830:(2), or spontaneously via their transmembrane domains (not shown in the figure). Lumenal proteins are exported across the thylakoid membrane into the lumen by either the Tat-dependent pathway (2) or the Sec-dependent pathway (3) and released by cleavage from the thylakoid targeting signal. The different pathways utilize different signals and energy sources. The Sec (secretory) pathway requires ATP as an energy source and consists of SecA, which binds to the imported protein and a Sec membrane complex to shuttle the protein across. Proteins with a twin
1117:
that encloses a single lumen (as in higher‐plant chloroplasts) and allows water‐soluble and lipid‐soluble molecules to diffuse through the entire membrane network. Moreover, perforations are often observed within the parallel thylakoid sheets. These gaps in the membrane allow for the traffic of particles of different sizes throughout the cell, including ribosomes, glycogen granules, and lipid bodies. The relatively large distance between the thylakoids provides space for the external light-harvesting antennae, the
422:
complex network of alternating helical membrane surfaces of different radii and pitch was shown to minimize the surface and bending energies of the membranes. This new model, the most extensive one generated to date, revealed that features from two, seemingly contradictory, older models coexist in the structure. Notably, similar arrangements of helical elements of alternating handedness, often referred to as "parking garage" structures, were proposed to be present in the
848:
826:) complexes. After entering the chloroplast, the first targeting peptide is cleaved off by a protease processing imported proteins. This unmasks the second targeting signal and the protein is exported from the stroma into the thylakoid in a second targeting step. This second step requires the action of protein translocation components of the thylakoids and is energy-dependent. Proteins are inserted into the membrane via the SRP-dependent pathway (1), the
318:
806:
62:
303:
1079:
839:) pathway. The chloroplast SRP can interact with its target proteins either post-translationally or co-translationally, thus transporting imported proteins as well as those that are translated inside the chloroplast. The SRP pathway requires GTP and the pH gradient as energy sources. Some transmembrane proteins may also spontaneously insert into the membrane from the stromal side without energy requirement.
458:(VIPP1). Plants cannot survive without this protein, and reduced VIPP1 levels lead to slower growth and paler plants with reduced ability to photosynthesize. VIPP1 appears to be required for basic thylakoid membrane formation, but not for the assembly of protein complexes of the thylakoid membrane. It is conserved in all organisms containing thylakoids, including cyanobacteria, green algae, such as
517:
295:
205:
357:. The thylakoid lipid bilayer shares characteristic features with prokaryotic membranes and the inner chloroplast membrane. For example, acidic lipids can be found in thylakoid membranes, cyanobacteria and other photosynthetic bacteria and are involved in the functional integrity of the photosystems. The thylakoid membranes of higher plants are composed primarily of
797:. Chloroplasts also need to balance the ratios of photosystem I and II for the electron transfer chain. The redox state of the electron carrier plastoquinone in the thylakoid membrane directly affects the transcription of chloroplast genes encoding proteins of the reaction centers of the photosystems, thus counteracting imbalances in the electron transfer chain.
593:
photosystems in the thylakoid membrane system, mobile electron carriers are required to shuttle electrons between them. These carriers are plastoquinone and plastocyanin. Plastoquinone shuttles electrons from photosystem II to the cytochrome b6f complex, whereas plastocyanin carries electrons from the cytochrome b6f complex to photosystem I.
821:
inside the chloroplast. Most thylakoid proteins encoded by a plant's nuclear genome need two targeting signals for proper localization: An N-terminal chloroplast targeting peptide (shown in yellow in the figure), followed by a thylakoid targeting peptide (shown in blue). Proteins are imported through
1116:
In contrast to the thylakoid network of higher plants, which is differentiated into grana and stroma lamellae, the thylakoids in cyanobacteria are organized into multiple concentric shells that split and fuse to parallel layers forming a highly connected network. This results in a continuous network
764:
took place. This results in the four major thylakoid protein complexes being encoded in part by the chloroplast genome and in part by the nuclear genome. Plants have developed several mechanisms to co-regulate the expression of the different subunits encoded in the two different organelles to assure
536:
consists of at least 335 different proteins. Out of these, 89 are in the lumen, 116 are integral membrane proteins, 62 are peripheral proteins on the stroma side, and 68 peripheral proteins on the lumenal side. Additional low-abundance lumenal proteins can be predicted through computational methods.
