17:
1462:
33:
113:
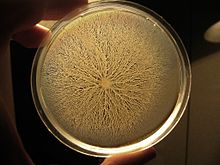
This multicellular behavior has been mostly observed in controlled laboratory conditions and relies on two critical elements: 1) the nutrient composition and 2) viscosity of culture medium (i.e. % agar). One particular feature of this type of motility is the formation of dendritic fractal-like
40:
were inoculated at the center of a dish with gelose containing nutrients. The bacteria start mass-migrating outwards about twelve hours after inoculation, forming dendrites which reach the border of the dish (diameter 90mm) within a few hours. Two days after inoculation the number of bacteria has
144:
Swarming bacteria undergo morphological differentiation that distinguish them from their planktonic state. Cells localized at migration front are typically hyperelongated, hyperflagellated and grouped in multicellular raft structures.
1203:"Single-cell analysis in situ in a Bacillus subtilis swarming community identifies distinct spatially separated subpopulations differentially expressing hag (flagellin), including specialized swarmers"
559:"rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids"
41:
increased so much that they diffuse light significantly and appear white. This picture was taken against a lightsource to make the dendrites (white branched structures) clearly stand out.
601:
Tremblay, Julien; Richardson, Anne-Pascale; Lépine, François; Déziel, Eric (2007). "Self-produced extracellular stimuli modulate the
Pseudomonas aeruginosa swarming motility behaviour".
49:
114:
patterns formed by migrating swarms moving away from an initial location. Although the majority of species can produce tendrils when swarming, some species like
1804:
510:"The Yersinia enterocolitica Motility Master Regulatory Operon, flhDC, Is Required for Flagellin Production, Swimming Motility, and Swarming Motility"
942:"Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells"
48:
is a rapid (2–10 μm/s) and coordinated translocation of a bacterial population across solid or semi-solid surfaces, and is an example of
1201:
Hamze, K.; Autret, S.; Hinc, K.; Laalami, S.; Julkowska, D.; Briandet, R.; Renault, M.; Absalon, C.; Holland, I. B. (2011-01-01).
1493:
1673:
1412:
1244:"Swarm-Cell Differentiation in Salmonellaenterica Serovar Typhimurium Results in Elevated Resistance to Multiple Antibiotics"
136:. Biosurfactant molecules are thought to act by lowering surface tension, thus permitting bacteria to move across a surface.
1162:
strain 3610: detection of different cellular morphologies and constellations of cells as the complex architecture develops"
153:
The fundamental role of swarming motility remains unknown. However, it has been observed that active swarming bacteria of
1422:
1309:
1615:
1512:
1058:
Matsuyama, Tohey; Takagi, Yuko; Nakagawa, Yoji; Itoh, Hiroto; Wakita, Junichi; Matsushita, Mitsugu (2000-01-15).
132:
to occur. Biosurfactant synthesis is usually under the control of an intercellular communication system called
701:"Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa"
1383:
1334:
649:"Swarming of Pseudomonas aeruginosa Is Dependent on Cell-to-Cell Signaling and Requires Flagella and Pili"
1630:
1583:
1339:
1517:
1488:
1388:
1326:
1668:
1001:"The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli"
897:
McCarter, Linda L. (2004). "Dual
Flagellar Systems Enable Motility under Different Circumstances".
1749:
1534:
56:. Swarming motility was first reported by Jorgen Henrichsen and has been mostly studied in genus
188:
Harshey, Rasika M. (2003-01-01). "Bacterial
Motility on a Surface: Many Ways to a Common Goal".
1769:
1429:
1302:
418:
Kirov, S.M.; Tassell, B.C.; Semmler, A.B.T.; O'Donovan, L.A.; Rabaan, A.A.; Shaw, J.G. (2002).
155:
22:
1825:
1578:
1483:
1060:"Dynamic Aspects of the Structured Cell Population in a Swarming Colony of Proteus mirabilis"
375:
Harshey, Rasika M. (1994). "Bees aren't the only ones: swarming in Gram-negative bacteria".
1789:
1754:
1592:
1539:
1446:
1012:
953:
820:
712:
610:
201:
8:
1799:
1774:
1764:
1354:
94:
1602:
1016:
957:
824:
716:
614:
159:
shows an elevated resistance to certain antibiotics compared to undifferentiated cells.
