demko L, Hazai E.
2146:
2446:
2008:Current Opinion in Structural Biology
1080:Current Opinion in Structural Biology
792:
513:
335:
78:but could also be another biopolymer.
917:protein may result in activation or
829:
332:of the overall system is minimized.
821:
546:
447:
13:
2377:10.1023/B:JCAM.0000017496.76572.6f
2096:Journal of Computational Chemistry
1431:Journal of Computational Chemistry
1388:Journal of Computational Chemistry
1347:Journal of Computational Chemistry
1312:Journal of Computational Chemistry
989:List of molecular graphics systems
817:the presence of induce-fit models.
808:docking accuracy (DA) calculation;
426:cryo-electron microscopy (cryo-EM)
281:ligands to the appropriate target
14:
2853:
2726:
313:analogy is more appropriate than
2002:Totrov M, Abagyan R (Apr 2008).
1908:Expert Opinion on Drug Discovery
909:A binding interaction between a
2699:10.4018/978-1-5225-2325-3.ch002
2680:
2610:
2575:
2540:
2491:
2440:
2399:
2348:
2224:
2175:
2140:
1892:
1851:
1568:
904:
874:
369:solvent-accessible surface area
177:to each other to form a stable
2549:Journal of Medicinal Chemistry
2277:Journal of Medicinal Chemistry
2149:Journal of Medicinal Chemistry
1680:Journal of Medicinal Chemistry
1149:
1115:Nature Reviews. Drug Discovery
845:
497:searches about rotatable bonds
1:
1920:10.1517/17460441.2015.1094458
1518:10.1093/bioinformatics/bti337
1478:10.1016/S1093-3263(01)00134-6
1269:Goldman BB, Wipke WT (2000).
1092:10.1016/S0959-440X(96)80061-3
1065:
1059:Scoring functions for docking
553:Scoring functions for docking
379:
2233:Journal of Molecular Biology
1877:10.1016/j.drudis.2018.05.006
1775:10.1371/journal.pone.0000820
1542:Journal of Molecular Biology
1236:Journal of Molecular Biology
7:
2517:10.1124/pharmrev.120.000246
2416:10.1007/978-1-4939-8630-9_5
2192:10.1007/978-1-0716-1209-5_3
984:Katchalski-Katzir algorithm
971:
869:G protein-coupled receptors
405:Docking flow-chart overview
275:structure-based drug design
112:intermolecular interactions
10:
2858:
2666:10.1016/j.jmgm.2007.05.005
1727:10.1016/j.jmgm.2006.06.002
892:Directory of Useful Decoys
796:
550:
451:
222:G-protein coupled receptor
154:
2469:10.1007/s10822-008-9189-4
2020:10.1016/j.sbi.2008.01.004
1554:10.1016/j.jmb.2007.01.086
1248:10.1016/j.jmb.2004.02.015
1170:10.1142/S0219633619500184
1024:Molecular design software
467:states) and all possible
432:but can also derive from
102:A candidate binding mode.
2631:10.1021/acs.jcim.5b00234
2596:10.1021/acs.jcim.0c00693
2334:10.1021/acs.jcim.5b00603
287:rational design of drugs
2822:Computational chemistry
2794:Ligand:Receptor Docking
2504:Pharmacological Reviews
1213:10.1126/science.1719636
933:— most drugs are small
305:which will open up the
258:play a central role in
135:Docking assessment (DA)
2245:10.1006/jmbi.1999.3371
994:Macromolecular docking
749:
406:
235:
197:
157:Macromolecular docking
1443:10.1002/jcc.540130311
1324:10.1002/jcc.540130412
881:X-ray crystallography
771:X-ray crystallography
750:
414:X-ray crystallography
404:
365:complementary surface
355:Shape complementarity
326:molecular recognition
293:Definition of problem
212:
195:
2408:Rational Drug Design
1864:Drug Discovery Today
1594:10.3390/ijms20184574
955:Reverse pharmacology
574:
531:Receptor flexibility
397:Mechanics of docking
2832:Medicinal chemistry
2817:Molecular modelling
2461:2008JCAMD..22..193I
2369:2003JCAMD..17..755B
2161:10.1021/jm00026a001
1821:1994JCAMD...8..583K
1766:2007PLoSO...2..820W
1640:1994JCAMD...8..565K
1205:1991Sci...254..954J
999:Molecular mechanics
563:molecular mechanics
260:signal transduction
2799:2019-02-02 at the
2777:2019-12-31 at the
2065:10.1002/prot.22189
1973:10.1002/prot.22146
1829:10.1007/BF00123667
1648:10.1007/BF00123666
793:Docking assessment
745:
514:Ligand flexibility
507:genetic algorithms
501:molecular dynamics
407:
336:Docking approaches
236:
198:
163:molecular modeling
46:, most commonly a
2827:Protein structure
2737:. Virtua Drug Ltd
2590:(12): 6544–6554.