365:
that are asymmetrically arranged along and across the membranes. Thylakoid membranes are richer in galactolipids rather than phospholipids; also they predominantly consist of hexagonal phase II forming monogalacotosyl diglyceride lipid. Despite this unique composition, plant thylakoid membranes have
1832:
Elena Aseeva; Friederich Ossenbühl; Claudia Sippel; Won K. Cho; Bernhard Stein; Lutz A. Eichacker; Jörg Meurer; Gerhard Wanner; Peter
Westhoff; Jürgen Soll; Ute C. Vothknecht (2007). "Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes".
421:
study of the thylakoid membranes has shown that the stroma lamellae are organized in wide sheets perpendicular to the grana stack axis and form multiple right-handed helical surfaces at the granal interface. Left-handed helical surfaces consolidate between the right-handed helices and sheets. This
997:
consumes two protons from the stroma. These are released in the lumen when the reduced plastoquinol is oxidized by the cytochrome b6f protein complex on the lumen side of the thylakoid membrane. From the plastoquinone pool, electrons pass through the cytochrome b6f complex. This integral membrane
877:
The first step in photosynthesis is the light-driven reduction (splitting) of water to provide the electrons for the photosynthetic electron transport chains as well as protons for the establishment of a proton gradient. The water-splitting reaction occurs on the lumenal side of the thylakoid
592:
Photosystem II is located mostly in the grana thylakoids, whereas photosystem I and ATP synthase are mostly located in the stroma thylakoids and the outer layers of grana. The cytochrome b6f complex is distributed evenly throughout thylakoid membranes. Due to the separate location of the two
697:
The cytochrome b6f complex is part of the thylakoid electron transport chain and couples electron transfer to the pumping of protons into the thylakoid lumen. Energetically, it is situated between the two photosystems and transfers electrons from photosystem II-plastoquinone to
324:
The prevailing model of the granum-stroma assembly is stacks of granal thylakoids wrapped by right-handed helical stromal thylakoids which are connected to large parallel sheets of stromal thylakoids and adjacent right-handed helices by left-handed helical structures. (Based on
712:
The thylakoid ATP synthase is a CF1FO-ATP synthase similar to the mitochondrial ATPase. It is integrated into the thylakoid membrane with the CF1-part sticking into the stroma. Thus, ATP synthesis occurs on the stromal side of the thylakoids where the ATP is needed for the
1045:
due to charge separation, thylakoid membranes lack a charge gradient. To compensate for this, the 10,000 fold proton concentration gradient across the thylakoid membrane is much higher compared to a 10 fold gradient across the inner membrane of mitochondria. The resulting
915:
Photosystem I uses light energy to reduce NADP to NADPH + H, and is active in both noncyclic and cyclic electron transport. In cyclic mode, the energized electron is passed down a chain that ultimately returns it (in its base state) to the chlorophyll that energized
834:
motif in their thylakoid signal peptide are shuttled through the Tat (twin arginine translocation) pathway, which requires a membrane-bound Tat complex and the pH gradient as an energy source. Some other proteins are inserted into the membrane via the SRP
1113:, and thylakoid membranes each have specialized roles in the cyanobacterial cell. Understanding the organization, functionality, protein composition, and dynamics of the membrane systems remains a great challenge in cyanobacterial cell biology.
729:
is present in the lumen and shuttles electrons from the cytochrome b6f protein complex to photosystem I. While plastoquinones are lipid-soluble and therefore move within the thylakoid membrane, plastocyanin moves through the thylakoid lumen.
946:
A major function of the thylakoid membrane and its integral photosystems is the establishment of chemiosmotic potential. The carriers in the electron transport chain use some of the electron's energy to actively transport protons from the
366:
been shown to assume largely lipid-bilayer dynamic organization. Lipids forming the thylakoid membranes, richest in high-fluidity linolenic acid are synthesized in a complex pathway involving exchange of lipid precursors between the
683:). The P is short for pigment and the number is the specific absorption peak in nanometers for the chlorophyll molecules in each reaction center. This is the green pigment present in plants that is not visible to unaided eyes.