1779:
1640:
1635:
922:
874:
857:
784:
759:
400:
388:
168:
16:
1268:
1243:
444:
419:
298:
273:
249:
224:
1744:
1663:
1650:
1610:
1295:
1273:
1259:
1224:
1183:
1138:
1097:
1092:
1079:
1059:
1040:
1035:
1000:
981:
976:
941:
914:
879:
838:
833:
808:
789:
775:
740:
735:
700:
678:
673:
664:
648:
626:
622:
583:
539:
534:
509:
490:
485:
468:
449:
392:
352:
347:
322:
303:
254:
205:
116:
926:
525:
404:
338:
289:
1625:
1573:
1556:
1478:
1470:
1263:
1255:
1214:
1173:
1128:
1087:
1071:
1030:
1020:
971:
961:
906:
869:
828:
779:
771:
730:
720:
668:
660:
618:
573:
529:
521:
480:
439:
431:
384:
342:
334:
293:
285:
244:
240:
236:
197:
1075:
435:
1441:
1318:
1133:
1116:
53:
1784:
1718:
1695:
1529:
1524:
1507:
1500:
1393:
1005:
Proceedings of the
National Academy of Sciences of the United States of America
946:
Proceedings of the
National Academy of Sciences of the United States of America
705:
Proceedings of the
National Academy of Sciences of the United States of America
133:
1461:
1819:
1658:
1400:
1371:
1349:
1083:
129:
966:
760:"Rhamnolipids Modulate Swarming Motility Patterns of Pseudomonas aeruginosa"
1739:
1713:
1703:
1680:
1561:
1277:
1228:
1219:
1202:
1187:
1142:
1101:
1025:
918:
842:
793:
744:
725:
682:
630:
587:
558:
543:
494:
453:
356:
274:"Differentiation of Serratia marcescens 274 into swimmer and swarmer cells"
209:
1178:
1157:
1044:
985:
883:
578:
396:
307:
258:
32:
1759:
1708:
1549:
1405:
858:"Surface-induced swarmer cell differentiation of Vibrio parahaemoiyticus"
106:
88:
1566:
1544:
64:
1158:"Branched swarming patterns on a synthetic medium formed by wild-type
910:
1794:
1434:
70:
1723:
1685:
1417:
1359:
647:
Köhler, T; Curty, LK; Barja, F; Van Delden, C; Pechère, JC (2000).
128:
In some species, swarming motility requires the self-production of
82:
76:
58:
1344:
417:
225:"Bacterial surface translocation: a survey and a classification"
120:
do form concentric circles motif instead of dendritic patterns.
1378:
1155:
100:
600:
1620:
1156:
Julkowska, D.; Obuchowski, M; Holland, IB; Séror, SJ (2004).
420:"Lateral Flagella and Swarming Motility in Aeromonas Species"
123:
1366:
1287:
1114:
1057:
646:
556:
1115:
Daniels, Ruth; Vanderleyden, Jos; Michiels, Jan (2004).
757:
507:
469:"Swarming motility in undomesticated Bacillus subtilis"
1242:
Kim, W.; Killam, T.; Sood, V.; Surette, M. G. (2003).
1200:
508:
Young, GM; Smith, MJ; Minnich, SA; Miller, VL (1999).
998:
758:
Caiazza, N.C.; Shanks, R.M.Q.; O'Toole, G.A. (2005).
1241:
557:
Déziel, E; Lépine, F; Milot, S; Villemur, R (2003).
1117:"Quorum sensing and swarming migration in bacteria"
899:
Journal of
Molecular Microbiology and Biotechnology
809:"Swarmer cell differentiation in Proteus mirabilis"
1805:Task allocation and partitioning of social insects
855:
320:
1817:
939:
323:"Surface Motility of Serratia liquefaciens MG1"
698:
271:
1303:
999:Burkart, M; Toguchi, A; Harshey, RM (1998).
466:
467:Kearns, Daniel B.; Losick, Richard (2004).
139:
1310:
1296:
222:
148:
124:Biosurfactant, quorum sensing and swarming
1267:
1218:
1177:
1132:
1091:
1034:
1024:
975:
965:
873:
832:
783:
734:
724:
694:
692:
672:
642:
640:
577:
533:
484:
443:
370:
368:
366:
346:
297:
248:
896:
31:
15:
374:
321:Eberl, L; Molin, S; Givskov, M (1999).
187:
1818:
806:
689:
637:
363:
202:10.1146/annurev.micro.57.030502.091014
1413:Patterns of self-organization in ants
1291:
856:McCarter, L.; Silverman, M. (1990).
