1241:
however, it does not reveal the numerous folding pathways that are possible. A different molecule of the same exact protein may be able to follow marginally different folding pathways, seeking different lower energy intermediates, as long as the same native structure is reached. Different pathways may have different frequencies of utilization depending on the thermodynamic favorability of each pathway. This means that if one pathway is found to be more thermodynamically favorable than another, it is likely to be used more frequently in the pursuit of the native structure. As the protein begins to fold and assume its various conformations, it always seeks a more thermodynamically favorable structure than before and thus continues through the energy funnel. Formation of secondary structures is a strong indication of increased stability within the protein, and only one combination of secondary structures assumed by the polypeptide backbone will have the lowest energy and therefore be present in the native state of the protein. Among the first structures to form once the polypeptide begins to fold are alpha helices and beta turns, where alpha helices can form in as little as 100 nanoseconds and beta turns in 1 microsecond.
519:. Chaperones exist in all cellular compartments and interact with the polypeptide chain in order to allow the native three-dimensional conformation of the protein to form; however, chaperones themselves are not included in the final structure of the protein they are assisting in. Chaperones may assist in folding even when the nascent polypeptide is being synthesized by the ribosome. Molecular chaperones operate by binding to stabilize an otherwise unstable structure of a protein in its folding pathway, but chaperones do not contain the necessary information to know the correct native structure of the protein they are aiding; rather, chaperones work by preventing incorrect folding conformations. In this way, chaperones do not actually increase the rate of individual steps involved in the folding pathway toward the native structure; instead, they work by reducing possible unwanted aggregations of the polypeptide chain that might otherwise slow down the search for the proper intermediate and they provide a more efficient pathway for the polypeptide chain to assume the correct conformations. Chaperones are not to be confused with folding
803:
one must have a suitable solvent for crystallization, obtain a pure protein at supersaturated levels in solution, and precipitate the crystals in solution. Once a protein is crystallized, X-ray beams can be concentrated through the crystal lattice which would diffract the beams or shoot them outwards in various directions. These exiting beams are correlated to the specific three-dimensional configuration of the protein enclosed within. The X-rays specifically interact with the electron clouds surrounding the individual atoms within the protein crystal lattice and produce a discernible diffraction pattern. Only by relating the electron density clouds with the amplitude of the X-rays can this pattern be read and lead to assumptions of the phases or phase angles involved that complicate this method. Without the relation established through a mathematical basis known as
840:
different but discrete protein states, i.e. native state, intermediate states, unfolded state, depends on the denaturant value; therefore, the global fluorescence signal of their equilibrium mixture also depends on this value. One thus obtains a profile relating the global protein signal to the denaturant value. The profile of equilibrium unfolding may enable one to detect and identify intermediates of unfolding. General equations have been developed by Hugues
Bedouelle to obtain the thermodynamic parameters that characterize the unfolding equilibria for homomeric or heteromeric proteins, up to trimers and potentially tetramers, from such profiles. Fluorescence spectroscopy can be combined with fast-mixing devices such as
983:
1258:
446:
832:
proteins, at the interface between two protein domains, or at the interface between subunits of oligomeric proteins. In this apolar environment, they have high quantum yields and therefore high fluorescence intensities. Upon disruption of the protein's tertiary or quaternary structure, these side chains become more exposed to the hydrophilic environment of the solvent, and their quantum yields decrease, leading to low fluorescence intensities. For Trp residues, the wavelength of their maximal fluorescence emission also depend on their environment.
1249:
transition state. The transition state can be referred to as a variant or premature form of the native state rather than just another intermediary step. The folding of the transition state is shown to be rate-determining, and even though it exists in a higher energy state than the native fold, it greatly resembles the native structure. Within the transition state, there exists a nucleus around which the protein is able to fold, formed by a process referred to as "nucleation condensation" where the structure begins to collapse onto the nucleus.
1237:. The description of protein folding by the leveling free-energy landscape is also consistent with the 2nd law of thermodynamics. Physically, thinking of landscapes in terms of visualizable potential or total energy surfaces simply with maxima, saddle points, minima, and funnels, rather like geographic landscapes, is perhaps a little misleading. The relevant description is really a high-dimensional phase space in which manifolds might take a variety of more complicated topological forms.
1187:
959:(NMR) is able to collect protein structural data by inducing a magnet field through samples of concentrated protein. In NMR, depending on the chemical environment, certain nuclei will absorb specific radio-frequencies. Because protein structural changes operate on a time scale from ns to ms, NMR is especially equipped to study intermediate structures in timescales of ps to s. Some of the main techniques for studying proteins structure and non-folding protein structural changes include
419:
193:
conformation. The amino acid composition is not as important as the sequence. The essential fact of folding, however, remains that the amino acid sequence of each protein contains the information that specifies both the native structure and the pathway to attain that state. This is not to say that nearly identical amino acid sequences always fold similarly. Conformations differ based on environmental factors as well; similar proteins fold differently based on where they are found.
689:-like structures which can cause degenerative disorders and cell death. The amyloids are fibrillary structures that contain intermolecular hydrogen bonds which are highly insoluble and made from converted protein aggregates. Therefore, the proteasome pathway may not be efficient enough to degrade the misfolded proteins prior to aggregation. Misfolded proteins can interact with one another and form structured aggregates and gain toxicity through intermolecular interactions.
220:
319:
789:
28:
20:
466:, or the inward folding of the hydrophobic groups. The hydrophobic collapse introduces entropy back to the system via the breaking of the water cages which frees the ordered water molecules. The multitude of hydrophobic groups interacting within the core of the globular folded protein contributes a significant amount to protein stability after folding, because of the vastly accumulated van der Waals forces (specifically
979:. NOE is especially useful because magnetization transfers can be observed between spatially proximal hydrogens are observed. Different NMR experiments have varying degrees of timescale sensitivity that are appropriate for different protein structural changes. NOE can pick up bond vibrations or side chain rotations, however, NOE is too sensitive to pick up protein folding because it occurs at larger timescale.
462:
aqueous environment, the water molecules tend to aggregate around the hydrophobic regions or side chains of the protein, creating water shells of ordered water molecules. An ordering of water molecules around a hydrophobic region increases order in a system and therefore contributes a negative change in entropy (less entropy in the system). The water molecules are fixed in these water cages which drives the
500:
1001:
thermodynamics and kinetics between the excited and ground. Saturation
Transfer measures changes in signal from the ground state as excited states become perturbed. It uses weak radio frequency irradiation to saturate the excited state of a particular nuclei which transfers its saturation to the ground state. This signal is amplified by decreasing the magnetization (and the signal) of the ground state.
208:
1019:, excited intermediates were studied with relaxation dispersion and Saturation transfer. SOD1 had been previously tied to many disease causing mutants which were assumed to be involved in protein aggregation, however the mechanism was still unknown. By using Relaxation Dispersion and Saturation Transfer experiments many excited intermediate states were uncovered misfolding in the SOD1 mutants.
1296:. Because of computational cost, ab initio MD folding simulations with explicit water are limited to peptides and small proteins. MD simulations of larger proteins remain restricted to dynamics of the experimental structure or its high-temperature unfolding. Long-time folding processes (beyond about 1 millisecond), like folding of larger proteins (>150 residues) can be accessed using
1126:(vWF) is a protein with an essential role in blood clot formation process. It discovered – using single molecule optical tweezers measurement – that calcium-bound vWF acts as a shear force sensor in the blood. Shear force leads to unfolding of the A2 domain of vWF, whose refolding rate is dramatically enhanced in the presence of calcium. Recently, it was also shown that the simple src
453:
451:
449:
447:
595:, mechanical forces, and the presence of chemical denaturants can contribute to protein denaturation, as well. These individual factors are categorized together as stresses. Chaperones are shown to exist in increasing concentrations during times of cellular stress and help the proper folding of emerging proteins as well as denatured or misfolded ones.
452:
1210:, meaning that naturally evolved proteins have optimized their folding energy landscapes, and that nature has chosen amino acid sequences so that the folded state of the protein is sufficiently stable. In addition, the acquisition of the folded state had to become a sufficiently fast process. Even though nature has reduced the level of
450:
1330:. The longest published result of a simulation performed using Anton as of 2011 was a 2.936 millisecond simulation of NTL9 at 355 K. Such simulations are currently able to unfold and refold small proteins (<150 amino acids residues) in equilibrium and predict how mutations affect folding kinetics and stability.
1240:
The unfolded polypeptide chain begins at the top of the funnel where it may assume the largest number of unfolded variations and is in its highest energy state. Energy landscapes such as these indicate that there are a large number of initial possibilities, but only a single native state is possible;
577:
to humans, suggesting that they evolved very early and have an important function. Some proteins never fold in cells at all except with the assistance of chaperones which either isolate individual proteins so that their folding is not interrupted by interactions with other proteins or help to unfold
586:
amorphous aggregates. The external factors involved in protein denaturation or disruption of the native state include temperature, external fields (electric, magnetic), molecular crowding, and even the limitation of space (i.e. confinement), which can have a big influence on the folding of proteins.
1248:
for a particular protein is found. The transition state in the energy funnel diagram is the conformation that must be assumed by every molecule of that protein if the protein wishes to finally assume the native structure. No protein may assume the native structure without first passing through the
1000:
phenomenon. This technique exposes the target nuclei to a 90 pulse followed by one or more 180 pulses. As the nuclei refocus, a broad distribution indicates the target nuclei is involved in an intermediate excited state. By looking at
Relaxation dispersion plots the data collect information on the
831:
are high enough to give good fluorescence signals. Both Trp and Tyr are excited by a wavelength of 280 nm, whereas only Trp is excited by a wavelength of 295 nm. Because of their aromatic character, Trp and Tyr residues are often found fully or partially buried in the hydrophobic core of
802:
is one of the more efficient and important methods for attempting to decipher the three dimensional configuration of a folded protein. To be able to conduct X-ray crystallography, the protein under investigation must be located inside a crystal lattice. To place a protein inside a crystal lattice,
739:
or
Vyndaqel (a kinetic stabilizer of tetrameric transthyretin) for the treatment of transthyretin amyloid diseases. This suggests that the process of amyloid fibril formation (and not the fibrils themselves) causes the degeneration of post-mitotic tissue in human amyloid diseases. Misfolding and
539:
because they provide the protein with the aid needed to assume its proper alignments and conformations efficiently enough to become "biologically relevant". This means that the polypeptide chain could theoretically fold into its native structure without the aid of chaperones, as demonstrated by
192:
The primary structure of a protein, its linear amino-acid sequence, determines its native conformation. The specific amino acid residues and their position in the polypeptide chain are the determining factors for which portions of the protein fold closely together and form its three-dimensional
839:
of proteins by measuring the variation in the intensity of fluorescence emission or in the wavelength of maximal emission as functions of a denaturant value. The denaturant can be a chemical molecule (urea, guanidinium hydrochloride), temperature, pH, pressure, etc. The equilibrium between the
684:
that are organized in a supramolecular arrangement known as a cross-β structure. These β-sheet-rich assemblies are very stable, very insoluble, and generally resistant to proteolysis. The structural stability of these fibrillar assemblies is caused by extensive interactions between the protein
461:
Minimizing the number of hydrophobic side-chains exposed to water is an important driving force behind the folding process. The hydrophobic effect is the phenomenon in which the hydrophobic chains of a protein collapse into the core of the protein (away from the hydrophilic environment). In an
277:
The α-Helices and β-Sheets are commonly amphipathic, meaning they have a hydrophilic and a hydrophobic portion. This ability helps in forming tertiary structure of a protein in which folding occurs so that the hydrophilic sides are facing the aqueous environment surrounding the protein and the
1121:
have been used to stretch single protein molecules from their C- and N-termini and unfold them to allow study of the subsequent refolding. The technique allows one to measure folding rates at single-molecule level; for example, optical tweezers have been recently applied to study folding and
309:
Tertiary structure may give way to the formation of quaternary structure in some proteins, which usually involves the "assembly" or "coassembly" of subunits that have already folded; in other words, multiple polypeptide chains could interact to form a fully functional quaternary protein.
258:
to form a spiral shape (refer to figure on the right). The β pleated sheet is a structure that forms with the backbone bending over itself to form the hydrogen bonds (as displayed in the figure to the left). The hydrogen bonds are between the amide hydrogen and carbonyl oxygen of the
1356:
AlphaFold's protein structure prediction results at CASP were described as "transformational" and "astounding". Some researchers noted that the accuracy is not high enough for a third of its predictions, and that it does not reveal the physical mechanism of protein folding for the
263:. There exists anti-parallel β pleated sheets and parallel β pleated sheets where the stability of the hydrogen bonds is stronger in the anti-parallel β sheet as it hydrogen bonds with the ideal 180 degree angle compared to the slanted hydrogen bonds formed by parallel sheets.
1353:, a test that measures the degree of similarity between the structure, predicted by a computational program, and the empirical structure, determined experimentally in a lab. A score of 100 is considered a complete match, within the distance cutoff used for calculating GDT.
685:
monomers, formed by backbone hydrogen bonds between their β-strands. The misfolding of proteins can trigger the further misfolding and accumulation of other proteins into aggregates or oligomers. The increased levels of aggregated proteins in the cell leads to formation of
157:
proteins with lengths of up to a hundred amino acids typically fold in a single step. Time scales of milliseconds are the norm, and the fastest known protein folding reactions are complete within a few microseconds. The folding time scale of a protein depends on its size,
278:
hydrophobic sides are facing the hydrophobic core of the protein. Secondary structure hierarchically gives way to tertiary structure formation. Once the protein's tertiary structure is formed and stabilized by the hydrophobic interactions, there may also be
1040:
by determining the overall size of a monolayer of the protein and its density in real time at sub-Angstrom resolution, although real-time measurement of the kinetics of protein folding are limited to processes that occur slower than ~10 Hz. Similar to
478:
molecule containing a large hydrophobic region. The strength of hydrogen bonds depends on their environment; thus, H-bonds enveloped in a hydrophobic core contribute more than H-bonds exposed to the aqueous environment to the stability of the native state.
1164:
is a thought experiment based on the observation that if a protein were folded by sequential sampling of all possible conformations, it would take an astronomical amount of time to do so, even if the conformations were sampled at a rapid rate (on the
826:
is a highly sensitive method for studying the folding state of proteins. Three amino acids, phenylalanine (Phe), tyrosine (Tyr) and tryptophan (Trp), have intrinsic fluorescence properties, but only Tyr and Trp are used experimentally because their
1159:
In 1969, Cyrus
Levinthal noted that, because of the very large number of degrees of freedom in an unfolded polypeptide chain, the molecule has an astronomical number of possible conformations. An estimate of 3 or 10 was made in one of his papers.
995:
have become some of the primary techniques for NMR analysis of folding. In addition, both techniques are used to uncover excited intermediate states in the protein folding landscape. To do this, CPMG Relaxation dispersion takes advantage of the
3522:
Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW (December 2005). "Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses".
1150:
Computational studies of protein folding includes three main aspects related to the prediction of protein stability, kinetics, and structure. A 2013 review summarizes the available computational methods for protein folding.