863:, the pumping of protons across the thylakoid membranes coupled with the electron transport chain of the photosystems and cytochrome complex, and ATP synthesis by the ATP synthase utilizing the generated proton gradient.
480:. Disruption of isolated thylakoids, for example by mechanical shearing, releases the lumenal fraction. Peripheral and integral membrane fractions can be extracted from the remaining membrane fraction. Treatment with
446:
that contain semicrystalline membrane structures called prolamellar bodies. When exposed to light, these prolamellar bodies develop into thylakoids. This does not happen in seedlings grown in the dark, which undergo
658:
to harvest light at a variety of wavelengths. Each antenna complex has between 250 and 400 pigment molecules and the energy they absorb is shuttled by resonance energy transfer to a specialized chlorophyll
1919:
Liu C, Willmund F, Golecki J, Cacace S, Markert C, Heß B, Schroda M, Schroda M (2007). "The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in
Chlamydomonas".
528:
studies of thylakoid fractions have provided further details on the protein composition of the thylakoids. These data have been summarized in several plastid protein databases that are available online.
1093:
are photosynthetic prokaryotes with highly differentiated membrane systems. Cyanobacteria have an internal system of thylakoid membranes where the fully functional electron transfer chains of
744:
signal, the largest groups with known functions are 19% involved in protein processing (proteolysis and folding), 18% in photosynthesis, 11% in metabolism, and 7% redox carriers and defense.
1037:
of the PMF to generate the potential energy required for ATP synthesis. The PMF is the sum of a proton chemical potential (given by the proton concentration gradient) and a transmembrane
417:. Grana thylakoids and stroma thylakoids can be distinguished by their different protein composition. Grana contribute to chloroplasts' large surface area to volume ratio. A recent
2430:
Gutensohn M, Fan E, Frielingsdorf S, Hanner P, Hou B, Hust B, Klösgen R (2006). "Toc, Tic, Tat et al.: structure and function of protein transport machineries in chloroplasts".
1340:
565:
which play an important role in light-harvesting and the light-dependent reactions of photosynthesis. There are four major protein complexes in the thylakoid membrane:
667:
molecules at the reaction center absorb energy, an electron is excited and transferred to an electron-acceptor molecule. Photosystem I contains a pair of chlorophyll
1718:
Horowitz CJ; Berry DK; Briggs CM; Caplan ME; Cumming A; Schneider AS (2015). "Disordered nuclear pasta, magnetic field decay, and crust cooling in neutron stars".
789:
rate of chloroplast-encoded proteins is controlled by the presence or absence of assembly partners (control by epistasy of synthesis). This mechanism involves
2600:
Olive, J; Ajlani, G; Astier, C; Recouvreur, M; Vernotte, C (1997). "Ultrastructure and light adaptation of phycobilisome mutants of
Synechocystis PCC 6803".
2294:
Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman F (2001). "Assembly-controlled regulation of chloroplast gene translation".
426:
and in ultradense nuclear matter. This structural organization may constitute a fundamental geometry for connecting between densely packed layers or sheets.
413:) is a stack of thylakoid discs. Chloroplasts can have from 10 to 100 grana. Grana are connected by stroma thylakoids, also called intergranal thylakoids or
1105:. Cyanobacteria must be able to reorganize the membranes, synthesize new membrane lipids, and properly target proteins to the correct membrane system. The
2112:"In-Depth Analysis of the Thylakoid Membrane Proteome of Arabidopsis thaliana Chloroplasts: New Proteins, New Functions, and a Plastid Proteome Database"
740:
Lumenal proteins can be predicted computationally based on their targeting signals. In
Arabidopsis, out of the predicted lumenal proteins possessing the
1289:
Sato N (2004). "Roles of the acidic lipids sulfoquinovosyl diacylglycerol and phosphatidylglycerol in photosynthesis: their specificity and evolution".
537:
Of the thylakoid proteins with known functions, 42% are involved in photosynthesis. The next largest functional groups include proteins involved in
923:), and is only active in noncyclic transport. Electrons in this system are not conserved, but are rather continually entering from oxidized 2H
2017:"Central Functions of the Lumenal and Peripheral Thylakoid Proteome of Arabidopsis Determined by Experimentation and Genome-Wide Prediction"
911:
The noncyclic variety involves the participation of both photosystems, while the cyclic electron flow is dependent on only photosystem I.