13:
1423:symmetry breaking of escaping ants
940:Harshey, RM; Matsuyama, T (1994).
875:10.1111/j.1365-2958.1990.tb00678.x
389:10.1111/j.1365-2958.1994.tb00433.x
14:
1837:
1460:
1260:10.1128/JB.185.10.3111-3117.2003
834:10.1111/j.1462-2920.2005.00806.x
776:10.1128/JB.187.21.7351-7361.2005
699:Rashid, MH; Kornberg, A (2000).
665:10.1128/JB.182.21.5990-5996.2000
623:10.1111/j.1462-2920.2007.01396.x
486:10.1046/j.1365-2958.2003.03584.x
272:Alberti, L; Harshey, RM (1990).
1235:
1194:
1149:
1108:
1051:
992:
933:
890:
849:
800:
751:
594:
526:10.1128/JB.181.9.2823-2833.1999
339:10.1128/JB.181.6.1703-1712.1999
290:10.1128/jb.172.8.4322-4328.1990
550:
501:
460:
411:
314:
265:
241:10.1128/MMBR.36.4.478-503.1972
216:
181:
1:
1076:10.1128/JB.182.2.385-393.2000
436:10.1128/JB.184.2.547-555.2002
190:Annual Review of Microbiology
174:
1384:Mixed-species foraging flock
1335:Agent-based model in biology
1317:
1134:10.1016/j.femsre.2003.09.004
27:exhibiting swarming motility
7:
1631:Particle swarm optimization
162:
10:
1842:
1340:Collective animal behavior
813:Environmental Microbiology
807:Rather, Philip N. (2005).
603:Environmental Microbiology
50:bacterial multicellularity
1732:
1694:
1649:
1601:
1469:
1458:
1325:
1121:FEMS Microbiology Reviews
1669:Self-propelled particles
140:Cellular differentiation
36:Bacteria of the species
1750:Collective intelligence
1616:Ant colony optimization
1248:Journal of Bacteriology
1064:Journal of Bacteriology
967:10.1073/pnas.91.18.8631
764:Journal of Bacteriology
653:Journal of Bacteriology
514:Journal of Bacteriology
424:Journal of Bacteriology
327:Journal of Bacteriology
278:Journal of Bacteriology
229:Bacteriological Reviews
149:Ecological significance
1770:Microbial intelligence
1430:Shoaling and schooling
1220:10.1099/mic.0.047159-0
1026:10.1073/pnas.95.5.2568
862:Molecular Microbiology
726:10.1073/pnas.060030097
473:Molecular Microbiology
377:Molecular Microbiology
223:Henrichsen, J (1972).
156:Salmonella typhimurium
42:
29:
23:Pseudomonas aeruginosa
1179:10.1099/mic.0.27061-0
579:10.1099/mic.0.26154-0
35:
19:
1790:Spatial organization
1755:Decentralised system
1593:Sea turtle migration
1447:Swarming (honey bee)
1765:Group size measures
1327:Biological swarming
1017:1998PNAS...95.2568B
958:1994PNAS...91.8631H
825:2005EnvMi...7.1065R
717:2000PNAS...97.4885H
615:2007EnvMi...9.2622T
1780:Predator satiation
1641:Swarm (simulation)
1636:Swarm intelligence
1611:Agent-based models
1442:Swarming behaviour
169:Bacterial motility
43:
30:
1813:
1812:
1800:Military swarming
1745:Animal navigation
1664:Collective motion
1651:Collective motion
1518:reverse migration
1452:Swarming motility
1172:(Pt 6): 1839–49.
1160:Bacillus subtilis
911:10.1159/000077866
572:(Pt 8): 2005–13.
117:Proteus mirabilis
46:Swarming motility
38:Bacillus subtilis
1833:
1626:Crowd simulation
1603:Swarm algorithms
1574:Insect migration
1479:Animal migration
1471:Animal migration
1464:
1389:Mobbing behavior
1312:
1305:
1298:
1289:
1288:
1282:
1281:
1271:
1239:
1233:
1232:
1222:
1213:(9): 2456–2469.