1057:
The study of protein folding has been greatly advanced in recent years by the development of fast, time-resolved techniques. Experimenters rapidly trigger the folding of a sample of unfolded protein and observe the resulting
405:
Proteins will have limitations on their folding abilities by the restricted bending angles or conformations that are possible. These allowable angles of protein folding are described with a two-dimensional plot known as the
294:
arrangement in a native structure of a protein. Tertiary structure of a protein involves a single polypeptide chain; however, additional interactions of folded polypeptide chains give rise to quaternary structure formation.
1173:
scale). Based upon the observation that proteins fold much faster than this, Levinthal then proposed that a random conformational search does not occur, and the protein must, therefore, fold through a series of meta-stable
448:
731:. It is not completely clear whether the aggregates are the cause or merely a reflection of the loss of protein homeostasis, the balance between synthesis, folding, aggregation and protein turnover. Recently the
482:
In proteins with globular folds, hydrophobic amino acids tend to be interspersed along the primary sequence, rather than randomly distributed or clustered together. However, proteins that have recently been born
679:
if it cannot achieve its normal native state. This can be due to mutations in the amino acid sequence or a disruption of the normal folding process by external factors. The misfolded protein typically contains
986:
Timescale of protein structural changes matched with NMR experiments. For protein folding, CPMG Relaxation
Dispersion (CPMG RD) and chemical exchange saturation transfer (CEST) collect data in the appropriate
429:
Protein folding must be thermodynamically favorable within a cell in order for it to be a spontaneous reaction. Since it is known that protein folding is a spontaneous reaction, then it must assume a negative
69:
The folding of many proteins begins even during the translation of the polypeptide chain. The amino acids interact with each other to produce a well-defined three-dimensional structure, known as the protein's
1225:" landscape allows the protein to fold to the native state through any of a large number of pathways and intermediates, rather than being restricted to a single mechanism. The theory is supported by both
3789:
Ould-Abeih MB, Petit-Topin I, Zidane N, Baron B, Bedouelle H (June 2012). "Multiple folding states and disorder of ribosomal protein SA, a membrane receptor for laminin, anticarcinogens, and pathogens".
4162:
Cross GH, Freeman NJ, Swann MJ (2008). "Dual
Polarization Interferometry: A Real-Time Optical Technique for Measuring (Bio)molecular Orientation, Structure and Function at the Solid/Liquid Interface".
1537:
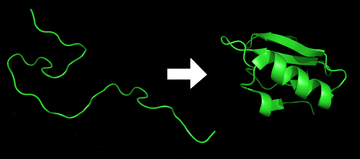
1265:, like the one diagrammed here, to model the possible shapes and folding pathways a protein can take as it condenses from its initial randomly coiled state (left) into its native 3D structure (right).
610:
have been found that grow at temperatures as high as 122 °C, which of course requires that their full complement of vital proteins and protein assemblies be stable at that temperature or above.
561:. Under certain conditions some proteins can refold; however, in many cases, denaturation is irreversible. Cells sometimes protect their proteins against the denaturing influence of heat with
724:. These age onset degenerative diseases are associated with the aggregation of misfolded proteins into insoluble, extracellular aggregates and/or intracellular inclusions including cross-β
1349:, a long-standing structureprediction contest The team achieved a level of accuracy much higher than any other group. It scored above 90% for around two-thirds of the proteins in CASP's
5036:
Schaefer M, Bartels C, Karplus M (December 1998). "Solution conformations and thermodynamics of structured peptides: molecular dynamics simulation with an implicit solvation model".
760:, where loss of function is the origin of the disorder. While protein replacement therapy has historically been used to correct the latter disorders, an emerging approach is to use
89:
are important. Failure to fold into a native structure generally produces inactive proteins, but in some instances, misfolded proteins have modified or toxic functionality. Several
925:(FT) instruments, provide powerful means for determining protein conformations in solution even for very large protein molecules. Such VCD studies of proteins can be combined with
881:
are chiral, and thus absorb such light. The absorption of this light acts as a marker of the degree of foldedness of the protein ensemble. This technique has been used to measure
2418:
Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW (July 2004). "Context-dependent contributions of backbone hydrogen bonding to beta-sheet folding energetics".
304:
2332:
Cui D, Ou S, Patel S (December 2014). "Protein-spanning water networks and implications for prediction of protein–protein interactions mediated through hydrophobic effects".
1117:
Single molecule techniques such as optical tweezers and AFM have been used to understand protein folding mechanisms of isolated proteins as well as proteins with chaperones.
142:, and other contexts. Residual structure present, if any, in the supposedly unfolded state may form a folding initiation site and guide the subsequent folding reactions.
5384:
Piana S, Piana S, Sarkar K, Lindorff-Larsen K, Guo M, Gruebele M, Shaw DE (2010). "Computational Design and
Experimental Testing of the Fastest-Folding β-Sheet Protein".
815:
use the presence of a heavy metal ion to diffract the X-rays into a more predictable manner, reducing the number of variables involved and resolving the phase problem.
550:
Along with its role in aiding native structure formation, chaperones are shown to be involved in various roles such as protein transport, degradation, and even allow
598:
Under some conditions proteins will not fold into their biochemically functional forms. Temperatures above or below the range that cells tend to live in will cause
6245:
425:. In the compact fold (to the right), the hydrophobic amino acids (shown as black spheres) collapse toward the center to become shielded from aqueous environment.
7049:
3750:"Dimeric tyrosyl-tRNA synthetase from Bacillus stearothermophilus unfolds through a monomeric intermediate. A quantitative analysis under equilibrium conditions"
964:
960:
442:. For a negative delta G to arise and for protein folding to become thermodynamically favorable, then either enthalpy, entropy, or both terms must be favorable.
1217:
A consequence of these evolutionarily selected sequences is that proteins are generally thought to have globally "funneled energy landscapes" (a term coined by
235:
is the first step in the folding process that a protein takes to assume its native structure. Characteristic of secondary structure are the structures known as
6498:
4930:"Transition-state structure as a unifying basis in protein-folding mechanisms: contact order, chain topology, stability, and the extended nucleus mechanism"
5635:
254:. Formation of intramolecular hydrogen bonds provides another important contribution to protein stability. α-helices are formed by hydrogen bonding of the
968:
546:; however, this process proves to be too inefficient or too slow to exist in biological systems; therefore, chaperones are necessary for protein folding
3428:
Hammarström P, Wiseman RL, Powers ET, Kelly JW (January 2003). "Prevention of transthyretin amyloid disease by changing protein misfolding energetics".
4624:
6043:
1036:
is a surface-based technique for measuring the optical properties of molecular layers. When used to characterize protein folding, it measures the
4005:
Vallurupalli P, Bouvignies G, Kay LE (May 2012). "Studying "invisible" excited protein states in slow exchange with a major state conformation".
153:, and must pass through a number of intermediate states, like checkpoints, before the process is complete. On the other hand, very small single-
535:
or interconversion between cis and trans stereoisomers of peptide group. Chaperones are shown to be critical in the process of protein folding
4289:
Park C, Marqusee S (March 2005). "Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding".
6908:
4677:
Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG (March 1995). "Funnels, pathways, and the energy landscape of protein folding: a synthesis".
3184:"Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation"
4195:
Bu Z, Cook J, Callaway DJ (September 2001). "Dynamic regimes and correlated structural dynamics in native and denatured alpha-lactalbumin".
6238:
5444:
3959:
Ortega, Gabriel; Pons, Miquel; Millet, Oscar (2013). "Protein
Functional Dynamics in Multiple Timescales as Studied by NMR Spectroscopy".
2174:
Zhang G, Ignatova Z (February 2011). "Folding at the birth of the nascent chain: coordinating translation with co-translational folding".
5420:
1214:
in proteins, some degree of it remains up to now as can be observed in the presence of local minima in the energy landscape of proteins.
897:(Tm) of the protein. As for fluorescence spectroscopy, circular-dichroism spectroscopy can be combined with fast-mixing devices such as
1138:
Biotin painting enables condition-specific cellular snapshots of (un)folded proteins. Biotin 'painting' shows a bias towards predicted
523:
proteins, which catalyze chemical reactions responsible for slow steps in folding pathways. Examples of folding catalysts are protein
2136:
3672:
Bedouelle H (February 2016). "Principles and equations for measuring and interpreting protein stability: From monomer to tetramer".
1070:. Among the many scientists who have contributed to the development of these techniques are Jeremy Cook, Heinrich Roder, Terry Oas,
6797:
6792:
6767:
6757:
6752:
6742:
5856:
5628:
5512:
Callaway, Ewen (30 November 2020). "'It will change everything': DeepMind's AI makes gigantic leap in solving protein structures".
3393:
Soto C, Estrada L, Castilla J (March 2006). "Amyloids, prions and the inherent infectious nature of misfolded protein aggregates".
653:
Some proteins have multiple native structures, and change their fold based on some external factors. For example, the KaiB protein
5490:
6813:
6747:
6231:
2916:
Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013). "Molecular chaperone functions in protein folding and proteostasis".
1415:
1004:
The main limitations in NMR is that its resolution decreases with proteins that are larger than 25 kDa and is not as detailed as
869:
is one of the most general and basic tools to study protein folding. Circular dichroism spectroscopy measures the absorption of
645:
chaperonin that is absolutely necessary for the folding and assembly in vivo of the bacteriophage T4 major capsid protein gp23.
7044:
6843:
3182:
Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, Hirayama H, Nakagawa S, Nunoura T, Horikoshi K (August 2008).
578:
misfolded proteins, allowing them to refold into the correct native structure. This function is crucial to prevent the risk of
4845:
4179:
3602:
2152:
2094:
1934:
1628:
1549:
1467:
1420:
3709:"Quantitative measurement of protein stability from unfolding equilibria monitored with the fluorescence maximum wavelength"
6141:
6036:
5621:
5587:
82:
7061:
1619:
Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2010). "Protein
Structure and Function".
1106:
1008:. Additionally, protein NMR analysis is quite difficult and can propose multiple solutions from the same NMR spectrum.
6146:
6136:
6088:
3976:
1139:
488:
982:
6126:
4599:
1033:
1028:
812:
701:
4332:
Mashaghi A, Kramer G, Lamb DC, Mayer MP, Tans SJ (January 2014). "Chaperone action at the single-molecule level".
3043:"Electric field-driven disruption of a native beta-sheet protein conformation and generation of a helix-structure"
6254:
6131:
6029:
5884:
1952:"The design and characterization of two proteins with 88% sequence identity but different structure and function"
1430:
713:
709:
7086:
6454:
918:
5205:"De novo prediction of protein folding pathways and structure using the principle of sequential stabilization"
2259:
554:
exposed to certain external denaturant factors an opportunity to refold into their correct native structures.
1195:
1012:
697:
81:
The correct three-dimensional structure is essential to function, although some parts of functional proteins
2873:
Hartl FU, Bracher A, Hayer-Hartl M (July 2011). "Molecular chaperones in protein folding and proteostasis".
780:
or folding of proteins and observing conformational changes using standard non-crystallographic techniques.
6667:
6093:
6083:
5849:
4655:
4533:
4475:"Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein"
3245:
1365:
and great progress towards a decades-old grand challenge of biology, predicting the structure of proteins.
1281:
1230:
145:
The duration of the folding process varies dramatically depending on the protein of interest. When studied
6612:
6156:
6073:
1405:
893:
of unfolding as well as the protein's m value, or denaturant dependence. A temperature melt measures the
551:
232:
202:
123:
7096:
6078:
5644:
2211:"Macromolecular crowding perturbs protein refolding kinetics: implications for folding inside the cell"
956:
885:
of the protein by measuring the change in this absorption as a function of denaturant concentration or
757:
474:
exists as a driving force in thermodynamics only if there is the presence of an aqueous medium with an
272:
63:
4564:
Compiani M, Capriotti E (December 2013). "Computational and theoretical methods for protein folding".
2110:
6212:
6068:
3918:
976:
890:
823:
761:
732:
654:
579:
528:
187:
5086:
2035:
1803:
1644:
Yagi-Utsumi M, Chandak MS, Yanaka S, Hiranyakorn M, Nakamura T, Kato K, Kuwajima K (November 2020).
7091:
6552:
5608:
6877:
5842:
3825:
Royer CA (May 2006). "Probing protein folding and conformational transitions with fluorescence".
1358:
1338:
1297:
1105:
is routinely used to probe the fraction unfolded under a wide range of solution conditions (e.g.
557:
A fully denatured protein lacks both tertiary and secondary structure, and exists as a so-called
467:
5287:
2953:"The denatured state (the other half of the folding equation) and its role in protein stability"
6918:
6113:
6103:
6052:
5333:
Lindorff-Larsen K, Piana S, Dror RO, Shaw DE (October 2011). "How fast-folding proteins fold".
2030:
1161:
992:
721:
705:
343:
339:
4525:
657:, acting as a clock for cyanobacteria. It has been estimated that around 0.5–4% of PDB (
6299:
6121:
5988:
5720:
4103:
Sekhar A, Rumfeldt JA, Broom HR, Doyle CM, Sobering RE, Meiering EM, Kay LE (November 2016).
3492:
2084:
1385:
1362:
1175:
1123:
1005:
894:
882:
870:
836:
799:
793:
777:
569:(a type of chaperone), which assist other proteins both in folding and in remaining folded.
491:, show the opposite pattern of hydrophobic amino acid clustering along the primary sequence.
170:
169:
Understanding and simulating the protein folding process has been an important challenge for
4600:"Structural Biochemistry/Proteins/Protein Folding - Wikibooks, open books for an open world"
3592:
2705:"A Shift in Aggregation Avoidance Strategy Marks a Long-Term Direction to Protein Evolution"
1646:"Residual Structure of Unfolded Ubiquitin as Revealed by Hydrogen/Deuterium-Exchange 2D NMR"
457:
Entropy is decreased as the water molecules become more orderly near the hydrophobic solute.
5879:
5568:
5521:
5342:
5216:
5157:
4941:
4883:
4832:. Progress in Molecular Biology and Translational Science. Vol. 84. pp. 161–202.
4802:
4747:
4639:
4486:
4429:
4243:
4116:
4056:
3631:
3437:
3288:
3195:
3054:
2828:
2772:
2559:
2492:
2427:
2384:
2086:
Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding
2022:
1963:
1876:
1815:
1577:
1350:
1071:
1037:
463:
422:
363:
355:
335:
244:
47:
4232:"Determining biophysical protein stability in lysates by a fast proteolysis assay, FASTpp"
4045:"Relaxation dispersion NMR spectroscopy as a tool for detailed studies of protein folding"
3562:
2929:
8:
6262:
6182:
6151:
5756:
3147:
Ellis RJ (July 2006). "Molecular chaperones: assisting assembly in addition to folding".
1395:
1375:
1327:
991:
Because protein folding takes place in about 50 to 3000 s CPMG Relaxation dispersion and
811:" would render predicting the diffraction patterns very difficult. Emerging methods like
524:
510:
399:
327:
150:
5564:
5525:
5346:
5220:
5161:
4945:
4887:
4806:
4751:
4643:
4490:
4433:
4247:
4120:
4060:
3963:. Advances in Protein Chemistry and Structural Biology. Vol. 92. pp. 219–251.
3635:
3479:
Chiti F, Dobson CM (2006). "Protein misfolding, functional amyloid, and human disease".