878:
membrane and is driven by the light energy captured by the photosystems. This oxidation of water conveniently produces the waste product O
2163:
Kleffmann T, Hirsch-Hoffmann M, Gruissem W, Baginsky S (2006). "plprot: a comprehensive proteome database for different plastid types".
1610:
Terasaki M, Shemesh T, Kasthuri N, Klemm R, Schalek R, Hayworth K, Hand A, Yankova M, Huber G, Lichtman J, Rapoport T, Kozlov M (2013).
451:. An underexposure to light can cause the thylakoids to fail. This causes the chloroplasts to fail resulting to the death of the plant.
309:
10-nm-thick STEM tomographic slice from a lettuce chloroplast. Grana stacks are interconnected by unstacked stromal thylakoids, called
442:
emerge from the ground. Thylakoid formation requires light. In the plant embryo and in the absence of light, proplastids develop into
397:. During the light-dependent reaction, protons are pumped across the thylakoid membrane into the lumen making it acidic down to pH 4.
1041:(given by charge separation across the membrane). Compared to the inner membranes of mitochondria, which have a significantly higher
1121:. This macrostructure, as in the case of higher plants, shows some flexibility during changes in the physicochemical environment.
2380:"Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts"
1017:
The proton gradient is also caused by the consumption of protons in the stroma to make NADPH from NADP+ at the NADP reductase.
2204:"Proteomics of the Chloroplast: Systematic Identification and Targeting Analysis of Lumenal and Peripheral Thylakoid Proteins"
1779:
Schneider AS; Berry DK; Caplan ME; Horowitz CJ; Lin Z (2016). "Effect of topological defects on "nuclear pasta" observables".
2729:
2708:
2681:
2535:
2628:
919:
Photosystem II uses light energy to oxidize water molecules, producing electrons (e), protons (H), and molecular oxygen (O
756:, which encodes a number of thylakoid proteins. However, during the course of plastid evolution from their cyanobacterial
2331:"Chloroplast Biogenesis of Photosystem II Cores Involves a Series of Assembly-Controlled Steps That Regulate Translation"
1178:
827:
741:
2627:
Nagy, G; Posselt, D; Kovács, L; Holm, JK; Szabó, M; Ughy, B; Rosta, L; Peters, J; Timmins, P; Garab, G (1 June 2011).
1563:"The Three-Dimensional Network of the Thylakoid Membranes in Plants: Quasihelical Model of the Granum-Stroma Assembly"
370:
and inner membrane of the plastid envelope and transported from the inner membrane to the thylakoids via vesicles.
1353:
Spraque SG (1987). "Structural and functional organization of galactolipids on thylakoid membrane organization".
17:
1514:"Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography"
2629:"Reversible membrane reorganizations during photosynthesis in vivo: revealed by small-angle neutron scattering"
1870:"Vipp1 deletion mutant of Synechocystis: A connection between bacterial phage shock and thylakoid biogenesis?"
679:
chlorophyll that absorbs 680 nm light best (note that these wavelengths correspond to deep red – see the
2251:
Vener AV, Ohad I, Andersson B (1998). "Protein phosphorylation and redox sensing in chloroplast thylakoids".
2011:
Peltier J, Emanuelsson O, Kalume D, Ytterberg J, Friso G, Rudella A, Liberles D, Söderberg L, Roepstorff P,
959:, as low as pH 4, compared to pH 8 in the stroma. This represents a 10,000 fold concentration gradient for
1195:
1025:
The molecular mechanism of ATP (Adenosine triphosphate) generation in chloroplasts is similar to that in
836:
493:
353:
embedded directly in the membrane. It is an alternating pattern of dark and light bands measuring each 1
1190:
714:
524:
Thylakoids contain many integral and peripheral membrane proteins, as well as lumenal proteins. Recent
193:
108:
90:
2748:
1067:
1010:
994:
856:
562:
505:
346:
224:
1955:
Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht U, Soll J, Westhoff P (2001).
1469:
Benning C, Xu C, Awai K (2006). "Non-vesicular and vesicular lipid trafficking involving plastids".