1198:
1192:
1191:
1181:
1153:
1147:
1146:
1136:
1112:
1106:
1105:
1095:
1055:
1049:
1048:
1038:
1028:
996:
990:
989:
979:
969:
937:
931:
930:
894:
888:
887:
877:
853:
847:
846:
836:
804:
798:
797:
787:
755:
749:
748:
738:
728:
696:
687:
686:
676:
644:
635:
634:
598:
592:
591:
581:
563:
554:
548:
547:
537:
505:
499:
498:
488:
464:
458:
457:
447:
415:
409:
408:
372:
361:
360:
350:
318:
312:
311:
301:
269:
263:
262:
252:
220:
214:
213:
185:
1841:
1840:
1836:
1835:
1834:
1832:
1831:
1830:
1816:
1815:
1814:
1809:
1728:
1690:
1645:
1597:
1465:
1456:
1321:
1316:
1286:
1285:
1240:
1236:
1199:
1195:
1154:
1150:
1113:
1109:
1056:
1052:
997:
993:
952:(18): 8631–35.
938:
934:
895:
891:
854:
850:
805:
801:
770:(21): 7351–61.
756:
752:
697:
690:
659:(21): 5990–96.
645:
638:
609:(10): 2622–30.
599:
595:
561:
555:
551:
506:
502:
465:
461:
416:
412:
373:
364:
319:
315:
270:
266:
221:
217:
186:
182:
177:
165:
151:
142:
126:
54:swarm behaviour
28:
26:
12:
11:
5:
1839:
1829:
1828:
1811:
1810:
1808:
1807:
1802:
1797:
1792:
1787:
1785:Quorum sensing
1782:
1777:
1772:
1767:
1762:
1757:
1752:
1747:
1742:
1736:
1734:
1733:Related topics
1730:
1729:
1727:
1726:
1721:
1719:Swarm robotics
1716:
1711:
1706:
1700:
1698:
1696:Swarm robotics
1692:
1691:
1689:
1688:
1683:
1678:
1677:
1676:
1666:
1661:
1655:
1653:
1647:
1646:
1644:
1643:
1638:
1633:
1628:
1623:
1618:
1613:
1607:
1605:
1599:
1598:
1596:
1595:
1590:
1589:
1588:
1587:
1586:
1571:
1570:
1569:
1564:
1554:
1553:
1552:
1547:
1542:
1537:
1530:Fish migration
1527:
1525:Cell migration
1522:
1521:
1520:
1515:
1508:Bird migration
1505:
1504:
1503:
1501:coded wire tag
1498:
1497:
1496:
1486:
1475:
1473:
1467:
1466:
1459:
1457:
1455:
1454:
1449:
1444:
1439:
1438:
1437:
1427:
1426:
1425:
1420:
1410:
1409:
1408:
1398:
1397:
1396:
1394:feeding frenzy
1386:
1381:
1376:
1375:
1374:
1364:
1363:
1362:
1357:
1347:
1342:
1337:
1331:
1329:
1323:
1322:
1315:
1314:
1307:
1300:
1292:
1284:
1283:
1254:(10): 3111–7.
1234:
1193:
1148:
1107:
1070:(2): 385–393.
1050:
1011:(5): 2568–73.
991:
932:
905:(1–2): 18–29.
889:
868:(7): 1057–62.
848:
819:(8): 1065–73.
799:
750:
711:(9): 4885–90.
688:
636:
593:
549:
520:(9): 2823–33.
500:
459:
410:
362:
333:(6): 1703–12.
313:
284:(8): 4322–28.
264:
235:(4): 478–503.