2561:10.1021/jm061277y
2425:978-1-4939-8629-3
2289:10.1021/jm0608356
2201:978-1-0716-1209-5
2118:10.1002/jcc.10295
1692:10.1021/jm0306430
1400:10.1002/jcc.10378
1353:(14): 1639–1662.
1054:Virtual screening
1004:Protein structure
951:virtual screening
888:Astex Diverse Set
830:Enrichment factor
787:Poisson-Boltzmann
764:Protein Data Bank
541:rotamer libraries
434:homology modeling
361:molecular surface
219:beta-2 adrenergic
215:crystal structure
210:
187:scoring functions
173:and a target are
153:
152:
149:
22:Docking glossary
2849:
2768:
2766:
2765:
2756:. Archived from
2748:
2743:
2742:
2721:
2720:
2684:
2678:
2677:
2649:
2643:
2642:
2614:
2608:
2607:
2579:
2573:
2572:
2544:
2538:
2537:
2519:
2495:
2489:
2488:
2455:(3–4): 193–199.
2444:
2438:
2437:
2403:
2397:
2396:
2352:
2346:
2345:
2317:
2311:
2310:
2300:
2283:(23): 6789–801.
2268:
2257:
2256:
2228:
2222:
2221:
2179:
2173:
2172:
2144:
2138:
2137:
2111:
2091:
2085:
2084:
2048:
2042:
2041:
2031:
1999:
1993:
1992:
1956:
1950:
1949:
1931:
1905:
1896:
1890:
1889:
1879:
1870:(8): 1530–1537.
1855:
1849:
1848:
1804:
1798:
1797:
1787:
1777:
1745:
1739:
1738:
1710:
1704:
1703:
1674:
1668:
1667:
1623:
1617:
1616:
1606:
1596:
1572:
1566:
1565:
1537:
1531:
1530:
1520:
1496:
1490:
1489:
1461:
1455:
1454:
1426:
1420:
1419:
1382:
1373:
1372:
1362:
1342:
1336:
1335:
1307:
1301:
1300:
1290:
1266:
1260:
1259:
1231:
1225:
1224:
1188:
1182:
1181:
1153:
1147:
1146:
1110:
1104:
1103:
1075:
943:scoring function
822:Docking accuracy
783:Generalized Born
754:
752:
751:
746:
744:
743:
722:
721:
717:
698:
697:
676:
675:
654:
653:
629:
628:
598:
597:
547:Scoring function
448:Search algorithm
442:scoring function
438:search algorithm
420:NMR spectroscopy
233:
211:
183:binding affinity
161:In the field of
145:
42:The "receiving"
19:
2857:
2856:
2852:
2851:
2850:
2848:
2847:
2846:
2807:
2806:
2801:Wayback Machine
2779:Wayback Machine
2763:
2761:
2752:Malinauskas T.
2740:
2738:
2729:
2724:
2709:
2685:
2681:
2650:
2646:
2615:
2611:
2580:
2576:
2545:
2541:
2496:
2492:
2445:
2441:
2426:
2404:
2400:
2363:(11): 755–763.
2353:
2349:
2318:
2314:
2269:
2260:
2229:
2225:
2202:
2180:
2176:
2155:(26): 4953–67.