1135:
1145:
1106:
895:
770:
597:
1279:"Photosynthesis" McGraw Hill Encyclopedia of Science and Technology, 10th ed. 2007. Vol. 13 p. 469
2753:
1130:
734:
2671:
2553:"Thylakoid membrane perforations and connectivity enable intracellular traffic in cyanobacteria"
2758:
2700:
1101:
reside. The presence of different membrane systems lends these cells a unique complexity among
1047:
692:
647:
617:
601:
581:
350:
786:
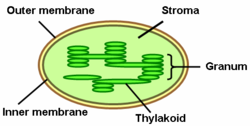
550:
476:
Thylakoids can be purified from plant cells using a combination of differential and gradient
423:
367:
2478:
2260:
2073:
1798:
1737:
1682:
1478:
1298:
1234:
1098:
1038:
904:
Noncyclic electron transport or non-cyclic photophosphorylation produces NADPH + H and ATP.
883:
675:, at its reaction center that maximally absorbs 700 nm light. Photosystem II contains
621:
464:
390:
80:
1957:"VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation"
8:
1030:
418:
99:
2482:
2264:
2077:
1831:
1802:
1741:
1686:
1482:
1302:
1238:
2577:
2552:
2404:
2379:
2355:
2330:
1814:
1788:
1761:
1727:
1700:
1672:
1636:
1611:
1587:
1562:
1538:
1513:
1420:
1398:"Magnetic resonance studies of dynamic organization of lipids in chloroplast membranes"
1378:
1322:
1257:
1222:
1042:
1034:
386:
148:
2693:
2613:
2501:
2466:
2272:
2228:
2203:
2136:
2111:
2041:
2016:
2763:
2725:
2704:
2677:
2651:
2582:
2531:
2506:
2447:
2409:
2360:
2311:
2276:
2233:
2202:
Peltier J, Friso G, Kalume D, Roepstorff P, Nilsson F, Adamska I, van Wijk K (2000).
2180:
2141:
2089:
2046:
2012:
1988:
1983:
1956:
1937:
1933:
1901:
1896:
1869:
1850:
1753:
1641:
1592:
1543:
1494:
1451:
1439:
1370:
1314:
1262:
818:
790:
777:. Biogenesis, stability and turnover of thylakoid protein complexes are regulated by
538:
158:
2672:
Heller, H. Craig; Orians, Gordan H.; Purves, William K. & Sadava, David (2004).
1818:
1765:
1704:
1397:
1326:
1221:
Bussi Y, Shimoni E, Weiner A, Kapon R, Charuvi D, Nevo R, Efrati E, Reich Z (2019).
405:
In higher plants thylakoids are organized into a granum-stroma membrane assembly. A
2643:
2609:
2572:
2564:
2496:
2486:
2439:
2399:
2391:
2350:
2342:
2303:
2268:
2223:
2215:
2172:
2131:
2123:
2081:
2036:
2028:
1978:
1968:
1929:
1891:
1881:
1842:
1806:
1749:
1745:
1690:
1631:
1623:
1582:
1574:
1533:
1525:
1486:
1412:
1382:
1362:
1306:
1252:
1242:
1155:
860:
793:
through binding of excess protein to the 5' untranslated region of the chloroplast
680:
655:
546:
481:
1612:"Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs"
1424:
251:
thylakoids, which join granum stacks together as a single functional compartment.
2085:
1846:
1200:
1110:
952:
823:
778:
639:
542:
501:
381:
172:
138:
31:
2443:
2162:
2110:
Friso G, Giacomelli L, Ytterberg A, Peltier J, Rudella A, Sun Q, Wijk K (2004).
773:
of nuclear genes encoding parts of the photosynthetic apparatus is regulated by
1810:
1695:
1660:
1627:
1160:
1118:
1094:
900:
Two different variations of electron transport are used during photosynthesis:
886:. The molecular oxygen formed by the reaction is released into the atmosphere.