215:
179:
178:
176:
173:
172:
171:
164:
161:
150:
147:
141:
138:
134:quorum sensing
125:
122:
20:
9:
6:
4:
3:
2:
1838:
1827:
1824:
1823:
1821:
1806:
1803:
1801:
1798:
1796:
1793:
1791:
1788:
1786:
1783:
1781:
1778:
1776:
1773:
1771:
1768:
1766:
1763:
1761:
1758:
1756:
1753:
1751:
1748:
1746:
1743:
1741:
1738:
1737:
1735:
1731:
1725:
1722:
1720:
1717:
1715:
1712:
1710:
1707:
1705:
1702:
1701:
1699:
1697:
1693:
1687:
1684:
1682:
1679:
1675:
1672:
1671:
1670:
1667:
1665:
1662:
1660:
1659:Active matter
1657:
1656:
1654:
1652:
1648:
1642:
1639:
1637:
1634:
1632:
1629:
1627:
1624:
1622:
1619:
1617:
1614:
1612:
1609:
1608:
1606:
1604:
1600:
1594:
1591:
1585:
1582:
1581:
1580:
1577:
1576:
1575:
1572:
1568:
1565:
1563:
1560:
1559:
1558:
1555:
1551:
1548:
1546:
1543:
1541:
1538:
1536:
1535:diel vertical
1533:
1532:
1531:
1528:
1526:
1523:
1519:
1516:
1514:
1511:
1510:
1509:
1506:
1502:
1499:
1495:
1492:
1491:
1490:
1487:
1485:
1482:
1481:
1480:
1477:
1476:
1474:
1472:
1468:
1463:
1453:
1450:
1448:
1445:
1443:
1440:
1436:
1433:
1432:
1431:
1428:
1424:
1421:
1419:
1416:
1415:
1414:
1411:
1407:
1404:
1403:
1402:
1399:
1395:
1392:
1391:
1390:
1387:
1385:
1382:
1380:
1377:
1373:
1372:herd behavior
1370:
1369:
1368:
1365:
1361:
1358:
1356:
1353:
1352:
1351:
1348:
1346:
1343:
1341:
1338:
1336:
1333:
1332:
1330:
1328:
1324:
1320:
1313:
1308:
1306:
1301:
1299:
1294:
1293:
1290:
1279:
1275:
1270:
1265:
1261:
1257:
1253:
1249:
1245:
1238:
1230:
1226:
1221:
1216:
1212:
1208:
1204:
1197:
1189:
1185:
1180:
1175:
1171:
1167:
1163:
1161:
1152:
1144:
1140:
1135:
1130:
1127:(3): 261–89.
1126:
1122:
1118:
1111:
1103:
1099:
1094:
1089:
1085:
1081:
1077:
1073:
1069:
1065:
1061:
1054:
1046:
1042:
1037:
1032:
1027:
1022:
1018:
1014:
1010:
1006:
1002:
995:
987:
983:
978:
973:
968:
963:
959:
955:
951:
947:
943:
936:
928:
924:
920:
916:
912:
908:
904:
900:
893:
885:
881:
876:
871:
867:
863:
859:
852:
844:
840:
835:
830:
826:
822:
818:
814:
810:
803:
795:
791:
786:
781:
777:
773:
769:
765:
761:
754:
746:
742:
737:
732:
727:
722:
718:
714:
710:
706:
702:
695:
693:
684:
680:
675:
670:
666:
662:
658:
654:
650:
643:
641:
632:
628:
624:
620:
616:
612:
608:
604:
597:
589:
585:
580:
575:
571:
567:
560:
553:
545:
541:
536:
531:
527:
523:
519:
515:
511:
504:
496:
492:
487:
482:
479:(3): 581–90.
478:
474:
470:
463:
455:
451:
446:
441:
437:
433:
430:(2): 547–55.
429:
425:
421:
414:
406:
402:
398:
394:
390:
386:
383:(3): 389–94.
382:
378:
371:
369:
367:
358:
354:
349:
344:
340:
336:
332:
328:
324:
317:
309:
305:
300:
295:
291:
287:
283:
279:
275:
268:
260:
256:
251:
246:
242:
238:
234:
230:
226:
219:
211:
207:
203:
199:
196:(1): 249–73.
195:
191:
184:
180:
170:
167:
166:
160:
158:
157:
146:
137:
135:
131:
130:biosurfactant
121:
119:
118:
111:
109:
108:
103:
102:
97:
96:
91:
90:
85:
84:
79:
78:
73:
72:
67:
66:
61:
60:
55:
51:
47:
39:
34:
25:
24:
18:
1826:Bacteriology
1740:Allee effect
1714:Nanorobotics
1704:Ant robotics
1681:Vicsek model
1451:
1251:
1247:
1237:
1210:
1207:Microbiology
1206:
1196:
1169:
1166:Microbiology
1165:
1159:
1151:
1124:
1120:
1110:
1067:
1063:
1053:
1008:
1004:
994:
949:
945:
935:
902:
898:
892:
865:
861:
851:
816:
812:
802:
767:
763:
753:
708:
704:
656:
652:
606:
602:
596:
569:
566:Microbiology
565:
552:
517:
513:
503:
476:
472:
462:
427:
423:
413:
380:
376:
330:
326:
316:
281:
277:
267:
232:
228:
218:
193:
189:
183:
154:
152:
143:
127:
115:
112:
105:
99:
93:
87:
81:
75:
69:
63:
57:
45:
44:
37:
21:
1760:Eusociality
1709:Microbotics
1579:butterflies
1550:sardine run
1484:altitudinal
1406:pack hunter
107:Escherichia
89:Pseudomonas
1674:clustering
1567:philopatry
1545:salmon run
1540:Lessepsian
175:References
65:Salmonella
1795:Stigmergy
1775:Mutualism
1435:bait ball
1084:0021-9193
71:Aeromonas
1820:Category
1724:Symbrion
1686:BIO-LGCA
1489:tracking
1418:ant mill
1360:sort sol
1355:flocking
1319:Swarming
1278:12730171
1229:21602220
1188:15184570
1143:15449604
1102:10629184
927:21963003
919:15170400
843:16011745
794:16237018
745:10758151
683:11029417
631:17803784
588:12904540
544:10217774
495:12864845
454:11751834
405:21068729
357:10074060
210:14527279
163:See also
83:Yersinia
77:Bacillus
59:Serratia
1584:monarch
1513:flyways
1494:history
1345:Droving
1045:9482927
1013:Bibcode
986:8078935
954:Bibcode
884:2233248
821:Bibcode
785:1273001
713:Bibcode
611:Bibcode
397:7997156
308:2198253
259:4631369
95:Proteus
1557:Homing
1379:Locust
1276:
1269:154059
1266:
1227:
1186:
1141:
1100:
1090:
1082:
1043:
1033:
984:
974:
925:
917:
882:
841:
792:
782:
743:
733:
681:
671:
629:
586:
542:
532:
493:
452:
445:139559
442:
403:
395:
355:
345:
306:
299:213257
296:
257:
250:408329
247:
208:
101:Vibrio
1621:Boids
1562:natal
1350:Flock
1093:94287
1036:19416
977:44660
923:S2CID
736:18327
674:94731
562:(PDF)
535:93725
401:S2CID
348:93566
1401:Pack
1367:Herd
1274:PMID
1225:PMID
1184:PMID
1139:PMID
1098:PMID
1080:ISSN
1041:PMID
982:PMID
915:PMID
880:PMID
839:PMID
790:PMID
741:PMID
679:PMID
627:PMID
584:PMID
540:PMID
491:PMID
450:PMID
393:PMID
353:PMID
304:PMID
255:PMID
206:PMID
104:and
52:and
1264:PMC
1256:doi
1252:185
1215:doi
1211:157
1174:doi
1170:150
1129:doi
1088:PMC
1072:doi
1068:182
1031:PMC
1021:doi
972:PMC
962:doi
907:doi
870:doi
829:doi
780:PMC
772:doi
768:187
731:PMC
721:doi
669:PMC
661:doi
657:182
619:doi
574:doi
570:149
530:PMC
522:doi
518:181
481:doi
440:PMC
432:doi
428:184
385:doi
343:PMC
335:doi
331:181
294:PMC
286:doi
282:172
245:PMC
237:doi
198:doi
1822::
1272:.
1262:.
1250:.
1246:.
1223:.
1209:.
1205:.
1182:.
1168:.
1164:.
1137:.
1125:28
1123:.
1119:.
1096:.
1086:.
1078:.
1066:.
1062:.
1039:.
1029:.
1019:.
1009:95
1007:.
1003:.
980:.
970:.
960:.
950:91
948:.
944:.
921:.
913:.
901:.
878:.
864:.
860:.
837:.
827:.
815:.
811:.
788:.
778:.
766:.
762:.
739:.
729:.
719:.
709:97
707:.
703:.
691:^
677:.
667:.
655:.
651:.
639:^
625:.
617:.
605:.
582:.
568:.
564:.
538:.
528:.
516:.
512:.
489:.
477:49
475:.
471:.
448:.
438:.
426:.
422:.
399:.
391:.
381:13
379:.
365:^
351:.
341:.
329:.
325:.
302:.
292:.
280:.
276:.
253:.
243:.
233:36
231:.
227:.
204:.
194:57
192:.
110:.
98:,
92:,
86:,
80:,
74:,
68:,
62:,
1311:e
1304:t
1297:v
1280:.
1258::
1231:.
1217::
1190:.
1176::
1145:.
1131::
1104:.
1074::
1047:.
1023::
1015::
988:.
964::
956::
929:.
909::
903:7
886:.
872::
866:4
845:.
831::
823::
817:7
796:.
774::
747:.
723::
715::
685:.
663::
633:.
621::
613::
607:9
590:.
576::
546:.
524::
497:.
483::
456:.
434::
407:.
387::
359:.
337::
310:.
288::
261:.
239::
212:.
200::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.