3441:
3292:
3199:
3058:
2832:
2776:
2563:
2496:
2431:
2388:
2026:
1967:
1880:
1819:
1706:
1581:
6276:
5545:
5495:
5462:
5366:
5239:
5204:
5180:
5145:
5013:
4988:
4907:
4712:
4686:
4509:
4474:
4450:
4417:
4393:
4368:
4314:
4266:
4231:
4139:
4105:"Probing the free energy landscapes of ALS disease mutants of SOD1 by NMR spectroscopy"
4104:
4077:
4044:
3968:
3504:
3461:
3372:
3311:
3276:
3218:
3183:
3124:
3099:
3075:
3042:
2982:
2898:
2852:
2796:
2729:
2704:
2680:
2655:
2631:
2606:
2549:
2513:
2482:
2470:
2451:
2375:
Tanford C (June 1978). "The hydrophobic effect and the organization of living matter".
2357:
2314:
2055:
2010:
1986:
1951:
1849:
1693:
Kim PS, Baldwin RL (1990). "Intermediates in the folding reactions of small proteins".
1670:
1645:
1601:
1509:
1484:
1400:
1285:
1262:
1063:
1042:
866:
861:
849:
773:
676:
570:
566:
504:
484:
471:
224:
149:, the slowest folding proteins require many minutes or hours to fold, primarily due to
5072:
4837:
3885:
2504:
2235:
2210:
1741:
6578:
6060:
5920:
5894:
5776:
5668:
5549:
5537:
5466:
5401:
5358:
5244:
5185:
5126:
5053:
5018:
4989:"Using simulations to provide the framework for experimental protein folding studies"
4969:
4964:
4929:
4899:
4851:
4841:
4775:
4770:
4735:
4704:
4581:
4514:
4455:
4398:
4349:
4306:
4271:
4212:
4175:
4144:
4082:
4022:
3982:
3972:
3938:
3890:
3842:
3807:
3771:
3730:
3689:
3649:
3598:
3540:
3496:
3453:
3410:
3364:
3359:
3342:
3316:
3257:
3223:
3164:
3129:
3080:
3023:
2974:
2933:
2890:
2844:
2788:
2734:
2685:
2636:
2587:
2582:
2537:
2518:
2443:
2400:
2349:
2306:
2240:
2191:
2148:
2141:
2090:
2060:
1991:
1930:
1892:
1853:
1841:
1784:
1745:
1710:
1675:
1624:
1593:
1545:
1514:
1463:
1361:
to be considered solved. Nevertheless, it is considered a significant achievement in
1293:
1190:
The energy funnel by which an unfolded polypeptide chain assumes its native structure
938:
926:
922:
902:
804:
658:
607:
431:
407:
90:
75:
5370:
4736:"Protein folding funnels: a kinetic approach to the sequence-structure relationship"
4716:
4318:
3508:
3465:
3376:
2986:
2361:
2318:
606:
turn opaque). Protein thermal stability is far from constant, however; for example,
6838:
6542:
6207:
5998:
5956:
5951:
5946:
5771:
5694:
5529:
5452:
5393:
5350:
5234:
5224:
5175:
5165:
5116:
5045:
5008:
5000:
4959:
4949:
4911:
4891:
4874:
Dill KA, MacCallum JL (November 2012). "The protein-folding problem, 50 years on".
4833:
4810:
4793:
Sharma V, Kaila VR, Annila A (2009). "Protein folding as an evolutionary process".
4765:
4755:
4696:
4647:
4573:
4544:
4504:
4494:
4445:
4437:
4388:
4380:
4341:
4298:
4261:
4251:
4204:
4167:
4134:
4124:
4072:
4064:
4014:
3964:
3930:
3880:
3834:
3799:
3761:
3720:
3681:
3639:
3532:
3488:
3445:
3402:
3354:
3306:
3296:
3213:
3203:
3156:
3119:
3111:
3070:
3062:
3013:
2964:
2925:
2902:
2882:
2856:
2836:
2800:
2780:
2724:
2716:
2675:
2667:
2626:
2618:
2577:
2567:
2508:
2500:
2455:
2435:
2392:
2341:
2296:
2230:
2222:
2183:
2050:
2040:
1981:
1971:
1884:
1831:
1823:
1776:
1737:
1702:
1665:
1657:
1605:
1585:
1504:
1496:
1410:
1390:
1319:
1292:. First equilibrium folding simulations were done using implicit solvent model and
1245:
1199:
1118:
1067:
1059:
972:
620:
387:
291:
163:
86:
39:
1091:
603:
6223:
5904:
5797:
5761:
4256:
4171:
3685:
3018:
3001:
1888:
1867:
Anfinsen CB (July 1973). "Principles that govern the folding of protein chains".
1226:
1075:
753:
717:
599:
109:
5121:
5104:
4814:
4731:
3934:
2969:
2952:
2720:
2671:
2301:
2284:
1270:
1218:
6591:
6586:
6177:
6098:
5925:
5802:
5730:
5699:
5658:
5533:
5209:
Proceedings of the National Academy of Sciences of the United States of America
5150:
Proceedings of the National Academy of Sciences of the United States of America
4934:
Proceedings of the National Academy of Sciences of the United States of America
4740:
Proceedings of the National Academy of Sciences of the United States of America
4479:
Proceedings of the National Academy of Sciences of the United States of America
4109:
Proceedings of the National Academy of Sciences of the United States of America
4068:
3406:
3188:
Proceedings of the National Academy of Sciences of the United States of America
3160:
3115:
2542:
Proceedings of the National Academy of Sciences of the United States of America
2226:
2015:
Proceedings of the National Academy of Sciences of the United States of America
1956:
Proceedings of the National Academy of Sciences of the United States of America
1455:
1234:
1222:
1079:
532:
330:
that is mainly guided by hydrophobic interactions, formation of intramolecular
283:
255:
154:
5457:
5397:
5103:
Kmiecik S, Gront D, Kolinski M, Wieteska L, Dawid AE, Kolinski A (July 2016).
5004:
3644:
3619:
3066:
2187:
1780:
1661:
1257:
776:, typically, experimental techniques for studying protein folding rely on the
7080:
5983:
5978:
5766:
5613:
4651:
3766:
3749:
3725:
3708:
1533:
1451:
1203:
828:
808:
383:
371:
367:
331:
279:
251:
247:
159:
139:
113:
104:
formed by misfolded proteins, the infectious varieties of which are known as
5354:
5229:
5170:
4895:
4760:
4499:
4129:
3449:
3301:
3208:
2656:"Gene Birth Contributes to Structural Disorder Encoded by Overlapping Genes"
2622:
2572:
2045:
1976:
418:
6947:
6929:
6192:
5684:
5541:
5405:
5362:
5248:
5189:
5130:
5049:
5022:
4973:
4954:
4903:
4855:
4700:
4585:
4518:
4459:
4402:
4353:
4310:
4275:
4216:
4208:
4148:
4086:
4026:
3986:
3942:
3846:
3811:
3734:
3693:
3653:
3544:
3500:
3457:
3414:
3368:
3320:
3227:
3168:
3133:
3084:
3027:
2937:
2894:
2792:
2738:
2689:
2640:
2522:
2447:
2396:
2353:
2244:
2195:
2064:
1995:
1845:
1788:
1679:
1597:
1380:
1308:
1304:
1186:
1083:
906:
898:
845:
841:
740:
excessive degradation instead of folding and function leads to a number of
260:
71:
5057:
4779:
4708:
3894:
3868:
3775:
3261:
2978:
2848:
2819:
Hartl FU (June 1996). "Molecular chaperones in cellular protein folding".
2591:
2310:
1896:
1749:
1714:
1518:
513:
are a class of proteins that aid in the correct folding of other proteins
219:
7023:
6891:
6021:
6003:
5993:
5725:
5689:
5421:"DeepMind solves 50-year-old 'grand challenge' with protein folding A.I."
2487:
2404:
1425:
1102:
1087:
1046:
950:
886:
878:
874:
745:
741:
670:
641:
particles during infection. Like GroES, gp31 forms a stable complex with
625:
558:
475:
395:
236:
212:
127:
59:
5146:"Characterization of protein-folding pathways by reduced-space modeling"
4691:
4418:"Calcium modulates force sensing by the von Willebrand factor A2 domain"
3343:"Protein-misfolding diseases and chaperone-based therapeutic approaches"
2886:
2784:
2554:
2439:
1836:
1589:
66:. This structure permits the protein to become biologically functional.
6853:
6281:
6187:
5930:
5807:
5751:
5735:
5704:
4441:
3100:"Effects of macromolecular crowding on protein folding and aggregation"
2345:
1827:
1170:
1166:
1127:
767:
681:
351:
347:
240:
117:
55:
4577:
4384:
4345:
4018:
3838:
3803:
3536:
1500:
1322:, a massively parallel supercomputer designed and built around custom
1229:
and experimental studies, and it has been used to improve methods for
318:
6601:
6547:
6008:
5823:
5792:
5308:
5262:
4302:
2840:
1334:
1289:
1276:
1244:
There exists a saddle point in the energy funnel landscape where the
997:
749:
736:
583:
520:
362:; however, a protein molecule may fold spontaneously during or after
2538:"Evidence for nonrandom hydrophobicity structures in protein chains"
1764:
788:
27:
6881:
6514:
6506:
5961:
5899:
4549:
1802:
Scalvini, Barbara; Sheikhhassani, Vahid; Mashaghi, Alireza (2021).
1643:
1342:
664:
574:
542:
435:
434:
value. Gibbs free energy in protein folding is directly related to
359:
290:
residues. These non-covalent and covalent contacts take a specific
287:
146:
51:
1623:(Third ed.). New York, NY: Garland Science. pp. 120–70.
944:
637:
and able to substitute for it in the assembly of bacteriophage T4
19:
6632:
6519:
6479:
6410:
6405:
6400:
6395:
6390:
6345:
6172:
5971:
5966:
5865:
5572:
5491:'The game has changed.' AI triumphs at solving protein structures
4473:
Jagannathan B, Elms PJ, Bustamante C, Marqusee S (October 2012).
4472:
3869:"Protein structure determination in solution by NMR spectroscopy"
2285:"Forces contributing to the conformational stability of proteins"
725:
686:
615:
588:
515:
439:
375:
131:
112:
are caused by the incorrect folding of some proteins because the
101:
98:
94:
43:
4534:"Biotinylation by proximity labelling favours unfolded proteins"
3788:
2260:"Torsion Angles and the Ramachnadran Plot in Protein Structures"
1728:
Jackson SE (1998). "How do small single-domain proteins fold?".
6848:
6823:
6818:
6808:
6772:
6762:
6737:
6722:
6717:
6712:
6707:
6702:
6697:
6692:
6687:
6652:
6637:
6484:
6449:
6434:
6429:
6424:
6385:
6380:
6375:
6370:
6365:
6360:
6355:
6350:
6340:
6335:
6330:
6325:
6320:
5383:
2703:
Foy SG, Wilson BA, Bertram J, Cordes MH, Masel J (April 2019).
1312:
1284:
can be used for simulating various aspects of protein folding.
1052:
728:
562:
499:
4828:
Robson B, Vaithilingam A (2008). "Protein Folding Revisited".
3243:
1318:
Long continuous-trajectory simulations have been performed on
207:
7039:
7029:
7019:
7004:
6986:
6981:
6976:
6971:
6903:
6863:
6828:
6802:
6787:
6782:
6777:
6732:
6727:
6682:
6677:
6672:
6642:
6627:
6524:
6493:
6474:
6469:
6464:
6459:
6444:
6439:
6419:
6315:
6308:
6304:
6294:
6289:
4676:
3427:
3244:
Marusich, EI; Kurochkina, LP; Mesyanzhinov, VV (April 1998).
2763:
Dobson CM (December 2003). "Protein folding and misfolding".
2417:
1801:
1568:
Selkoe DJ (December 2003). "Folding proteins in fatal ways".
1288:(MD) was used in simulations of protein folding and dynamics
930:
912:
693:
642:
638:
634:
602:
proteins to unfold or denature (this is why boiling makes an
379:
105:
74:. This structure is determined by the amino-acid sequence or
5332:
5102:
2009:
Rose GD, Fleming PJ, Banavar JR, Maritan A (November 2006).
1066:, ultrafast mixing of solutions, photochemical methods, and
629:) appears to be structurally and functionally homologous to
7054:
7034:
7014:
7009:
6959:
6942:
6898:
6886:
6868:
6858:
6657:
6647:
6622:
6617:
6562:
6557:
5834:
3521:
2209:
van den Berg B, Wain R, Dobson CM, Ellis RJ (August 2000).
2208:
1950:
Alexander PA, He Y, Chen Y, Orban J, Bryan PN (July 2007).
1454:, Johnson A, Lewis J, Raff M, Roberts K, Walters P (2002).
1346:
1323:
1221:) that are largely directed toward the native state. This "
1145:
1016:
772:
While inferences about protein folding can be made through
135:
126:
of proteins is a process of transition from a folded to an
4632:
Journal de Chimie Physique et de Physico-Chimie Biologique
4415:
3563:"Phase Problem in X-ray Crystallography, and Its Solution"
2283:
Pace CN, Shirley BA, McNutt M, Gajiwala K (January 1996).
835:
Fluorescence spectroscopy can be used to characterize the
410:, depicted with psi and phi angles of allowable rotation.
6202:
6197:
4416:
Jakobi AJ, Mashaghi A, Tans SJ, Huizinga EG (July 2011).
4004:
3181:
2915:
2282:
2008:
4102:
1618:
716:, as well as intracellular aggregation diseases such as
4331:
3097:
2872:
1450:
1341:(AI) protein structure prediction program developed by
592:
391:
5105:"Coarse-Grained Protein Models and Their Applications"
5035:
4531:
2702:
2535:
1762:
1485:"The formation and stabilization of protein structure"
1181:
398:, the possible presence of cofactors and of molecular
5445:"Structural biology: How proteins got their close-up"
4795:
Physica A: Statistical Mechanics and Its Applications
4042:
3098:
van den Berg B, Ellis RJ, Dobson CM (December 1999).
1122:
unfolding of proteins involved in blood coagulation.
1112:
704:(mad cow disease), amyloid-related illnesses such as
5202:
4729:
1949:
1303:
Several large-scale computational projects, such as
1198:
of a protein during folding can be visualized as an
768:
Experimental techniques for studying protein folding
764:
to fold mutated proteins to render them functional.
313:
5288:"The Folding@home Consortium (FAHC) – Folding@home"
4366:
4229:
3923:
Progress in Nuclear Magnetic Resonance Spectroscopy
3706:
3572:. Macmillan Publishers Ltd, Nature Publishing Group
3392:
3275:Porter, Lauren L.; Looger, Loren L. (5 June 2018).
2536:Irbäck A, Peterson C, Potthast F (September 1996).
1763:Kubelka J, Hofrichter J, Eaton WA (February 2004).
1022:
921:(VCD) techniques for proteins, currently involving
6253:
5485:
5483:
5203:Adhikari AN, Freed KF, Sosnick TR (October 2012).
4827:
3916:
2604:
2471:"On hydrophobicity correlations in protein chains"
2140:
1531:
1130:accesses multiple unfolding pathways under force.