814:
663:
at the reaction center of each photosystem. When either of the two chlorophyll
576:
477:
394:
330:
307:
Scanning transmission electron microscope (STEM) imaging of thylakoid membranes
271:
248:
228:
220:
186:
165:
1490:
1310:
2742:
2568:
1661:""Parking-garage" structures in nuclear astrophysics and cellular biophysics"
1223:"Fundamental helical geometry consolidates the plant photosynthetic membrane"
1186:
1150:
1090:
1002:
990:
766:
572:
459:
358:
1560:
1247:
2655:
2586:
2551:
Nevo R, Charuvi D, Shimoni E, Schwarz R, Kaplan A, Ohad I, Reich Z (2007).
2451:
2413:
2395:
2364:
2346:
2315:
2280:
2237:
2184:
2145:
2093:
2050:
1992:
1973:
1941:
1905:
1886:
1854:
1757:
1645:
1596:
1578:
1547:
1529:
1498:
1318:
1266:
1140:
1059:
1055:
1051:
1026:
948:
941:
907:
Cyclic electron transport or cyclic photophosphorylation produces only ATP.
761:
757:
726:
707:
613:
586:
362:
333:. A stack of thylakoids is called a granum and resembles a stack of coins.
42:
2510:
2491:
1455:
1374:
2219:
2176:
2010:
651:
643:
638:
These photosystems are light-driven redox centers, each consisting of an
633:
569:
254:
In thylakoid membranes, chlorophyll pigments are found in packets called
216:
51:
2699:(7th ed.). New York: W.H. Freeman and Company Publishers. pp.
1778:
239:. Chloroplast thylakoids frequently form stacks of disks referred to as
2647:
2429:
2307:
2127:
2109:
2032:
1954:
1416:
1366:
1006:
975:
931:+ 4 H + 4 e) and exiting with NADP when it is finally reduced to NADPH.
872:
847:
733:
The lumen of the thylakoids is also the site of water oxidation by the
525:
448:
435:
255:
2467:"ATP formation caused by acid-base transition of spinach chloroplasts"
805:
760:
ancestors, extensive gene transfer from the chloroplast genome to the
317:
2328:
2293:
1717:
1440:"13C NMR studies of lipid fatty-acyl chains of chloroplast membranes"
851:
Light-dependent reactions of photosynthesis at the thylakoid membrane
497:
443:
354:
329:
Thylakoids are membrane-bound structures embedded in the chloroplast
2599:
2201:
1609:
302:
2719:
2525:
1867:
1793:
1677:
1658:
1102:
1058:. As the protons travel back down the gradient through channels in
983:
831:
533:
439:
179:
61:
2690:
1732:
1511:
859:
of photosynthesis. These include light-driven water oxidation and
2550:
809:
Schematic representation of thylakoid protein targeting pathways.
516:
1918:
1659:
Berry DK; Caplan ME; Horowitz CJ; Huber G; Schneider AS (2016).
1078:
27:
Membrane enclosed compartments in chloroplasts and cyanobacteria
2717:
2626:
2523:
1220:
979:
960:
956:
782:
753:
2151:
2190:
1179:
1070:
are coupled to the synthesis of ATP via the proton gradient.
774:
609:
605:
258:. Each quantasome contains 230 to 250 chlorophyll molecules.
204:
2721:
2527:
2464:
1561:
Mustárdy, L.; Buttle, K.; Steinbach, G.; Garab, G. (2008).
794:
676:
672:
596:
Together, these proteins make use of light energy to drive
1512:
Shimoni E, Rav-Hon O, Ohad I, Brumfeld V, Reich Z (2005).
971:
The protons in the lumen come from three primary sources.
2691:
Raven, Peter H.; Ray F. Evert; Susan E. Eichhorn (2005).
2250:
1341:
Encyclopædia
Britannica 2006 Ultimate Reference Suite DVD
813:
Thylakoid proteins are targeted to their destination via
294:
1013:
also transfers two protons from the stroma to the lumen.
1073:
800:
769:
and assembly of these protein complexes. For example,
2377:
520:
Thylakoid disc with embedded and associated proteins.
30:"Granum" redirects here. For the town in Canada, see
2329:
Minai L, Wostrikoff K, Wollman F, Choquet Y (2006).
737:
associated with the lumenal side of photosystem II.
2602:
Biochimica et
Biophysica Acta (BBA) - Bioenergetics
1468:
2692:
1868:Westphal S, Heins L, Soll J, Vothknecht U (2001).