2145:
2141:
2109:10.1.1.147.1131
2102:(13): 1637–56.
2092:
2088:
2049:
2045:
2000:
1996:
1957:
1953:
1914:(12): 1301–13.
1903:
1897:
1893:
1856:
1852:
1805:
1801:
1746:
1742:
1711:
1707:
1675:
1671:
1624:
1620:
1573:
1569:
1538:
1534:
1511:(10): 2347–55.
1497:
1493:
1462:
1458:
1427:
1423:
1383:
1376:
1360:10.1.1.471.5900
1343:
1339:
1308:
1304:
1267:
1263:
1232:
1228:
1199:(5034): 954–5.
1189:
1185:
1154:
1150:
1127:10.1038/nrd1549
1111:
1107:
1076:
1072:
1068:
1063:
974:
907:
877:
859:
855:
848:
832:
824:
801:
795:
733:
729:
713:
709:
705:
687:
683:
665:
661:
640:
636:
606:
602:
584:
580:
575:
572:
571:
555:
549:
533:
516:
456:
450:
399:
382:
357:
338:
311:“hand-in-glove”
295:
225:
200:
159:
76:small molecules
17:
12:
11:
5:
2855:
2845:
2844:
2842:Drug discovery
2839:
2837:Bioinformatics
2834:
2829:
2824:
2819:
2805:
2804:
2791:
2788:Click2Drug.org
2785:
2769:
2749:
2728:
2727:External links
2725:
2723:
2722:
2708:978-1799812043
2707:
2679:
2644:
2625:(1): 188–200.
2609:
2574:
2539:
2510:(4): 527–565.
2490:
2439:
2424:
2398:
2347:
2312:
2258:
2239:(2): 337–356.
2223:
2200:
2174:
2139:
2086:
2043:
1994:
1967:(1): 192–206.
1951:
1891:
1850:
1815:(5): 583–606.
1799:
1740:
1721:(1): 198–212.
1705:
1686:(7): 1739–49.
1669:
1618:
1567:
1548:(1): 283–301.
1532:
1505:Bioinformatics
1491:
1456:
1437:(3): 380–397.
1421:
1374:
1337:
1318:(4): 505–524.
1302:
1261:
1242:(5): 1161–82.
1226:
1183:
1148:
1121:(11): 935–49.
1105:
1069:
1067:
1064:
1062:
1061:
1056:
1051:
1046:
1041:
1036:
1031:
1026:
1021:
1016:
1011:
1009:Protein design
1006:
1001:
996:
991:
986:
981:
975:
973:
970:
969:
968:
965:bioremediation
962:
958:
913:ligand and an
911:small molecule
906:
903:
876:
873:
857:
853:
847:
844:
831:
828:
823:
820:
819:
818:
815:
812:
809:
794:
791:
776:false positive
742:
739:
736:
732:
728:
725:
720:
716:
712:
708:
704:
701:
696:
693:
690:
686:
682:
679:
674:
671:
668:
664:
660:
657:
652:
649:
646:
643:
639:
635:
632:
627:
624:
621:
618:
615:
612:
609:
605:
601:
596:
593:
590:
587:
583:
579:
551:Main article:
548:
545:
532:
529:
515:
512:
511:
510:
504:
498:
490:systematic or
452:Main article:
449:
446:
430:
429:
423:
417:
398:
395:
381:
378:
356:
353:
349:
348:
345:
337:
334:
315:“lock-and-key”
299:“lock-and-key”
294:
291:
279:small molecule
151:
150:
142:
141:
140:
139:
136:
133:
126:
123:
116:hydrogen bonds
108:
103:
100:
97:
90:
87:
84:
79:
68:
55:
40:
24:
23:
15:
9:
6:
4:
3:
2:
2854:
2843:
2840:
2838:
2835:
2833:
2830:
2828:
2825:
2823:
2820:
2818:
2815:
2814:
2812:
2802:
2798:
2795:
2792:
2789:
2786:
2784:
2780:
2776:
2773:
2770:
2760:on 2009-02-26
2759:
2755:
2750:
2747:
2736:
2731:
2730:
2718:
2714:
2710:
2704:
2700:
2696:
2692:
2691:
2683:
2675:
2671:
2667:
2663:
2659:
2655:
2648:
2640:
2636:
2632:
2628:
2624:
2620:
2613:
2605:
2601:
2597:
2593:
2589:
2585:
2578:
2570:
2566:
2562:
2558:
2555:(4): 726–41.