1045:, the stimulus for folding can be a denaturant or
844:, to measure protein folding kinetics, generate a
16:Change of a linear protein chain to a 3D structure
4792:
4563:
4161:
3336:
3334:
3332:
3330:
3002:"Molecular chaperones in protein quality control"
243:that fold rapidly because they are stabilized by
7078:
5565:"CASP14 #s just came out and they're astounding"
4155:
3958:
3667:
3665:
3663:
2605:Wilson BA, Foy SG, Neme R, Masel J (June 2017).
665:Protein misfolding and neurodegenerative disease
97:are believed to result from the accumulation of
5507:
5505:
5480:
5143:
4043:Neudecker P, Lundström P, Kay LE (March 2009).
3281:Proceedings of the National Academy of Sciences
3277:"Extant fold-switching proteins are widespread"
3239:
3237:
3040:
945:Protein nuclear magnetic resonance spectroscopy
227:displaying hydrogen bonding within the backbone
5643:
4986:
4923:
4921:
4194:
3556:
3554:
3327:
2758:
2756:
2754:
2752:
2750:
2748:
2468:
2078:
2076:
2074:
1252:
573:have been found in all species examined, from
6239:
6037:
5850:
5629:
4873:
4672:
4670:
4668:
3747:
3660:
3340:
3006:Journal of Biochemistry and Molecular Biology
2868:
2866:
2814:
2812:
2810:
2173:
1924:
1460:Molecular Biology of the Cell; Fourth Edition
176:
5562:
5502:
5412:
5377:
5326:
5073:"Fragment-based Protein Folding Simulations"
4869:
4867:
4865:
4830:Molecular Biology of Protein Folding, Part B
4325:
4288:
3707:Monsellier E, Bedouelle H (September 2005).
3515:
3421:
3388:
3386:
3274:
3234:
2134:
2011:"A backbone-based theory of protein folding"
1920:
1918:
1916:
1914:
1912:
1910:
1908:
1906:
1053:Studies of folding with high time resolution
933:data for protein solutions in heavy water (D
818:
4918:
4821:
4532:Minde DP, Ramakrishna M, Lilley KS (2018).
4369:"Protein folding and unfolding under force"
4367:Jagannathan B, Marqusee S (November 2013).
3713:Protein Engineering, Design & Selection
3594:Principles of Protein X-Ray Crystallography
3551:
3478:
3472:
2745:
2653:
2411:
2071:
1804:"Topological principles of protein folding"
1563:
1561:
1227:computational simulations of model proteins
6246:
6232:
6051:
6044:
6030:
5857:
5843:
5636:
5622:
4665:
2863:
2807:
2331:
1692:
913:Vibrational circular dichroism of proteins
5456:
5238:
5228:
5179:
5169:
5120:
5012:
4963:
4953:
4862:
4769:
4759:
4690:
4625:"Are there pathways for protein folding?"
4622:
4548:
4508:
4498:
4449:
4392:
4265:
4255:
4230:Minde DP, Maurice MM, Rüdiger SG (2012).
4138:
4128:
4076:
3884:
3765:
3724:
3671:
3643:
3597:. Springer Science & Business Media.
3383:
3358:
3310:
3300:
3246:"Chaperones in bacteriophage T4 assembly"
3217:
3207:
3123:
3074:
3017:
2968:
2728:
2679:
2630:
2581:
2571:
2553:
2512:
2486:
2300:
2234:
2054:
2044:
2034:
1985:
1975:
1903:
1860:
1835:
1669:
1508:
322:All forms of protein structure summarized
5511:
5442:
5436:
5196:
4188:
4007:Journal of the American Chemical Society
3866:
3493:10.1146/annurev.biochem.75.101304.123901
2993:
2257:
1866:
1558:
1482:
1462:. New York and London: Garland Science.
1333:In 2020 a team of researchers that used
1256:
1185:
1146:Computational studies of protein folding
1011:In a study focused on the folding of an
981:
787:
783:
692:Aggregated proteins are associated with
498:
444:
417:
350:of the protein begins to fold while the
317:
218:
206:
26:
18:
5137:
4993:Archives of Biochemistry and Biophysics
4980:
2999:
2950:
2944:
2374:
1727:
1476:
1416:Protein misfolding cyclic amplification
298:
7079:
4927:
3961:Dynamics of Proteins and Nucleic Acids
3917:Zhuravleva A, Korzhnev DM (May 2017).
3617:
3590:
3560:
2762:
2469:Irbäck A, Sandelin E (November 2000).
2082:
1567:
1444:
1154:
623:, and the phage encoded gp31 protein (
540:protein folding experiments conducted
354:portion of the protein is still being
342:. The process of folding often begins
196:
6227:
6025:
5838:
5617:
5585:
5418:
4223:
4098:
4096:
4038:
4036:
4000:
3998:
3996:
3954:
3952:
3912:
3910:
3908:
3906:
3904:
3862:
3860:
3858:
3856:
3824:
3146:
3034:
2930:10.1146/annurev-biochem-060208-092442
2818:
2176:Current Opinion in Structural Biology
1769:Current Opinion in Structural Biology
1456:"The Shape and Structure of Proteins"
1421:Protein structure prediction software
1202:. According to Joseph Bryngelson and
993:chemical exchange saturation transfer
855:
531:that may be involved in formation of
413:
266:
5443:Stoddart, Charlotte (1 March 2022).
4282:
3091:
3041:Ojeda-May P, Garcia ME (July 2010).
2654:Willis S, Masel J (September 2018).
2276:
2002:
181:
5563:@MoAlQuraishi (November 30, 2020).
5144:Kmiecik S, Kolinski A (July 2007).
4987:Rizzuti B, Daggett V (March 2013).
4164:Handbook of Biosensors and Biochips
3873:The Journal of Biological Chemistry
3754:The Journal of Biological Chemistry
3341:Chaudhuri TK, Paul S (April 2006).
1808:Physical Chemistry Chemical Physics
1765:"The protein folding 'speed limit'"
1707:10.1146/annurev.bi.59.070190.003215
1538:"3. Protein Structure and Function"
1182:Energy landscape of protein folding
1068:laser temperature jump spectroscopy
374:", the process also depends on the
13:
7062:Prokaryotic ubiquitin-like protein
5586:Balls, Phillip (9 December 2020).
4093:
4033:
3993:
3969:10.1016/b978-0-12-411636-8.00006-7
3949:
3901:
3853:
3748:Park YC, Bedouelle H (July 1998).
1925:Voet D, Voet JG, Pratt CW (2016).
1133:
1113:Single-molecule force spectroscopy
1107:fast parallel proteolysis (FASTpp)
873:. In proteins, structures such as
14:
7108:
5602:
5588:"Behind the screens of AlphaFold"
5070:
1140:Intrinsically disordered proteins
1062:. Fast techniques in use include
889:. A denaturant melt measures the
648:
314:Driving forces of protein folding
3624:Acta Crystallographica Section D
3360:10.1111/j.1742-4658.2006.05181.x
1544:. San Francisco: W. H. Freeman.
1208:principle of minimal frustration
1034:Dual polarisation interferometry
1029:Dual-polarization interferometry
1023:Dual-polarization interferometry
917:The more recent developments of
813:multiple isomorphous replacement
702:bovine spongiform encephalopathy
655:switches fold throughout the day
250:, as was first characterized by
120:for certain protein structures.
23:Protein before and after folding
5885:Post-translational modification
5669:Structure determination methods
5579:
5556:
5301:
5280:
5255:
5096:
5079:
5064:
5029:
4786:
4723:
4616:
4592:
4557:
4466:
4409:
4360:
3818:
3782:
3741:
3700:
3611:
3584:
3268:
3175:
3140:
2909:
2696:
2647:
2598:
2529:
2462:
2368:
2325:
2251:
2202:
2167:
2128:
2103:
1943:
1795:
1431:Time-resolved mass spectrometry
710:familial amyloid cardiomyopathy
6592:Mitochondrial targeting signal
6255:Posttranslational modification
5609:Human Proteome Folding Project
3395:Trends in Biochemical Sciences
3149:Trends in Biochemical Sciences
2611:Nature Ecology & Evolution
1756:
1721:
1686:
1637:
1612:
1525:
1097:
919:vibrational circular dichroism
675:A protein is considered to be
503:Example of a small eukaryotic
1:
4838:10.1016/S0079-6603(08)00405-4
3886:10.1016/S0021-9258(18)45665-7
3570:Encyclopedia of Life Sciences
3525:Accounts of Chemical Research
3481:Annual Review of Biochemistry
2918:Annual Review of Biochemistry
2505:10.1016/S0006-3495(00)76472-1
1742:10.1016/S1359-0278(98)00033-9
1695:Annual Review of Biochemistry
1437:
1280:techniques for computational
1078:, Brian Dyer, William Eaton,
1013:amyotrophic lateral sclerosis
494:
305:Protein quarternary structure
6668:Ubiquitin-conjugating enzyme
5864:
5075:. University College London.
5038:Journal of Molecular Biology
4257:10.1371/journal.pone.0046147
4197:Journal of Molecular Biology
4172:10.1002/9780470061565.hbb055
3867:Wüthrich K (December 1990).
3686:10.1016/j.biochi.2015.11.013
3019:10.5483/BMBRep.2005.38.3.259
1889:10.1126/science.181.4096.223
1282:protein structure prediction
1231:protein structure prediction
7:
6956:E2 SUMO-conjugating enzyme
6613:Ubiquitin-activating enzyme
5122:10.1021/acs.chemrev.6b00163
4928:Fersht AR (February 2000).
4815:10.1016/j.physa.2008.12.004
3935:10.1016/j.pnmrs.2016.10.002
2970:10.1096/fasebj.10.1.8566543
2721:10.1534/genetics.118.301719
2672:10.1534/genetics.118.301249
2302:10.1096/fasebj.10.1.8566551
2135:Pratt C, Cornely K (2004).
1406:Potential energy of protein
1368:
1253:Modeling of protein folding
929:data for protein crystals,
901:to measure protein folding
696:-related illnesses such as
203:Protein secondary structure
64:three-dimensional structure
58:, changes from an unstable
10:
7113:
6939:E1 SUMO-activating enzyme
5926:Protein structural domains
5645:Protein tertiary structure
5534:10.1038/d41586-020-03348-4
4069:10.1016/j.bpj.2008.12.3907
3407:10.1016/j.tibs.2006.01.002
3161:10.1016/j.tibs.2006.05.001
2951:Shortle D (January 1996).
1927:Principles of Biochemistry
1351:global distance test (GDT)
1315:, target protein folding.
1026:
957:nuclear magnetic resonance
948:
871:circularly polarized light
859:
758:lysosomal storage diseases
668:
608:hyperthermophilic bacteria
529:peptidyl-prolyl isomerases
302:
273:Protein tertiary structure
270:
200:
185:
177:Process of protein folding
31:Results of protein folding
6997:
6928:
6917:
6600:
6577:
6535:
6275:
6261:
6213:Nucleic acid double helix
6165:
6112:
6059:
5939:
5913:
5872:
5816:
5785:
5744:
5713:
5677:
5651:
5458:10.1146/knowable-022822-1
5419:Shead, Sam (2020-11-30).
5398:10.1016/j.jmb.2010.10.023
5005:10.1016/j.abb.2012.12.015
3645:10.1107/S0907444903017815
3067:10.1016/j.bpj.2010.04.040
2264:www.proteinstructures.com
2188:10.1016/j.sbi.2010.10.008
1929:(Fifth ed.). Wiley.
1781:10.1016/j.sbi.2004.01.013
1662:10.1016/j.bpj.2020.10.003
1483:Anfinsen CB (July 1972).
824:Fluorescence spectroscopy
819:Fluorescence spectroscopy
762:pharmaceutical chaperones
733:European Medicines Agency
698:Creutzfeldt–Jakob disease
661:) proteins switch folds.
188:Protein primary structure
6553:Survival of motor neuron
3919:"Protein folding by NMR"
3767:10.1074/jbc.273.29.18052
3250:Biochemistry. Biokhimiia
3116:10.1093/emboj/18.24.6927
2227:10.1093/emboj/19.15.3870
895:denaturation temperature
489:intrinsically disordered
468:London Dispersion forces
386:), the concentration of
6919:Ubiquitin-like proteins
6878:Deubiquitinating enzyme
5355:10.1126/science.1208351
5230:10.1073/pnas.1209000109
5171:10.1073/pnas.0702265104
5090:(by Molecular Dynamics)
4896:10.1126/science.1219021
4761:10.1073/pnas.89.18.8721
4500:10.1073/pnas.1201800109
4130:10.1073/pnas.1611418113
3591:Drenth J (2007-04-05).
3450:10.1126/science.1079589
3302:10.1073/pnas.1800168115
3209:10.1073/pnas.0712334105
3000:Lee S, Tsai FT (2005).
2623:10.1038/s41559-017-0146
2573:10.1073/pnas.93.18.9533
2046:10.1073/pnas.0606843103
1977:10.1073/pnas.0700922104
1489:The Biochemical Journal
1359:protein folding problem
1339:artificial intelligence
587:High concentrations of
338:, and it is opposed by
6053:Biomolecular structure
5050:10.1006/jmbi.1998.2172
4955:10.1073/pnas.97.4.1525
4730:Leopold PE, Montal M,
4701:10.1002/prot.340210302
4652:10.1051/jcp/1968650044
4209:10.1006/jmbi.2001.5006
3726:10.1093/protein/gzi046
2397:10.1126/science.653353
2143:Essential Biochemistry
1621:Essential cell biology
1532:Berg JM, Tymoczko JL,
1266:
1206:, proteins follow the
1191:
988:
796:
507:
458:
426:
340:conformational entropy
323:
228:
216:
173:since the late 1960s.
32:
24:
7087:Biochemical reactions
5989:Photoreceptor protein
5721:Immunoglobulin domain
4422:Nature Communications
1386:Denaturation midpoint
1363:computational biology
1326:and interconnects by
1298:coarse-grained models
1260:
1189:
1124:von Willebrand factor
1006:X-ray crystallography
985:
883:equilibrium unfolding
837:equilibrium unfolding
800:X-ray crystallography
794:X-ray crystallography
791:
784:X-ray crystallography
502:
456:
421:
321:
222:
210:
171:computational biology
151:proline isomerization
116:does not produce the
54:as a linear chain of
30:
22:
5880:Protein biosynthesis
4623:Levinthal C (1968).
2607:"De Novo Gene Birth"
1730:Folding & Design
1094:and Lars Konermann.
939:quantum computations
735:approved the use of
525:disulfide isomerases
511:Molecular chaperones
464:hydrophobic collapse
423:Hydrophobic collapse
370:may be regarded as "
336:van der Waals forces
299:Quaternary structure
62:into a more ordered
6277:Heat shock proteins
6183:Protein engineering
5757:Leucine-rich repeat
5526:2020Natur.588..203C
5489:Robert F. Service,
5347:2011Sci...334..517L
5221:2012PNAS..10917442A
5162:2007PNAS..10412330K
4946:2000PNAS...97.1525F
4888:2012Sci...338.1042D
4807:2009PhyA..388..851S
4752:1992PNAS...89.8721L
4644:1968JCP....65...44L
4491:2012PNAS..10917820J
4434:2011NatCo...2..385J
4248:2012PLoSO...746147M
4121:2016PNAS..113E6939S
4115:(45): E6939–E6945.