989:The transfer of electrons from photosystem II to
2740:
2718:Herrero, Antonia; Flores, Enrique, eds. (2008).
2524:Herrero, Antonia; Flores, Enrique, eds. (2008).
1339:"photosynthesis."Encyclopædia Britannica. 2008.
1054:is high enough to drive ATP synthesis using the
822:the translocon of the outer and inner membrane (
310:
240:
73:
2006:
2004:
2002:
1033:(PMF). However, chloroplasts rely more on the
471:
385:is a continuous aqueous phase enclosed by the
1444:Indian Journal of Biochemistry and Biophysics
556:
400:
2425:
2423:
1082:Thylakoids (green) inside a cyanobacterium (
966:
889:
208:Thylakoids (dark green) inside a chloroplast
2195:
1999:
1066:are combined into ATP. In this manner, the
955:. During photosynthesis, the lumen becomes
454:Thylakoid formation requires the action of
2063:
1352:
532:According to these studies, the thylakoid
2676:(7th ed.). Sinauer Associates, Inc.
2576:
2544:
2500:
2490:
2420:
2403:
2354:
2227:
2156:
2135:
2064:van Wijk K (2004). "Plastid proteomics".
2040:
1982:
1972:
1895:
1885:
1792:
1772:
1731:
1694:
1676:
1635:
1586:
1537:
1256:
1246:
686:
247:). Grana are connected by intergranal or
2724:(1st ed.). Caister Academic Press.
2530:(1st ed.). Caister Academic Press.
2105:
2103:
1711:
1671:(5). American Physical Society: 055801.
1603:
1554:
1077:
846:
804:
616:uses the chemiosmotic potential to make
515:
316:
301:
293:
203:
2517:
2287:
1505:
1437:
1395:
1216:
1214:
1212:
1210:
1208:
1029:and takes the required energy from the
215:are membrane-bound compartments inside
14:
2741:
1462:
1346:
1288:
456:vesicle-inducing protein in plastids 1
2465:Jagendorf A. T. and E. Uribe (1966).
2371:
2322:
2244:
2100:
1652:
747:
2057:
1205:
1074:Thylakoid membranes in cyanobacteria
978:by photosystem II oxidises water to
2665:
1282:
866:
855:The thylakoids are the site of the
801:Protein targeting to the thylakoids
67:Components of a typical chloroplast
24:
2384:Philos Trans R Soc Lond B Biol Sci
1172:
998:assembly resembles cytochrome bc1.
604:across the thylakoid membrane and
286:means "sac-like" or "pouch-like".
282:, meaning "sac" or "pouch". Thus,
25:
2775:
1020:
720:
389:. It plays an important role for
2378:Allen J, Pfannschmidt T (2000).
1934:10.1111/j.1365-313X.2007.03047.x
785:in the thylakoid membranes. The
322:Granum-stroma assembly structure
60:
2620:
2593:
2458:
1948:
1912:
1861:
1825:
963:across the thylakoid membrane.
935:
725:The electron transport protein
701:
627:
1750:10.1103/PhysRevLett.114.031102
1431:
1389:
1333:
1273:
13:
1:
2614:10.1016/S0005-2728(96)00168-5
2273:10.1016/S1369-5266(98)80107-6
2152:The Plastid Proteome Database
1166:
995:non-cyclic electron transport
462:, and higher plants, such as
232:
2674:LIFE: The Science of Biology
2086:10.1016/j.plaphy.2004.10.015
1847:10.1016/j.plaphy.2007.01.005
752:Chloroplasts have their own
698:plastocyanin-photosystem I.
608:, a product of the terminal
561:Thylakoid membranes contain
494:peripheral membrane proteins
429:
289:
261:
7:
2444:10.1016/j.jplph.2005.11.009
1124:
842:
837:signal recognition particle
715:light-independent reactions
511:
472:Isolation and fractionation
349:of photosynthesis with the
336:
223:. They are the site of the
10:
2780:
2471:Proc. Natl. Acad. Sci. USA
1811:10.1103/PhysRevC.93.065806
1696:10.1103/PhysRevC.94.055801
1628:10.1016/j.cell.2013.06.031
1180:
939:
893:
870:
705:
690:
631:
563:integral membrane proteins
557:Integral membrane proteins
506:integral membrane proteins
434:Chloroplasts develop from
401:Granum and stroma lamellae
236:
231:. Thylakoids consist of a
29:
1491:10.1016/j.pbi.2006.03.012
1311:10.1007/s10265-004-0183-1
1068:light-dependent reactions
1011:cyclic electron transport
967:Source of proton gradient
890:Electron transport chains
857:light-dependent reactions
598:electron transport chains
496:, whereas treatment with
347:light-dependent reactions
313:. Scalebar = 200 nm. See.