2554:
2550:
2543:
2535:
2531:
2527:
2523:
2518:
2513:
2509:
2505:
2501:
2494:
2486:
2482:
2478:
2474:
2470:
2466:
2462:
2458:
2454:
2450:
2443:
2435:
2431:
2427:
2421:
2417:
2413:
2409:
2402:
2394:
2390:
2386:
2382:
2378:
2374:
2370:
2366:
2362:
2358:
2351:
2343:
2339:
2335:
2331:
2327:
2323:
2316:
2308:
2304:
2299:
2294:
2290:
2286:
2282:
2278:
2274:
2267:
2265:
2263:
2254:
2250:
2246:
2242:
2238:
2234:
2227:
2219:
2215:
2211:
2207:
2203:
2197:
2193:
2189:
2185:
2178:
2170:
2166:
2162:
2158:
2154:
2150:
2143:
2135:
2131:
2127:
2123:
2119:
2115:
2110:
2105:
2101:
2097:
2090:
2082:
2078:
2074:
2070:
2066:
2062:
2059:(3): 712–26.
2058:
2054:
2047:
2039:
2035:
2030:
2025:
2021:
2017:
2014:(2): 178–84.
2013:
2009:
2005:
1998:
1990:
1986:
1982:
1978:
1974:
1970:
1966:
1962:
1955:
1947:
1943:
1939:
1935:
1930:
1925:
1921:
1917:
1913:
1909:
1902:
1895:
1887:
1883:
1878:
1873:
1869:
1865:
1861:
1854:
1846:
1842:
1838:
1834:
1830:
1826:
1822:
1818:
1814:
1810:
1803:
1795:
1791:
1786:
1781:
1776:
1771:
1767:
1763:
1759:
1755:
1751:
1744:
1736:
1732:
1728:
1724:
1720:
1716:
1709:
1701:
1697:
1693:
1689:
1685:
1681:
1673:
1665:
1661:
1657:
1653:
1649:
1645:
1641:
1637:
1634:(5): 565–82.
1633:
1629:
1622:
1614:
1610:
1605:
1600:
1595:
1590:
1586:
1582:
1578:
1571:
1563:
1559:
1555:
1551:
1547:
1543:
1536:
1528:
1524:
1519:
1514:
1510:
1506:
1502:
1495:
1487:
1483:
1479:
1475:
1472:(4): 313–28.
1471:
1467:
1460:
1452:
1448:
1444:
1440:
1436:
1432:
1425:
1417:
1413:
1409:
1405:
1401:
1397:
1394:(2): 265–84.
1393:
1389:
1381:
1379:
1370:
1366:
1361:
1356:
1352:
1348:
1341:
1333:
1329:
1325:
1321:
1317:
1313:
1306:
1298:
1294:
1289:
1284:
1280:
1276:
1272:
1265:
1257:
1253:
1249:
1245:
1241:
1237:
1230:
1222:
1218:
1214:
1210:
1206:
1202:
1198:
1194:
1187:
1179:
1175:
1171:
1167:
1163:
1159:
1152:
1144:
1140:
1136:
1132:
1128:
1124:
1120:
1116:
1109:
1101:
1097:
1093:
1089:
1085:
1081:
1074:
1070:
1060:
1057:
1055:
1052:
1050:
1047:
1045:
1044:ZINC database
1042:
1040:
1037:
1035:
1034:Exscalate4Cov
1032:
1030:
1027:
1025:
1022:
1020:
1017:
1015:
1012:
1010:
1007:
1005:
1002:
1000:
997:
995:
992:
990:
987:
985:
982:
980:
977:
976:
966:
963:
959:
956:
952:
948:
944:
940:
939:
938:
936:
932:
928:
924:
920:
916:
912:
902:
901:(LEADS-PEP).