4061:2009BpJ....96.2045N
4049:Biophysical Journal
3636:2003AcCrD..59.1881T
3620:"The phase problem"
3442:2003Sci...299..713H
3293:2018PNAS..115.5968P
3200:2008PNAS..10510949T
3059:2010BpJ....99..595O
3047:Biophysical Journal
2887:10.1038/nature10317
2833:1996Natur.381..571H
2785:10.1038/nature02261
2777:2003Natur.426..884D
2564:1996PNAS...93.9533I
2497:2000BpJ....79.2252I
2475:Biophysical Journal
2440:10.1038/nature02611
2432:2004Natur.430..101D
2389:1978Sci...200.1012T
2111:"Protein Structure"
2027:2006PNAS..10316623R
1968:2007PNAS..10411963A
1881:1973Sci...181..223A
1820:2021PCCP...2321316S
1814:(37): 21316–21328.
1650:Biophysical Journal
1590:10.1038/nature02264
1582:2003Natur.426..900S
1396:Folding (chemistry)
1328:D. E. Shaw Research
1263:Markov state models
1196:configuration space
1176:intermediate states
1162:Levinthal's paradox
1155:Levinthal's paradox
975:(T1 & T2), and
722:Parkinson's disease
706:Alzheimer's disease
571:Heat shock proteins
567:heat shock proteins
487:, which tend to be
328:spontaneous process
286:formed between two
233:secondary structure
197:Secondary structure
83:may remain unfolded
5499:, 30 November 2020
5267:boinc.bakerlab.org
4734:(September 1992).
4442:10.1038/ncomms1385
2346:10.1002/prot.24683
2117:. Nature Education
1828:10.1039/D1CP03390E
1401:Phi value analysis
1286:Molecular dynamics
1267:
1261:Folding@home uses
1192:
1064:neutron scattering
1043:circular dichroism
989:
867:Circular dichroism
862:Circular dichroism
856:Circular dichroism
850:Phi value analysis
797:
633:chaperone protein
600:thermally unstable
552:denatured proteins
508:
505:heat shock protein
472:hydrophobic effect
459:
427:
414:Hydrophobic effect
372:folding themselves
344:co-translationally
324:
267:Tertiary structure
229:
225:beta pleated sheet
217:
85:, indicating that
33:
25:
7097:Protein structure
7074:
7073:
7070:
7069:
6579:Protein targeting
6573:
6572:
6221:
6220:
6019:
6018:
5921:Protein structure
5895:Protein targeting
5832:
5831:
5777:Trefoil knot fold
5659:Structural domain
5520:(7837): 203–204.
5449:Knowable Magazine
5087:"Protein folding"
4847:978-0-12-374595-8
4578:10.1021/bi4001529
4385:10.1002/bip.22321
4346:10.1021/cr400326k
4181:978-0-470-01905-4
4019:10.1021/ja3001419
3839:10.1021/cr0404390
3804:10.1021/bi300335r
3618:Taylor G (2003).
3604:978-0-387-33746-3
3561:Cowtan K (2001).
3537:10.1021/ar020073i
3287:(23): 5968–5973.
2154:978-0-471-39387-0
2096:978-0-7167-3268-6
2083:Fersht A (1999).
1936:978-1-118-91840-1
1630:978-0-8153-4454-4
1551:978-0-7167-4684-3
1501:10.1042/bj1280737
1469:978-0-8153-3218-3
1294:umbrella sampling
1015:involved protein
927:X-ray diffraction
923:Fourier transform
805:Fourier transform
778:gradual unfolding
744:diseases such as
659:Protein Data Bank
454:
432:Gibbs free energy
408:Ramachandran plot
284:disulfide bridges
223:An anti-parallel
182:Primary structure
91:neurodegenerative
76:primary structure
7104:
6926:
6925:
6839:Ubiquitin ligase
6605:(ubiquitylation)
6543:Alpha crystallin
6273:
6272:
6248:
6241:
6234:
6225:
6224:
6208:Structural motif
6046:
6039:
6032:
6023:
6022:
5999:Phycobiliprotein
5957:Globular protein
5952:Membrane protein
5947:List of proteins
5859:
5852:
5845:
5836:
5835:
5817:Irregular folds:
5772:Thioredoxin fold
5695:Homeodomain fold
5638:
5631:
5624:
5615:
5614:
5596:
5595:
5583:
5577:
5576:
5560:
5554:
5553:
5509:
5500:
5487:
5478:
5477:
5475:
5473:
5460:
5440:
5434:
5433:
5431:
5430:
5416:
5410:
5409:
5381:
5375:
5374:
5341:(6055): 517–20.
5330:
5324:
5323:
5321:
5319:
5305:
5299:
5298:
5296:
5294:
5284:
5278:
5277:
5275:
5273:
5259:
5253:
5252:
5242:
5232:
5200:
5194:
5193:
5183:
5173:
5141:
5135:
5134:
5124:
5115:(14): 7898–936.
5109:Chemical Reviews
5100:
5094:
5093:
5091:
5083:
5077:
5076:
5068:
5062:
5061:
5033:
5027:
5026:
5016:
4984:
4978:
4977:
4967:
4957:
4925:
4916:
4915:
4882:(6110): 1042–6.
4871:
4860:
4859:
4825:
4819:
4818:
4790:
4784:
4783:
4773:
4763:
4727:
4721:
4720:
4694:
4674:
4663:
4662:
4660:
4654:. Archived from
4629:
4620:
4614:
4613:
4611:
4610:
4604:en.wikibooks.org
4596:
4590:
4589:
4561:
4555:
4554:
4552:
4538:
4529:
4523:
4522:
4512:
4502:
4470:
4464:
4463:
4453:
4413:
4407:
4406:
4396:
4364:
4358:
4357:
4334:Chemical Reviews
4329:
4323:
4322:
4303:10.1038/nmeth740
4286:
4280:
4279:
4269:
4259:
4227:
4221:
4220:
4192:
4186:
4185:
4159:
4153:
4152:
4142:
4132:
4100:
4091:
4090:
4080:
4040:
4031:
4030:
4002:
3991:
3990:
3956:
3947:
3946:
3914:
3899:
3898:
3888:
3879:(36): 22059–62.
3864:
3851:
3850:
3827:Chemical Reviews
3822:
3816:
3815:
3786:
3780:
3779:
3769:
3745:
3739:
3738:
3728:
3704:
3698:
3697:
3669:
3658:
3657:
3647:
3615:
3609:
3608:
3588:
3582:
3581:
3579:
3577:
3567:
3558:
3549:
3548:
3519:
3513:
3512:
3476:
3470:
3469:
3425:
3419:
3418:
3390:
3381:
3380:
3362:
3347:The FEBS Journal
3338:
3325:
3324:
3314:
3304:
3272:
3266:
3265:
3241:
3232:
3231:
3221:
3211:
3194:(31): 10949–54.
3179:
3173:
3172:
3144:
3138:
3137:
3127:
3104:The EMBO Journal
3095:
3089:
3088:
3078:
3038:
3032:
3031:
3021:
2997:
2991:
2990:
2972:
2948:
2942:
2941:
2913:
2907:
2906:
2881:(7356): 324–32.
2870:
2861:
2860:
2841:10.1038/381571a0
2816:
2805:
2804:
2771:(6968): 884–90.
2760:
2743:
2742:
2732:
2715:(4): 1345–1355.
2700:
2694:
2693:
2683:
2651:
2645:
2644:
2634:
2602:
2596:
2595:
2585:
2575:
2557:
2533:
2527:
2526:
2516:
2490:
2488:cond-mat/0010390
2466:
2460:
2459:
2415:
2409:
2408:
2383:(4345): 1012–8.
2372:
2366:
2365:
2329:
2323:
2322:
2304:
2280:
2274:
2273:
2271:
2270:
2258:Al-Karadaghi S.
2255:
2249:
2248:
2238:
2215:The EMBO Journal
2206:
2200:
2199:
2171:
2165:
2164:
2162:
2161:
2146:
2137:"Thermodynamics"
2132:
2126:
2125:
2123:
2122:
2107:
2101:
2100:
2080:
2069:
2068:
2058:
2048:
2038:
2021:(45): 16623–33.
2006:
2000:
1999:
1989:
1979:
1947:
1941:
1940:
1922:
1901:
1900:
1875:(4096): 223–30.
1864:
1858:
1857:
1839:
1799:
1793:
1792:
1760:
1754:
1753:
1725:
1719:
1718:
1690:
1684:
1683:
1673:
1641:
1635:
1634:
1616:
1610:
1609:
1565:
1556:
1555:
1529:
1523:
1522:
1512:
1480:
1474:
1473:
1448:
1411:Protein dynamics
1391:Downhill folding
1376:Anfinsen's dogma
1345:placed first in
1246:transition state
1200:energy landscape
1119:Optical tweezers
905:and to generate
774:mutation studies
628:
621:bacteriophage T4
619:is the host for
455:
280:covalent bonding
215:spiral formation
164:circuit topology
147:outside the cell
130:. It happens in
87:protein dynamics
40:physical process
7112:
7111:
7107:
7106:
7105:
7103:
7102:
7101:
7092:Protein folding
7077:
7076:
7075:
7066:
6993:
6968:E3 SUMO ligase
6932:
6921:
6913:
6604:
6596:
6569:
6531:
6510:
6502:
6280:
6268:protein folding
6266:
6257:
6252:
6222:
6217:
6161:
6108:
6055:
6050:
6020:
6015:
5979:Fibrous protein
5935:
5909:
5905:Protein methods
5890:Protein folding
5868:
5863:
5833:
5828:
5812:
5798:Ferredoxin fold
5781:
5762:Flavodoxin fold
5740:
5709:
5673:
5664:Protein folding
5647:
5642:
5605:
5600:
5599:
5592:Chemistry World
5584:
5580:
5561:
5557:
5510:
5503:
5488:
5481:
5471:
5469:
5441:
5437:
5428:
5426:
5417:
5413:
5382:
5378:
5331:
5327:
5317:
5315:
5307:
5306:
5302:
5292:
5290:
5286:
5285:
5281:
5271:
5269:
5261:
5260:
5256:
5215:(43): 17442–7.
5201:
5197:
5156:(30): 12330–5.
5142:
5138:
5101:
5097:
5089:
5085:
5084:
5080:
5069:
5065:
5034:
5030:
4999:(1–2): 128–35.
4985:
4981:
4926:
4919:
4872:
4863:
4848:
4826:
4822:
4791:
4787:
4728:
4724:
4692:chem-ph/9411008
4675:
4666:
4658:
4627:
4621:
4617:
4608:
4606:
4598:
4597:
4593:
4572:(48): 8601–24.
4562:
4558:
4536:
4530:
4526:
4485:(44): 17820–5.
4471:
4467:
4414:
4410:
4365:
4361:
4330:
4326:
4287:
4283:
4228:
4224:
4193:
4189:
4182:
4160:
4156:
4101:
4094:
4041:
4034:
4013:(19): 8148–61.
4003:
3994:
3979:
3957:
3950:
3915:
3902:
3865:
3854:
3823:
3819:
3798:(24): 4807–21.
3787:
3783:
3760:(29): 18052–9.
3746:
3742:
3705:
3701:
3670:
3661:
3630:(11): 1881–90.
3616:
3612:
3605:
3589:
3585:
3575:
3573:
3565:
3559:
3552:
3520:
3516:
3477:
3473:
3436:(5607): 713–6.
3426:
3422:
3391:
3384:
3339:
3328:
3273:
3269:
3242:
3235:
3180:
3176:
3145:
3141:
3110:(24): 6927–33.
3096:
3092:
3039:
3035:
2998:
2994:
2949:
2945:
2914:
2910:
2871:
2864:
2827:(6583): 571–9.
2817:
2808:
2761:
2746:
2701:
2697:
2652:
2648:
2617:(6): 0146–146.
2603:
2599:
2555:chem-ph/9512004
2534:
2530:
2467:
2463:
2426:(6995): 101–5.
2416:
2412:
2373:
2369:
2340:(12): 3312–26.
2330:
2326:
2281:
2277:
2268:
2266:
2256:
2252:
2207:
2203:
2172:
2168:
2159:
2157:
2155:
2133:
2129:
2120:
2118:
2109:
2108:
2104:
2097:
2081:
2072:
2036:10.1.1.630.5487
2007:
2003:
1962:(29): 11963–8.
1948:
1944:
1937:
1923:
1904:
1865:
1861:
1800:
1796:
1761:
1757:
1726:
1722:
1691:
1687:
1656:(10): 2029–38.
1642:
1638:
1631:
1617:
1613:
1576:(6968): 900–4.
1566:
1559:
1552:
1530:
1526:
1481:
1477:
1470:
1449:
1445:
1440:
1435:
1371:
1255:
1184:
1157:
1148:
1136:
1134:Biotin painting
1115:
1100:
1076:Martin Gruebele
1055:
1031:
1025:
973:time relaxation
953:
947:
936:
915:
864:
858:
821:
786:
770:
754:cystic fibrosis
673:
667:
651:
624:
533:disulfide bonds
497:
445:
416:
316:
307:
301:
282:in the form of
275:
269:
231:Formation of a
205:
199:
190:
184:
179:
140:proteinopathies
36:Protein folding
17:
12:
11:
5:
7110:
7100:
7099:
7094:
7089:
7072:
7071:
7068:
7067:
7065:
7064:
7058:
7057:
7052:
7047:
7042:
7037:
7032:
7027:
7017:
7012:
7007:
7001:
6999:
6995:
6994:
6992:
6991:
6990:
6989:
6984:
6979:
6974:
6965:
6964:
6963:
6962:
6953:
6952:
6951:
6950:
6945:
6936:
6934:
6923:
6915:
6914:
6912:
6911:
6906:
6901:
6895:
6894:
6889:
6884:
6874:
6873:
6872:
6871:
6866:
6861:
6856:
6851:
6846:
6834:
6833:
6832:
6831:
6826:
6821:
6816:
6811:
6806:
6800:
6795:
6790:
6785:
6780:
6775:
6770:
6765:
6760:
6755:
6750:
6745:
6740:
6735:
6730:
6725:
6720:
6715:
6710:
6705:
6700:
6695:
6690:
6685:
6680:
6675:
6663:
6662:
6661:
6660:
6655:
6650:
6645:
6640:
6635:
6630:
6625:
6620:
6608:
6606:
6598:
6597:
6595:
6594:
6589:
6587:Signal peptide
6583:
6581:
6575:
6574:
6571:
6570:
6568:
6567:
6566:
6565:
6560:
6550:
6545:
6539:
6537:
6533:
6532:
6530:
6529:
6528:
6527:
6522:
6517:
6512:
6508:
6504:
6500:
6490:
6489:
6488:
6487:
6482:
6477:
6472:
6467:
6462:
6457:
6452:
6447:
6442:
6437:
6432:
6427:
6416:
6415:
6414:
6413:
6408:
6403:
6398:
6393:
6388:
6383:
6378:
6373:
6368:
6363:
6358:
6353:
6348:
6343:
6338:
6333:
6328:
6323:
6312:
6311:
6302:
6297:
6292:
6286:
6284:
6270:
6259:
6258:
6251:
6250:
6243:
6236:
6228:
6219:
6218:
6216:
6215:
6210:
6205:
6200:
6195:
6190:
6185:
6180:
6178:Protein domain
6175:
6169:
6167:
6163:
6162:
6160:
6159:
6157:Thermodynamics
6154:
6149:
6144:
6139:
6134:
6129:
6124:
6118:
6116:
6110:
6109:
6107:
6106:
6104:Thermodynamics
6101:
6096:
6091:
6086:
6081:
6076:
6071:
6065:
6063:
6057:
6056:
6049:
6048:
6041:
6034:
6026:
6017:
6016:
6014:
6013:
6012:
6011:
6006:
6001:
5991:
5986:
5981:
5976:
5975:
5974:
5969:
5964:
5954:
5949:
5943:
5941:
5937:
5936:
5934:
5933:
5928:
5923:
5917:
5915:
5911:
5910:
5908:
5907:
5902:
5897:
5892:
5887:
5882:
5876:
5874:
5870:
5869:
5862:
5861:
5854:
5847:
5839:
5830:
5829:
5827:
5826:
5820:
5818:
5814:
5813:
5811:
5810:
5805:
5803:Ribonuclease A
5800:
5795:
5789:
5787:
5783:
5782:
5780:
5779:
5774:
5769:
5764:
5759:
5754:
5748:
5746:
5742:
5741:
5739:
5738:
5733:
5731:Beta-propeller
5728:
5723:
5717:
5715:
5711:
5710:
5708:
5707:
5702:
5700:Alpha solenoid
5697:
5692:
5687:
5681:
5679:
5675:
5674:
5672:
5671:
5666:
5661:
5655:
5653:
5649:
5648:
5641:
5640:
5633:
5626:
5618:
5612:
5611:
5604:
5603:External links
5601:
5598:
5597:
5578:
5571:) – via
5555:
5501:
5479:
5435:
5411:
5376:
5325:
5300:
5279:
5263:"Rosetta@home"
5254:
5195:
5136:
5095:
5078:
5063:
5028:
4979:
4917:
4861:
4846:
4820:
4785:
4746:(18): 8721–5.
4722:
4664:
4661:on 2009-09-02.
4615:
4591:
4556:
4550:10.1101/274761
4524:
4465:
4408:
4359:
4324:
4291:Nature Methods
4281:
4242:(10): e46147.
4222:
4187:
4180:
4154:
4092:
4055:(6): 2045–54.
4032:
3992:
3977:
3948:
3900:
3852:
3833:(5): 1769–84.
3817:
3781:
3740:
3699:
3659:
3610:
3603:
3583:
3550:
3531:(12): 911–21.
3514:
3471:
3420:
3382:
3353:(7): 1331–49.
3326:
3267:
3256:(4): 399–406.
3233:
3174:
3155:(7): 395–401.
3139:
3090:
3033:
2992:
2943:
2908:
2862:
2806:
2744:
2695:
2666:(1): 303–313.
2646:
2597:
2548:(18): 9533–8.
2528:
2461:
2410:
2367:
2324:
2275:
2250:
2221:(15): 3870–5.
2201:
2166:
2153:
2127:
2102:
2095:
2070:
2001:
1942:
1935:
1902:
1859:
1794:
1755:
1720:
1685:
1636:
1629:
1611:
1557:
1550:
1524:
1475:
1468:
1442:
1441:
1439:
1436:
1434:
1433:
1428:
1423:
1418:
1413:
1408:
1403:
1398:
1393:
1388:
1383:
1378:
1372:
1370:
1367:
1254:
1251:
1223:folding funnel
1183:
1180:
1156:
1153:
1147:
1144:
1135:
1132:
1114:
1111:
1099:
1096:
1080:Sheena Radford
1054:
1051:
1027:Main article:
1024:
1021:
949:Main article:
946:
943:
934:
914:
911:
860:Main article:
857:
854:
829:quantum yields
820:
817:
785:
782:
769:
766:
714:polyneuropathy
669:Main article:
666:
663:
650:
649:Fold switching
647:
613:The bacterium
591:, extremes of
496:
493:
415:
412:
368:macromolecules
366:. While these
346:, so that the
332:hydrogen bonds
315:
312:
303:Main article:
300:
297:
271:Main article:
268:
265:
248:hydrogen bonds
245:intramolecular
201:Main article:
198:
195:
186:Main article:
183:
180:
178:
175:
128:unfolded state
15:
9:
6:
4:
3:
2:
7109:
7098:
7095:
7093:
7090:
7088:
7085:
7084:
7082:
7063:
7060:
7059:
7056:
7053:
7051:
7048:
7046:
7043:
7041:
7038:
7036:
7033:
7031:
7028:
7025:
7021:
7018:
7016:
7013:
7011:
7008:
7006:
7003:
7002:
7000:
6996:
6988:
6985:
6983:
6980:
6978:
6975:
6973:
6970:
6969:
6967:
6966:
6961:
6958:
6957:
6955:
6954:
6949:
6946:
6944:
6941:
6940:
6938:
6937:
6935:
6933:(SUMOylation)
6931:
6927:
6924:
6920:
6916:
6910:
6907:
6905:
6902:
6900:
6897:
6896:
6893:
6890:
6888:
6885:
6883:
6879:
6876:
6875:
6870:
6867:
6865:
6862:
6860:
6857:
6855:
6852:
6850:
6847:
6845:
6842:
6841:
6840:
6836:
6835:
6830:
6827:
6825:
6822:
6820:
6817:
6815:
6812:
6810:
6807:
6804:
6801:
6799:
6796:
6794:
6791:
6789:
6786:
6784:
6781:
6779:
6776:
6774:
6771:
6769:
6766:
6764:
6761:
6759:
6756:
6754:
6751:
6749:
6746:
6744:
6741:
6739:
6736:
6734:
6731:
6729:
6726:
6724:
6721:
6719:
6716:
6714:
6711:
6709:
6706:
6704:
6701:
6699:
6696:
6694:
6691:
6689:
6686:
6684:
6681:
6679:
6676:
6674:
6671:
6670:
6669:
6665:
6664:
6659:
6656:
6654:
6651:
6649:
6646:
6644:
6641:
6639:
6636:
6634:
6631:
6629:
6626:
6624:
6621:
6619:
6616:
6615:
6614:
6610:
6609:
6607:
6603:
6599:
6593:
6590:
6588:
6585:
6584:
6582:
6580:
6576:
6564:
6561:
6559:
6556:
6555:
6554:
6551:
6549:
6546:
6544:
6541:
6540:
6538:
6534:
6526:
6523:
6521:
6518:
6516:
6513:
6511:
6505:
6503:
6497:
6496:
6495:
6492:
6491:
6486:
6483:
6481:
6478:
6476:
6473:
6471:
6468:
6466:
6463:
6461:
6458:
6456:
6453:
6451:
6448:
6446:
6443:
6441:
6438:
6436:
6433:
6431:
6428:
6426:
6423:
6422:
6421:
6418:
6417:
6412:
6409:
6407:
6404:
6402:
6399:
6397:
6394:
6392:
6389:
6387:
6384:
6382:
6379:
6377:
6374:
6372:
6369:
6367:
6364:
6362:
6359:
6357:
6354:
6352:
6349:
6347:
6344:
6342:
6339:
6337:
6334:
6332:
6329:
6327:
6324:
6322:
6319:
6318:
6317:
6314:
6313:
6310:
6306:
6303:
6301:
6298:
6296:
6293:
6291:
6288:
6287:
6285:
6283:
6278:
6274:
6271:
6269:
6264:
6260:
6256:
6249:
6244:
6242:
6237:
6235:
6230:
6229:
6226:
6214:
6211:
6209:
6206:
6204:
6201:
6199:
6196:
6194:
6191:
6189:
6186:
6184:
6181:
6179:
6176:
6174:
6171:
6170:
6168:
6164:
6158:
6155:
6153:
6150:
6148:
6145:
6143:
6142:Determination
6140:
6138:
6135:
6133:
6130:
6128:
6125:
6123:
6120:
6119:
6117:
6115:
6111:
6105:
6102:
6100:
6097:
6095:
6092:
6090:
6089:Determination
6087:
6085:
6082:
6080:
6077:
6075:
6072:
6070:
6067:
6066:
6064:
6062:
6058:
6054:
6047:
6042:
6040:
6035:
6033:
6028:
6027:
6024:
6010:
6007:
6005:
6002:
6000:
5997:
5996:
5995:
5992:
5990:
5987:
5985:
5984:Chromoprotein
5982:
5980:
5977:
5973:
5970:
5968:
5965:
5963:
5960:
5959:
5958:
5955:
5953:
5950:
5948:
5945:
5944:
5942:
5938:
5932:
5929:
5927:
5924:
5922:
5919:
5918:
5916:
5912:
5906:
5903:
5901:
5898:
5896:
5893:
5891:
5888:
5886:
5883:
5881:
5878:
5877:
5875:
5871:
5867:
5860:
5855:
5853:
5848:
5846:
5841:
5840:
5837:
5825:
5822:
5821:
5819:
5815:
5809:
5808:SH2-like fold
5806:
5804:
5801:
5799:
5796:
5794:
5791:
5790:
5788:
5784:
5778:
5775:
5773:
5770:
5768:
5767:Rossmann fold
5765:
5763:
5760:
5758:
5755:
5753:
5750:
5749:
5747:
5743:
5737:
5734:
5732:
5729:
5727:
5724:
5722:
5719:
5718:
5716:
5712:
5706:
5703:
5701:
5698:
5696:
5693:
5691:
5688:
5686:
5683:
5682:
5680:
5676:
5670:
5667:
5665:
5662:
5660:
5657:
5656:
5654:
5650:
5646:
5639:
5634:
5632:
5627:
5625:
5620:
5619:
5616:
5610:
5607:
5606:
5593:
5589:
5582:
5574:
5570:
5566:
5559:
5551:
5547:
5543:
5539:
5535:
5531:
5527:
5523:
5519:
5515:
5508:
5506:
5498:
5497:
5492:
5486:
5484:
5468:
5464:
5459:
5454:
5450:
5446:
5439:
5425:
5422:
5415:
5407:
5403:
5399:
5395:
5391:
5387:
5380:
5372:
5368:
5364:
5360:
5356:
5352:
5348:
5344:
5340:
5336:
5329:
5314:
5310:
5304:
5289:
5283:
5268:
5264:
5258:
5250:
5246:
5241:
5236:
5231:
5226:
5222:
5218:
5214:
5210:
5206:
5199:
5191:
5187:
5182:
5177:
5172:
5167:
5163:
5159:
5155:
5151:
5147:
5140:
5132:
5128:
5123:
5118:
5114:
5110:
5106:
5099:
5088:
5082:
5074:
5067:
5059:
5055:
5051:
5047:
5044:(3): 835–48.
5043:
5039:
5032:
5024:
5020:
5015:
5010:
5006:
5002:
4998:
4994:
4990:
4983:
4975:
4971:
4966:
4961:
4956:
4951:
4947:
4943:
4940:(4): 1525–9.
4939:
4935:
4931:
4924:
4922:
4913:
4909:
4905:
4901:
4897:
4893:
4889:
4885:
4881:
4877:
4870:
4868:
4866:
4857:
4853:
4849:
4843:
4839:
4835:
4831:
4824:
4816:
4812:
4808:
4804:
4801:(6): 851–62.
4800:
4796:
4789:
4781:
4777:
4772:
4767:
4762:
4757:
4753:
4749:
4745:
4741:
4737:
4733:
4726:
4718:
4714:
4710:
4706:
4702:
4698:
4693:
4688:
4685:(3): 167–95.
4684:
4680:
4673:
4671:
4669:
4657:
4653:
4649:
4645:
4641:
4637:
4633:
4626:
4619:
4605:
4601:
4595:
4587:
4583:
4579:
4575:
4571:
4567:
4560:
4551:
4546:
4542:
4535:
4528:
4520:
4516:
4511:
4506:
4501:
4496:
4492:
4488:
4484:
4480:
4476:
4469:
4461:
4457:
4452:
4447:
4443:
4439:
4435:
4431:
4427:
4423:
4419:
4412:
4404:
4400:
4395:
4390:
4386:
4382:
4379:(11): 860–9.
4378:
4374:
4370:
4363:
4355:
4351:
4347:
4343:
4340:(1): 660–76.
4339:
4335:
4328:
4320:
4316:
4312:
4308:
4304:
4300:
4297:(3): 207–12.
4296:
4292:
4285:
4277:
4273:
4268:
4263:
4258:
4253:
4249:
4245:
4241:
4237:
4233:
4226:
4218:
4214:
4210:
4206:
4203:(4): 865–73.
4202:
4198:
4191:
4183:
4177:
4173:
4169:
4165:
4158:
4150:
4146:
4141:
4136:
4131:
4126:
4122:
4118:
4114:
4110:
4106:
4099:
4097:
4088:
4084:
4079:
4074:
4070:
4066:
4062:
4058:
4054:
4050:
4046:
4039:
4037:
4028:
4024:
4020:
4016:
4012:
4008:
4001:
3999:
3997:
3988:
3984:
3980:
3978:9780124116368
3974:
3970:
3966:
3962:
3955:
3953:
3944:
3940:
3936:
3932:
3928:
3924:
3920:
3913:
3911:
3909:
3907:
3905:
3896:
3892:
3887:
3882:
3878:
3874:
3870:
3863:
3861:
3859:
3857:
3848:
3844:
3840:
3836:
3832:
3828:
3821:
3813:
3809:
3805:
3801:
3797:
3793:
3785:
3777:
3773:
3768:
3763:
3759:
3755:
3751:
3744:
3736:
3732:
3727:
3722:
3719:(9): 445–56.
3718:
3714:
3710:
3703:
3695:
3691:
3687:
3683:
3679:
3675:
3668:
3666:
3664:
3655:
3651:
3646:
3641:
3637:
3633:
3629:
3625:
3621:
3614:
3606:
3600:
3596:
3595:
3587:
3571:
3564:
3557:
3555:
3546:
3542:
3538:
3534:
3530:
3526:
3518:
3510:
3506:
3502:
3498:
3494:
3490:
3486:
3482:
3475:
3467:
3463:
3459:
3455:
3451:
3447:
3443:
3439:
3435:
3431:
3424:
3416:
3412:
3408:
3404:
3400:
3396:
3389:
3387:
3378:
3374:
3370:
3366:
3361:
3356:
3352:
3348:
3344:
3337:
3335:
3333:
3331:
3322:
3318:
3313:
3308:
3303:
3298:
3294:
3290:
3286:
3282:
3278:
3271:
3263:
3259:
3255:
3251:
3247:
3240:
3238:
3229:
3225:
3220:
3215:
3210:
3205:
3201:
3197:
3193:
3189:
3185:
3178:
3170:
3166:
3162:
3158:
3154:
3150:
3143:
3135:
3131:
3126:
3121:
3117:
3113:
3109:
3105:
3101:
3094:
3086:
3082:
3077:
3072:
3068:
3064:
3060:
3056:
3052:
3048:
3044:
3037:
3029:
3025:
3020:
3015:
3012:(3): 259–65.