900:
895:
893:
889:
884:
882:
872:
870:
866:
862:
856:
843:
841:
837:
827:
816:
813:
810:
807:
806:
805:
800:
790:
788:
784:
779:
777:
772:
767:
765:
759:
755:
740:
737:
734:
730:
726:
723:
718:
714:
710:
706:
702:
699:
694:
691:
688:
684:
680:
677:
672:
669:
666:
662:
658:
655:
650:
647:
644:
641:
637:
633:
630:
625:
622:
619:
616:
613:
610:
607:
603:
599:
594:
591:
588:
585:
581:
577:
569:
567:
564:
559:
554:
544:
542:
537:
528:
524:
522:
508:
505:
502:
499:
496:
493:
489:
488:
487:
484:
482:
478:
474:
473:translational
470:
465:
464:conformations
461:
455:
445:
443:
439:
435:
427:
424:
421:
418:
415:
412:
411:
410:
403:
394:
390:
389:abstraction.
386:
377:
375:
374:pharmacophore
370:
366:
362:
352:
346:
343:
342:
341:
333:
331:
327:
322:
320:
319:"induced-fit"
316:
312:
308:
304:
300:
290:
288:
284:
280:
276:
271:
269:
265:
261:
257:
253:
252:carbohydrates
249:
248:nucleic acids
245:
241:
232:
228:
223:
220:
216:
194:
190:
188:
184:
180:
176:
172:
168:
164:
158:
148:
144:
143:
137:
134:
131:
127:
124:
121:
117:
113:
109:
107:
104:
101:
98:
95:
91:
88:
85:
83:
80:
77:
73:
69:
66:
62:
59:
56:
53:
49:
45:
41:
38:
34:
31:
28:
27:
26:
25:
21:
20:
2772:Docking@GRID
2762:. Retrieved
2758:the original
2745:
2739:. Retrieved
2689:
2682:
2660:(5): 845–9.
2657:
2653:
2647:
2622:
2618:
2612:
2587:
2583:
2577:
2552:
2548:
2542:
2507:
2503:
2493:
2452:
2448:
2442:
2407:
2401:
2360:
2356:
2350:
2328:(1): 54–72.
2325:
2321:
2315:
2280:
2276:
2236:
2232:
2226:
2183:
2177:
2152:
2148:
2142:
2099:
2095:
2089:
2056:
2052:
2046:
2011:
2007:
1997:
1964:
1960:
1954:
1911:
1907:
1894:
1867:
1863:
1853:
1812:
1808:
1802:
1757:
1753:
1743:
1718:
1714:
1708:
1683:
1679:
1672:
1631:
1627:
1621:
1587:(18): 4574.
1584:
1580:
1570:
1545:
1541:
1535:
1508:
1504:
1494:
1469:
1465:
1459:
1434:
1430:
1424:
1391:
1387:
1350:
1346:
1340:
1315:
1311:
1305:
1281:(1): 79–94.
1278:
1274:
1264:
1239:
1235:
1229:
1196:
1192:
1186:
1161:
1157:
1151:
1118:
1114:
1108:
1086:(3): 402–6.
1083:
1079:
1073:
1029:Docking@Home
908:
905:Applications
898:
896:
891:
887:
885:
878:
875:Benchmarking
849:
833:
825:
802:
780:
768:
760:
756:
570:
566:force fields
560:
556:
538:
534:
525:
517:
485:
480:
460:search space
457:
431:
408:
391:
387:
383:
358:
350:
339:
323:
318:
314:
310:
306:
302:
298:
296:
283:binding site
272:
237:
166:
160:
94:conformation
89:Binding mode
81:
64:
60:
36:
32:
1760:(9): e820.
1049:Lead Finder
979:Drug design
931:drug design
846:Prospective
521:Force field
503:simulations
477:granularity
330:free energy
132:of binding.
130:free-energy
120:hydrophobic
2811:Categories
2764:2008-07-15
2741:2008-07-15
1929:1911/88215
1066:References
927:antagonism
919:inhibition
797:See also:
600:=△
492:stochastic
469:rotational
380:Simulation
268:antagonism
155:See also:
52:biopolymer
2534:245163594
2218:232340746
2104:CiteSeerX
1845:206768542
1355:CiteSeerX
1178:164985789
1039:Ibercivis
947:in silico
789:methods.
727:△
703:△
681:△
659:△
634:△
578:△
495:torsional
122:contacts.
50:or other
2797:Archived
2775:Archived
2717:63136337
2674:17606396
2639:26651532
2604:33289563
2569:17300160
2526:34907092
2485:26260725
2477:18273555
2434:30039402
2393:12569345
2385:15072435
2342:26682916
2307:17154509
2253:10623530
2210:33759120
2134:15814316
2126:12926007
2081:36088213
2073:18704939
2053:Proteins
2038:18302984
1989:36656063
1981:18618708
1961:Proteins
1938:26414598
1886:29733895
1794:17786192
1754:PLOS ONE
1735:16860582
1700:15027865
1613:31540192
1562:17337005
1527:15728116
1486:11858640
1451:42749294
1408:14648625
1332:97778840
1297:10651041
1275:Proteins
1256:15046985
1135:15520816
972:See also
961:analogs.
861:affinity
440:and the
244:peptides
240:proteins
234:)
114:such as
44:molecule
30:Receptor
2457:Bibcode
2365:Bibcode
2298:3383317
2169:8544170
2029:2396190
1946:6589810
1837:7876902
1817:Bibcode
1785:1959118
1762:Bibcode
1664:8834526
1656:7876901
1636:Bibcode
1604:6769580
1416:3191066
1221:1719636
1201:Bibcode
1193:Science
1143:1069493
1100:8804827
935:organic
923:agonism
865:potency
264:agonism
217:of the
179:complex
167:docking
125:Ranking
106:Scoring
82:Docking
48:protein
2715:
2705:
2672:
2637:
2602:
2567:
2532:
2524:
2483:
2475:
2432:
2422:
2391:
2383:
2340:
2305:
2295:
2251:
2216:
2208:
2198:
2167:
2132:
2124:
2106:
2079:
2071:
2036:
2026:
1987:
1979:
1944:
1936:
1884:
1843:
1835:
1792:
1782:
1733:
1698:
1662:
1654:
1611:
1601:
1560:
1525:
1484:
1449:
1414:
1406:
1357:
1330:
1295:
1254:
1219:
1176:
1141:
1133:
1098:
915:enzyme
307:“lock”
256:lipids
254:, and
171:ligand
58:Ligand
2713:S2CID
2530:S2CID
2481:S2CID
2389:S2CID
2214:S2CID
2130:S2CID
2077:S2CID
1985:S2CID
1942:S2CID
1904:(PDF)
1841:S2CID
1660:S2CID
1447:S2CID
1412:S2CID
1328:S2CID
1174:S2CID
1164:(4).
1139:S2CID
836:decoy
303:“key”
175:bound
147:edit
72:binds
63:guest
2703:ISBN
2670:PMID
2635:PMID
2600:PMID
2565:PMID
2522:PMID
2473:PMID
2430:PMID
2420:ISBN
2381:PMID
2338:PMID
2303:PMID
2249:PMID
2206:PMID
2196:ISBN
2165:PMID
2122:PMID
2069:PMID
2034:PMID
1977:PMID
1934:PMID
1882:PMID
1833:PMID
1790:PMID
1731:PMID
1696:PMID
1652:PMID
1609:PMID
1558:PMID
1523:PMID
1482:PMID
1404:PMID
1293:PMID
1252:PMID
1217:PMID
1131:PMID
1096:PMID
481:pose
471:and
458:The
231:3SN6
118:and
99:Pose
39:lock
35:host
2695:doi
2662:doi
2627:doi
2592:doi
2557:doi
2512:doi
2465:doi
2412:doi
2373:doi
2330:doi
2293:PMC
2285:doi
2241:doi
2237:295
2188:doi
2157:doi
2114:doi
2061:doi
2024:PMC
2016:doi
1969:doi
1924:hdl
1916:doi
1872:doi
1825:doi
1780:PMC
1770:doi
1723:doi
1688:doi
1644:doi
1599:PMC
1589:doi
1550:doi
1546:368
1513:doi
1474:doi
1439:doi
1396:doi
1365:doi
1320:doi
1283:doi
1244:doi
1240:337
1209:doi
1197:254
1166:doi
1162:178
1123:doi
1088:doi
953:).