3011:
3007:
3003:
2996:
2988:
2984:
2980:
2976:
2971:
2966:
2962:
2958:
2957:FASEB Journal
2954:
2947:
2939:
2935:
2931:
2927:
2923:
2919:
2912:
2904:
2900:
2896:
2892:
2888:
2884:
2880:
2876:
2869:
2867:
2858:
2854:
2850:
2846:
2842:
2838:
2834:
2830:
2826:
2822:
2815:
2813:
2811:
2802:
2798:
2794:
2790:
2786:
2782:
2778:
2774:
2770:
2766:
2759:
2757:
2755:
2753:
2751:
2749:
2740:
2736:
2731:
2726:
2722:
2718:
2714:
2710:
2706:
2699:
2691:
2687:
2682:
2677:
2673:
2669:
2665:
2661:
2657:
2650:
2642:
2638:
2633:
2628:
2624:
2620:
2616:
2612:
2608:
2601:
2593:
2589:
2584:
2579:
2574:
2569:
2565:
2561:
2556:
2551:
2547:
2543:
2539:
2532:
2524:
2520:
2515:
2510:
2506:
2502:
2498:
2494:
2489:
2484:
2481:(5): 2252–8.
2480:
2476:
2472:
2465:
2457:
2453:
2449:
2445:
2441:
2437:
2433:
2429:
2425:
2421:
2414:
2406:
2402:
2398:
2394:
2390:
2386:
2382:
2378:
2371:
2363:
2359:
2355:
2351:
2347:
2343:
2339:
2335:
2328:
2320:
2316:
2312:
2308:
2303:
2298:
2294:
2290:
2289:FASEB Journal
2286:
2279:
2265:
2261:
2254:
2246:
2242:
2237:
2232:
2228:
2224:
2220:
2216:
2212:
2205:
2197:
2193:
2189:
2185:
2181:
2177:
2170:
2156:
2150:
2145:
2144:
2138:
2131:
2116:
2112:
2106:
2098:
2092:
2089:. Macmillan.
2088:
2087:
2079:
2077:
2075:
2066:
2062:
2057:
2052:
2047:
2042:
2037:
2032:
2028:
2024:
2020:
2016:
2012:
2005:
1997:
1993:
1988:
1983:
1978:
1973:
1969:
1965:
1961:
1957:
1953:
1946:
1938:
1932:
1928:
1921:
1919:
1917:
1915:
1913:
1911:
1909:
1907:
1898:
1894:
1890:
1886:
1882:
1878:
1874:
1870:
1863:
1855:
1851:
1847:
1843:
1838:
1833:
1829:
1825:
1821:
1817:
1813:
1809:
1805:
1798:
1790:
1786:
1782:
1778:
1774:
1770:
1766:
1759:
1751:
1747:
1743:
1739:
1736:(4): R81-91.
1735:
1731:
1724:
1716:
1712:
1708:
1704:
1700:
1696:
1689:
1681:
1677:
1672:
1667:
1663:
1659:
1655:
1651:
1647:
1640:
1632:
1626:
1622:
1615:
1607:
1603:
1599:
1595:
1591:
1587:
1583:
1579:
1575:
1571:
1564:
1562:
1553:
1547:
1543:
1539:
1535:
1528:
1520:
1516:
1511:
1506:
1502:
1498:
1495:(4): 737–49.
1494:
1490:
1486:
1479:
1471:
1465:
1461:
1457:
1453:
1447:
1443:
1432:
1429:
1427:
1424:
1422:
1419:
1417:
1414:
1412:
1409:
1407:
1404:
1402:
1399:
1397:
1394:
1392:
1389:
1387:
1384:
1382:
1379:
1377:
1374:
1373:
1366:
1364:
1360:
1354:
1352:
1348:
1344:
1340:
1336:
1331:
1329:
1325:
1321:
1316:
1314:
1310:
1306:
1301:
1299:
1295:
1291:
1287:
1283:
1279:
1278:
1273:
1272:
1264:
1259:
1250:
1247:
1242:
1238:
1236:
1232:
1228:
1224:
1220:
1215:
1213:
1209:
1205:
1204:Peter Wolynes
1201:
1197:
1188:
1179:
1177:
1172:
1168:
1163:
1152:
1143:
1141:
1131:
1129:
1125:
1120:
1110:
1108:
1104:
1095:
1093:
1092:Bengt Nölting
1089:
1085:
1081:
1077:
1073:
1069:
1065:
1061:
1050:
1048:
1044:
1039:
1035:
1030:
1020:
1018:
1014:
1009:
1007:
1002:
999:
994:
984:
980:
978:
974:
970:
966:
962:
958:
952:
942:
940:
932:
928:
924:
920:
910:
908:
907:chevron plots
904:
900:
896:
892:
888:
884:
880:
876:
875:alpha helices
872:
868:
863:
853:
851:
848:and derive a
847:
843:
838:
833:
830:
825:
816:
814:
810:
809:phase problem
806:
801:
795:
790:
781:
779:
775:
765:
763:
759:
755:
751:
747:
743:
738:
734:
730:
727:
723:
719:
715:
711:
707:
703:
699:
695:
690:
688:
683:
678:
672:
662:
660:
656:
646:
644:
640:
636:
632:
627:
622:
618:
617:
611:
609:
605:
601:
596:
594:
590:
585:
581:
580:precipitation
576:
572:
568:
564:
560:
555:
553:
549:
545:
544:
538:
534:
530:
526:
522:
518:
517:
512:
506:
501:
492:
490:
486:
480:
477:
473:
469:
465:
443:
441:
437:
433:
424:
420:
411:
409:
403:
401:
397:
393:
389:
385:
384:lipid bilayer
381:
377:
373:
369:
365:
361:
357:
353:
349:
345:
341:
337:
333:
329:
326:Folding is a
320:
311:
306:
296:
293:
289:
285:
281:
274:
264:
262:
257:
253:
252:Linus Pauling
249:
246:
242:
238:
237:alpha helices
234:
226:
221:
214:
209:
204:
194:
189:
174:
172:
167:
165:
161:
160:contact order
156:
152:
148:
143:
141:
137:
133:
129:
125:
121:
119:
115:
114:immune system
111:
107:
103:
100:
96:
92:
88:
84:
79:
77:
73:
67:
65:
61:
57:
53:
49:
45:
41:
37:
29:
21:
6930:SUMO protein
6267:
6193:Nucleic acid
6114:Nucleic acid
5889:
5714:All-β folds:
5685:Helix bundle
5678:All-α folds:
5663:
5591:
5581:
5558:
5517:
5513:
5494:
5470:. Retrieved
5448:
5438:
5427:. Retrieved
5423:
5414:
5392:(1): 43–48.
5389:
5386:J. Mol. Biol
5385:
5379:
5338:
5334:
5328:
5316:. Retrieved
5312:
5303:
5291:. Retrieved
5282:
5270:. Retrieved
5266:
5257:
5212:
5208:
5198:
5153:
5149:
5139:
5112:
5108:
5098:
5081:
5066:
5041:
5037:
5031:
4996:
4992:
4982:
4937:
4933:
4879:
4875:
4829:
4823:
4798:
4794:
4788:
4743:
4739:
4725:
4682:
4678:
4656:the original
4635:
4631:
4618:
4607:. Retrieved
4603:
4594:
4569:
4566:Biochemistry
4565:
4559:
4540:
4527:
4482:
4478:
4468:
4425:
4421:
4411:
4376:
4372:
4362:
4337:
4333:
4327:
4294:
4290:
4284:
4239:
4235:
4225:
4200:
4196:
4190:
4163:
4157:
4112:
4108:
4052:
4048:
4010:
4006:
3960:
3926:
3922:
3876:
3872:
3830:
3826:
3820:
3795:
3792:Biochemistry
3791:
3784:
3757:
3753:
3743:
3716:
3712:
3702:
3677:
3673:
3627:
3623:
3613:
3593:
3586:
3574:. Retrieved
3569:
3528:
3524:
3517:
3484:
3480:
3474:
3433:
3429:
3423:
3401:(3): 150–5.
3398:
3394:
3350:
3346:
3284:
3280:
3270:
3253:
3249:
3191:
3187:
3177:
3152:
3148:
3142:
3107:
3103:
3093:
3053:(2): 595–9.
3050:
3046:
3036:
3009:
3005:
2995:
2963:(1): 27–34.
2960:
2956:
2946:
2921:
2917:
2911:
2878:
2874:
2824:
2820:
2768:
2764:
2712:
2708:
2698:
2663:
2659:
2649:
2614:
2610:
2600:
2545:
2541:
2531:
2478:
2474:
2464:
2423:
2419:
2413:
2380:
2376:
2370:
2337:
2333:
2327:
2295:(1): 75–83.
2292:
2288:
2278:
2267:. Retrieved
2263:
2253:
2218:
2214:
2204:
2182:(1): 25–31.
2179:
2175:
2169:
2158:. Retrieved
2142:
2130:
2119:. Retrieved
2114:
2105:
2085:
2018:
2014:
2004:
1959:
1955:
1945:
1926:
1872:
1868:
1862:
1837:1887/3277889
1811:
1807:
1797:
1775:(1): 76–88.
1772:
1768:
1758:
1733:
1729:
1723:
1698:
1694:
1688:
1653:
1649:
1639:
1620:
1614:
1573:
1569:
1542:Biochemistry
1541:
1527:
1492:
1488:
1478:
1459:
1446:
1381:Chevron plot
1355:
1332:
1317:
1309:Folding@home
1305:Rosetta@home
1302:
1275:
1269:
1268:
1243:
1239:
1219:José Onuchic
1216:
1211:
1207:
1193:
1158:
1149:
1137:
1116:
1101:
1084:Chris Dobson
1056:
1038:conformation
1032:
1010:
1003:
990:
954:
916:
899:stopped flow
865:
846:chevron plot
842:stopped flow
834:
822:
798:
771:
748:-associated
718:Huntington's
691:
674:
652:
630:
614:
612:
597:
556:
547:
541:
536:
514:
509:
481:
460:
428:
404:
364:biosynthesis
325:
308:
276:
261:peptide bond
230:
191:
168:
144:
124:Denaturation
122:
80:
72:native state
68:
35:
34:
7024:neddylation
6290:Hsp10/GroES
6282:Chaperonins
6004:Phytochrome
5994:Biliprotein
5726:Beta barrel
5690:Globin fold
4373:Biopolymers
3576:November 3,
1426:Proteopathy
1212:frustration
1103:Proteolysis
1098:Proteolysis
1088:Alan Fersht
1047:temperature
951:Protein NMR
891:free energy
887:temperature
879:beta sheets
746:antitrypsin
742:proteopathy
671:Proteopathy
559:random coil
476:amphiphilic
396:temperature
356:synthesized
292:topological
241:beta sheets
213:alpha helix
60:random coil
56:amino acids
42:by which a
7081:Categories
6316:Hsp40/DnaJ
6263:Chaperones
6188:Proteasome
6147:Prediction
6137:Quaternary
6094:Prediction
6084:Quaternary
5931:Proteasome
5914:Structures
5786:α+β folds:
5752:TIM barrel
5745:α/β folds:
5736:Beta helix
5705:Death fold
5429:2020-11-30
4732:Onuchic JN
4609:2016-11-05
3487:: 333–66.
2924:: 323–55.
2269:2016-11-26
2160:2016-11-26
2121:2016-11-26
1701:: 631–60.
1438:References
1171:picosecond
1167:nanosecond
1128:SH3 domain
1072:Harry Gray
987:timescale.
495:Chaperones
400:chaperones
352:C-terminal
348:N-terminus
118:antibodies
93:and other
6602:Ubiquitin
6548:Clusterin
6127:Secondary
6074:Secondary
6009:Lipocalin
5873:Processes
5824:Conotoxin
5793:DNA clamp
5550:227243204
5467:247206999
5071:Jones D.
4638:: 44–45.
3929:: 52–77.
3680:: 29–37.
3674:Biochimie
2147:. Wiley.
2031:CiteSeerX
1854:237583577
1452:Alberts B
1335:AlphaFold
1290:in silico
1277:ab initio
998:spin echo
792:Steps of
750:emphysema
737:Tafamidis
677:misfolded
604:egg white
584:insoluble
565:known as
110:allergies
48:synthesis
6882:Ataxin 3
6166:See also
6132:Tertiary
6079:Tertiary
5962:Globulin
5900:Proteome
5866:Proteins
5542:33257889
5472:25 March
5406:20974152
5371:27988268
5363:22034434
5318:14 March
5309:"Foldit"
5293:14 March
5272:14 March
5249:23045636
5190:17636132
5131:27333362
5023:23266569
4974:10677494
4904:23180855
4856:19121702
4717:13838095
4679:Proteins
4586:24187909
4519:22949695
4460:21750539
4403:23784721
4354:24001118
4319:21364478
4311:15782190
4276:23056252
4236:PLOS ONE
4217:11575938
4149:27791136
4087:19289032
4027:22554188
3987:23954103
3943:28552172
3847:16683754
3812:22640394
3735:16087653
3694:26607240
3654:14573942
3545:16359163
3509:23797549
3501:16756495
3466:30829998
3458:12560553
3415:16473510
3377:23370420
3369:16689923
3321:29784778
3228:18664583
3169:16716593
3134:10601015
3085:20643079
3028:15943899
2987:24066207
2938:23746257
2895:21776078
2793:14685248
2739:30692195
2709:Genetics
2690:30026186
2660:Genetics
2641:28642936
2523:11053106
2448:15229605
2362:27113763
2354:25204743
2334:Proteins
2319:20021399
2245:10921869
2196:21111607
2115:Scitable
2065:17075053
1996:17609385
1846:34545868
1789:15102453
1680:33142107
1598:14685251
1536:(2002).
1534:Stryer L
1369:See also
1343:DeepMind
1060:dynamics
955:Protein
903:kinetics
756:and the
682:β-sheets
575:bacteria
548:in vivo.
543:in vitro
521:catalyst
436:enthalpy
360:ribosome
288:cysteine
256:backbone
95:diseases
52:ribosome
46:, after
6805:(CDC34)
6173:Protein
6122:Primary
6069:Primary
6061:Protein
5972:Albumin
5967:Edestin
5652:General
5573:Twitter
5522:Bibcode
5496:Science
5343:Bibcode
5335:Science
5313:fold.it
5240:3491489
5217:Bibcode
5181:1941469
5158:Bibcode
5058:9826519
5014:4084838
4942:Bibcode
4912:5756068
4884:Bibcode
4876:Science
4803:Bibcode
4780:1528885
4748:Bibcode
4709:7784423
4640:Bibcode
4541:bioRxiv
4510:3497811
4487:Bibcode
4451:3144584
4430:Bibcode
4428:: 385.