925:or
863:or
785:or
766:).
266:vs
227:PDB
67:key
65:or
61:or
37:or
33:or
2813::
2744:.
2711:.
2701:.
2668:.
2658:26
2656:.
2633:.
2623:56
2621:.
2598:.
2588:60
2586:.
2563:.
2553:50
2551:.
2528:.
2520:.
2508:73
2506:.
2502:.
2479:.
2471:.
2463:.
2453:22
2451:.
2428:.
2418:.
2387:.
2379:.
2371:.
2361:17
2359:.
2336:.
2326:56
2324:.
2301:.
2291:.
2281:49
2279:.
2275:.
2261:^
2247:.
2235:.
2212:.
2204:.
2194:.
2163:.
2153:38
2151:.
2128:.
2120:.
2112:.
2100:24
2098:.
2075:.
2067:.
2057:74
2055:.
2032:.
2022:.
2012:18
2010:.
2006:.
1983:.
1975:.
1965:74
1963:.
1940:.
1932:.
1922:.
1912:10
1910:.
1906:.
1880:.
1868:23
1866:.
1862:.
1839:.
1831:.
1823:.
1811:.
1788:.
1778:.
1768:.
1756:.
1752:.
1729:.
1719:26
1717:.
1694:.
1684:47
1682:.
1658:.
1650:.
1642:.
1630:.
1607:.
1597:.
1585:20
1583:.
1579:.
1556:.
1544:.
1521:.
1509:21
1507:.
1503:.
1480:.
1470:20
1468:.
1445:.
1435:13
1433:.
1410:.
1402:.
1392:25
1390:.
1377:^
1363:.
1351:19
1349:.
1326:.
1316:13
1314:.
1291:.
1279:38
1277:.
1273:.
1250:.
1238:.
1215:.
1207:.
1195:.
1172:.
1160:.
1137:.
1129:.
1117:.
1094:.
1082:.
854:50
852:IC
483:.
444:.
422:or
321:.
250:,
246:,
242:,
229::
189:.
165:,
2767:.
2719:.
2697::
2676:.
2664::
2641:.
2629::
2606:.
2594::
2571:.
2559::
2536:.
2514::
2487:.
2467::
2459::
2436:.
2414::
2395:.
2375::
2367::
2344:.
2332::
2309:.
2287::
2255:.
2243::
2220:.
2190::
2171:.
2159::
2136:.
2116::
2083:.
2063::
2040:.
2018::
1991:.
1971::
1948:.
1926::
1918::
1888:.
1874::
1847:.
1827::
1819::
1813:8
1796:.
1772::
1764::
1758:2
1737:.
1725::
1702:.
1690::
1666:.
1646::
1638::
1632:8
1615:.
1591::
1564:.
1552::
1529:.
1515::
1488:.
1476::
1453:.
1441::
1418:.
1398::
1371:.
1367::
1334:.
1322::
1299:.
1285::
1258:.
1246::
1223:.
1211::
1203::
1180:.
1168::
1145:.
1125::
1119:3
1102:.
1090::
1084:6
858:,
741:b
738:i
735:v
731:G
724:+
719:t
715:/
711:t
707:G
700:+
695:t
692:o
689:r
685:G
678:+
673:t
670:n
667:i
663:G
656:+
651:f
648:n
645:o
642:c
638:G
631:+
626:t
623:n
620:e
617:v
614:l
611:o
608:s
604:G
595:d
592:n
589:i
586:b
582:G
428:,
416:,
363:/
224:(
54:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.