4394:4065244
4267:3463568
4244:Bibcode
4140:5111666
4117:Bibcode
4078:2717354
4057:Bibcode
3895:2266107
3776:9660761
3632:Bibcode
3438:Bibcode
3430:Science
3312:6003340
3289:Bibcode
3262:9556522
3219:2490668
3196:Bibcode
3125:1171756
3076:2905109
3055:Bibcode
2979:8566543
2903:4337671
2857:4347271
2849:8637592
2829:Bibcode
2801:1036192
2773:Bibcode
2730:6456324
2681:6116962
2632:5476217
2592:8790365
2560:Bibcode
2514:1301114
2493:Bibcode
2456:4315026
2428:Bibcode
2385:Bibcode
2377:Science
2311:8566551
2056:1636505
2023:Bibcode
1987:1906725
1964:Bibcode
1897:4124164
1877:Bibcode
1869:Science
1816:Bibcode
1750:9710577
1715:2197986
1671:7732725
1606:6451881
1578:Bibcode
1519:4565129
1510:1173893
1271:De novo
967:,
937:O), or
807:, the "
729:fibrils
726:amyloid
687:amyloid
631:E. coli
616:E. coli
589:solutes
563:enzymes
537:in vivo
516:in vivo
485:de novo
470:). The
440:entropy
376:solvent
358:by the
132:cooking
108:. Many
102:fibrils
99:amyloid
44:protein
38:is the
6849:Cullin
6152:Design
6099:Design
5548:
5540:
5514:Nature
5465:
5404:
5369:
5361:
5247:
5237:
5188:
5178:
5129:
5056:
5021:
5011:
4972:
4962:
4910:
4902:
4854:
4844:
4778:
4768:
4715:
4707:
4584:
4517:
4507:
4458:
4448:
4401:
4391:
4352:
4317:
4309:
4274:
4264:
4215:
4178:
4147:
4137:
4085:
4075:
4025:
3985:
3975:
3941:
3893:
3845:
3810:
3774:
3733:
3692:
3652:
3601:
3543:
3507:
3499:
3464:
3456:
3413:
3375:
3367:
3319:
3309:
3260:
3226:
3216:
3167:
3132:
3122:
3083:
3073:
3026:
2985:
2977:
2936:
2901:
2893:
2875:Nature
2855:
2847:
2821:Nature
2799:
2791:
2765:Nature
2737:
2727:
2688:
2678:
2639:
2629:
2590:
2580:
2521:
2511:
2454:
2446:
2420:Nature
2405:653353
2403:
2360:
2352:
2317:
2309:
2243:
2236:306593
2233:
2194:
2151:
2093:
2063:
2053:
2033:
1994:
1984:
1933:
1895:
1852:
1844:
1787:
1748:
1713:
1678:
1668:
1627:
1604:
1596:
1570:Nature
1548:
1517:
1507:
1466:
1313:Foldit
1235:design
626:P17313
394:, the
390:, the
162:, and
155:domain
106:prions
7040:ATG12
7030:FAT10
7020:NEDD8
7005:ISG15
6998:Other
6987:PIAS4
6982:PIAS3
6977:PIAS2
6972:PIAS1
6922:(UBL)
6904:BIRC6
6864:FANCL
6536:Other
6525:TRAP1
6494:Hsp90
6420:Hsp70
6309:GroEL
6305:HSP60
6300:Hsp47
6295:Hsp27
5940:Types
5569:Tweet
5546:S2CID
5463:S2CID
5367:S2CID
4965:26468
4908:S2CID
4771:49992
4713:S2CID
4687:arXiv
4659:(PDF)
4628:(PDF)
4537:(PDF)
4315:S2CID
3566:(PDF)
3505:S2CID
3462:S2CID
3373:S2CID
2983:S2CID
2899:S2CID
2853:S2CID
2797:S2CID
2583:38463
2550:arXiv
2483:arXiv
2452:S2CID
2358:S2CID
2315:S2CID
1850:S2CID
1602:S2CID
1337:, an
1324:ASICs
1320:Anton
965:TOCSY
931:FT-IR
694:prion
643:GroEL
639:virus
635:GroES
582:into
388:salts
380:water
136:burns
50:by a
7055:UBL5
7045:FUB1
7035:ATG8
7015:UFM1
7010:URM1
6960:UBC9
6948:SAE2
6943:SAE1
6909:UFC1
6899:ATG3
6892:CYLD
6887:USP6
6869:UBR1
6859:MDM2
6658:SAE1
6653:NAE1
6648:ATG7
6643:UBA7
6638:UBA6
6633:UBA5
6628:UBA3
6623:UBA2
6618:UBA1
6563:SMN2
6558:SMN1
5538:PMID
5474:2022
5424:CNBC
5402:PMID
5359:PMID
5320:2023
5295:2023
5274:2023
5245:PMID
5186:PMID
5127:PMID
5054:PMID
5019:PMID
4970:PMID
4900:PMID
4852:PMID
4842:ISBN
4776:PMID
4705:PMID
4582:PMID
4515:PMID
4456:PMID
4399:PMID
4350:PMID
4307:PMID
4272:PMID
4213:PMID
4176:ISBN
4145:PMID
4083:PMID
4023:PMID
3983:PMID
3973:ISBN
3939:PMID
3891:PMID
3843:PMID
3808:PMID
3772:PMID
3731:PMID
3690:PMID
3650:PMID
3599:ISBN
3578:2016
3541:PMID
3497:PMID
3454:PMID
3411:PMID
3365:PMID
3317:PMID
3258:PMID
3224:PMID
3165:PMID
3130:PMID
3081:PMID
3024:PMID
2975:PMID
2934:PMID
2891:PMID
2845:PMID
2789:PMID
2735:PMID
2686:PMID
2637:PMID
2588:PMID
2519:PMID
2444:PMID
2401:PMID
2350:PMID
2307:PMID
2241:PMID
2192:PMID
2149:ISBN
2091:ISBN
2061:PMID
1992:PMID
1931:ISBN
1893:PMID
1842:PMID
1785:PMID
1746:PMID
1711:PMID
1676:PMID
1625:ISBN
1594:PMID
1546:ISBN
1515:PMID
1464:ISBN
1347:CASP
1311:and
1233:and
1194:The
1017:SOD1
969:HSQC
961:COSY
877:and
720:and
708:and
527:and
438:and
239:and
211:The
7050:MUB
6854:CBL
6844:VHL
6837:E3
6666:E2
6611:E1
6480:12A
6411:C19
6406:C14
6401:C13
6396:C11
6391:C10
6346:B11
6203:RNA
6198:DNA
5530:doi
5518:588
5453:doi
5394:doi
5390:405
5351:doi
5339:334
5235:PMC
5225:doi
5213:109
5176:PMC
5166:doi
5154:104
5117:doi
5113:116
5046:doi
5042:284
5009:PMC
5001:doi
4997:531
4960:PMC
4950:doi
4892:doi
4880:338
4834:doi
4811:doi
4799:388
4766:PMC
4756:doi
4697:doi
4648:doi
4574:doi
4545:doi
4505:PMC
4495:doi
4483:109
4446:PMC
4438:doi
4389:PMC
4381:doi
4342:doi
4338:114
4299:doi
4262:PMC
4252:doi
4205:doi
4201:312
4168:doi
4135:PMC
4125:doi
4113:113
4073:PMC
4065:doi
4015:doi
4011:134
3965:doi
3931:doi
3927:100
3881:doi
3877:265
3835:doi
3831:106
3800:doi
3762:doi
3758:273
3721:doi
3682:doi
3678:121
3640:doi
3533:doi
3489:doi
3446:doi
3434:299
3403:doi
3355:doi
3351:273
3307:PMC
3297:doi
3285:115
3214:PMC
3204:doi
3192:105
3157:doi
3120:PMC
3112:doi
3071:PMC
3063:doi
3014:doi
2965:doi
2926:doi
2883:doi
2879:475
2837:doi
2825:381
2781:doi
2769:426
2725:PMC
2717:doi
2713:211
2676:PMC
2668:doi
2664:210
2627:PMC
2619:doi
2578:PMC
2568:doi
2509:PMC
2501:doi
2436:doi
2424:430
2393:doi
2381:200
2342:doi
2297:doi
2231:PMC
2223:doi
2184:doi
2051:PMC
2041:doi
2019:103
1982:PMC
1972:doi
1960:104
1885:doi
1873:181
1832:hdl
1824:doi
1777:doi
1738:doi
1703:doi
1666:PMC
1658:doi
1654:119
1586:doi
1574:426
1505:PMC
1497:doi
1493:128
1274:or
1169:or
977:NOE
712:or
382:or
7083::
6880::
6824:V2
6819:V1
6809:R2
6803:R1
6798:Q2
6793:Q1
6773:L6
6768:L4
6763:L3
6758:L2
6753:L1
6743:J2
6738:J1
6723:G2
6718:G1
6713:E3
6708:E2
6703:E1
6698:D3
6693:D2
6688:D1
6520:ER
6485:14
6450:4L
6435:1L
6430:1B
6425:1A
6386:C7
6381:C6
6376:C5
6371:C3
6366:C1
6361:B9
6356:B6
6351:B4
6341:B2
6336:B1
6331:A3
6326:A2
6321:A1
5590:.
5544:.
5536:.
5528:.
5516:.
5504:^
5493:,
5482:^
5461:.
5451:.
5447:.
5400:.
5388:.
5365:.
5357:.
5349:.
5337:.
5311:.
5265:.
5243:.
5233:.
5223:.
5211:.
5207:.
5184:.
5174:.
5164:.
5152:.
5148:.
5125:.
5111:.
5107:.
5052:.
5040:.
5017:.
5007:.
4995:.
4991:.
4968:.
4958:.
4948:.
4938:97
4936:.
4932:.
4920:^
4906:.
4898:.
4890:.
4878:.
4864:^
4850:.
4840:.
4809:.
4797:.
4774:.
4764:.
4754:.
4744:89
4742:.
4738:.
4711:.
4703:.
4695:.
4683:21
4681:.
4667:^
4646:.
4636:65
4634:.
4630:.
4602:.
4580:.
4570:52
4568:.
4543:.
4539:.
4513:.
4503:.
4493:.
4481:.
4477:.
4454:.
4444:.
4436:.
4424:.
4420:.
4397:.
4387:.
4377:99
4375:.
4371:.
4348:.
4336:.
4313:.
4305:.
4293:.
4270:.
4260:.
4250:.
4238:.
4234:.
4211:.
4199:.
4174:.
4166:.
4143:.
4133:.
4123:.
4111:.
4107:.
4095:^
4081:.
4071:.
4063:.
4053:96
4051:.
4047:.
4035:^
4021:.
4009:.
3995:^
3981:.
3971:.
3951:^
3937:.
3925:.
3921:.
3903:^
3889:.
3875:.
3871:.
3855:^
3841:.
3829:.
3806:.
3796:51
3794:.
3770:.
3756:.
3752:.
3729:.
3717:18
3715:.
3711:.
3688:.
3676:.
3662:^
3648:.
3638:.
3628:59
3626:.
3622:.
3568:.
3553:^
3539:.
3529:38
3527:.
3503:.
3495:.
3485:75
3483:.
3460:.
3452:.
3444:.
3432:.
3409:.
3399:31
3397:.
3385:^
3371:.
3363:.
3349:.
3345:.
3329:^
3315:.
3305:.
3295:.
3283:.
3279:.
3254:63
3252:.
3248:.
3236:^
3222:.
3212:.
3202:.
3190:.
3186:.
3163:.
3153:31
3151:.
3128:.
3118:.
3108:18
3106:.
3102:.
3079:.
3069:.
3061:.
3051:99
3049:.
3045:.
3022:.
3010:38
3008:.
3004:.
2981:.
2973:.
2961:10
2959:.
2955:.
2932:.
2922:82
2920:.
2897:.
2889:.
2877:.
2865:^
2851:.
2843:.
2835:.
2823:.
2809:^
2795:.
2787:.
2779:.
2767:.
2747:^
2733:.
2723:.
2711:.
2707:.
2684:.
2674:.
2662:.
2658:.
2635:.
2625:.
2613:.
2609:.
2586:.
2576:.
2566:.
2558:.
2546:93
2544:.
2540:.
2517:.
2507:.
2499:.
2491:.
2479:79
2477:.
2473:.
2450:.
2442:.
2434:.
2422:.
2399:.
2391:.
2379:.
2356:.
2348:.
2338:82
2336:.
2313:.
2305:.
2293:10
2291:.
2287:.
2262:.
2239:.
2229:.
2219:19
2217:.
2213:.
2190:.
2180:21
2178:.
2139:.
2113:.
2073:^
2059:.
2049:.
2039:.
2029:.
2017:.
2013:.
1990:.
1980:.
1970:.
1958:.
1954:.
1905:^
1891:.
1883:.
1871:.
1848:.
1840:.
1830:.
1822:.
1812:23
1810:.
1806:.
1783:.
1773:14
1771:.
1767:.
1744:.
1732:.
1709:.
1699:59
1697:.
1674:.
1664:.
1652:.
1648:.
1600:.
1592:.
1584:.
1572:.
1560:^
1540:.
1513:.
1503:.
1491:.
1487:.
1458:.
1307:,
1300:.
1178:.
1142:.
1109:.
1090:,
1086:,
1082:,
1074:,
1049:.
971:,
963:,
941:.
909:.
852:.
752:,
700:,
593:pH
402:.
392:pH
334:,
166:.
138:,
134:,
78:.
7026:)
7022:(
6829:Z
6814:S
6788:O
6783:N
6778:M
6748:K
6733:I
6728:H
6683:C
6678:B
6673:A
6515:β
6509:2
6507:α
6501:1
6499:α
6475:9
6470:8
6465:7
6460:6
6455:5
6445:4
6440:2
6307:/
6279:/
6265:/
6247:e
6240:t
6233:v
6045:e
6038:t
6031:v
5858:e
5851:t
5844:v
5637:e
5630:t
5623:v
5594:.
5575:.
5567:(
5552:.
5532::
5524::
5476:.
5455::
5432:.
5408:.
5396::
5373:.
5353::
5345::
5322:.
5297:.
5276:.
5251:.
5227::
5219::
5192:.
5168::
5160::
5133:.
5119::
5092:.
5060:.
5048::
5025:.
5003::
4976:.
4952::
4944::
4914:.
4894::
4886::
4858:.
4836::
4817:.
4813::
4805::
4782:.
4758::
4750::
4719:.
4699::
4689::
4650::
4642::
4612:.
4588:.
4576::
4553:.
4547::
4521:.
4497::
4489::
4462:.
4440::
4432::
4426:2
4405:.
4383::
4356:.
4344::
4321:.
4301::
4295:2
4278:.
4254::
4246::
4240:7
4219:.
4207::
4184:.
4170::
4151:.
4127::
4119::
4089:.
4067::
4059::
4029:.
4017::
3989:.
3967::
3945:.
3933::
3897:.
3883::
3849:.
3837::
3814:.
3802::
3778:.
3764::
3737:.
3723::
3696:.
3684::
3656:.
3642::
3634::
3607:.
3580:.
3547:.
3535::
3511:.
3491::
3468:.
3448::
3440::
3417:.
3405::
3379:.
3357::
3323:.
3299::
3291::
3264:.
3230:.
3206::
3198::
3171:.
3159::
3136:.
3114::
3087:.
3065::
3057::
3030:.
3016::
2989:.
2967::
2940:.
2928::
2905:.
2885::
2859:.
2839::
2831::
2803:.
2783::
2775::
2741:.
2719::
2692:.
2670::
2643:.
2621::
2615:1
2594:.
2570::
2562::
2552::
2525:.
2503::
2495::
2485::
2458:.
2438::
2430::
2407:.
2395::
2387::
2364:.
2344::
2321:.
2299::
2272:.
2247:.
2225::
2198:.
2186::
2163:.
2124:.
2099:.
2067:.
2043::
2025::
1998:.
1974::
1966::
1939:.
1899:.
1887::
1879::
1856:.
1834::
1826::
1818::
1791:.
1779::
1752:.
1740::
1734:3
1717:.
1705::
1682:.
1660::
1633:.
1608:.
1588::
1580::
1554:.
1521:.
1499::
1472:.
935:2
378:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.