225:light-dependent reactions
59:
50:
41:
2569:10.1038/sj.emboj.7601594
2191:Plastid Protein Database
1146:Electrochemical gradient
896:electron transport chain
373:
2636:The Biochemical Journal
1248:10.1073/pnas.1905994116
1196:A Greek–English Lexicon
1131:Arthur Meyer (botanist)
735:oxygen evolving complex
648:photosynthetic pigments
351:photosynthetic pigments
2396:10.1098/rstb.2000.0697
2347:10.1105/tpc.105.037705
2015:, van Wijk KJ (2002).
1974:10.1073/pnas.061500998
1961:Proc Natl Acad Sci USA
1887:10.1073/pnas.061501198
1874:Proc Natl Acad Sci USA
1579:10.1105/tpc.108.059147
1530:10.1105/tpc.105.035030
1438:YashRoy, R.C. (1987).
1405:Journal of Biosciences
1396:YashRoy, R.C. (1990).
1227:Proc Natl Acad Sci USA
1087:
1050:between the lumen and
1048:chemiosmotic potential
852:
810:
693:Cytochrome b6f complex
687:Cytochrome b6f complex
671:molecules, designated
602:chemiosmotic potential
582:Cytochrome b6f complex
521:
326:
314:
299:
209:
2492:10.1073/pnas.55.1.170
2066:Plant Physiol Biochem
1835:Plant Physiol Biochem
1187:Liddell, Henry George
1081:
850:
828:Tat-dependent pathway
817:and prokaryotic-type
808:
519:
424:endoplasmic reticulum
368:endoplasmic reticulum
320:
305:
297:
207:
2253:Curr Opin Plant Biol
2220:10.1105/tpc.12.3.319
1471:Curr Opin Plant Biol
1039:electrical potential
884:cellular respiration
781:via redox-sensitive
622:photophosphorylation
465:Arabidopsis thaliana
391:photophosphorylation
298:Thylakoid structures
81:Chloroplast envelope
2483:1966PNAS...55..170J
2265:1998COPB....1..217V
2078:2004PlPB...42..963V
1803:2016PhRvC..93f5806S
1742:2015PhRvL.114c1102H
1687:2016PhRvC..94e5801B
1483:2006COPB....9..241B
1355:J Bioenerg Biomembr
1303:2004JPlR..117..495S
1239:2019PNAS..11622366B
1233:(44): 22366–22375.
1031:proton motive force
717:of photosynthesis.
419:electron tomography
345:is the site of the
173:Nucleoid (DNA ring)
100:Intermembrane space
2648:10.1042/BJ20110180
2308:10.1042/BST0290421
2177:10.1093/pcp/pcj005
2165:Plant Cell Physiol
2128:10.1105/tpc.017814
2033:10.1105/tpc.010304
1417:10.1007/bf02702669
1367:10.1007/BF00762303
1088:
1043:membrane potential
1035:chemical potential
882:that is vital for
853:
819:secretory pathways
811:
748:Protein expression
549:response (9%) and
522:
387:thylakoid membrane
343:thylakoid membrane
327:
315:
300:
233:thylakoid membrane
210:
149:Thylakoid membrane
2731:978-1-904455-15-8
2710:978-0-7167-1007-3
2695:Biology of Plants
2683:978-0-7167-9856-9
2537:978-1-904455-15-8
2296:Biochem Soc Trans
1781:Physical Review C
1665:Physical Review C
1573:(10): 2552–2557.
1001:The reduction of
791:negative feedback
656:phycobiliproteins
541:, processing and
539:protein targeting
202:
201:
159:Stromal thylakoid
16:(Redirected from
2771:
2749:Membrane biology
2735:
2714:
2698:
2687:
2666:Textbook sources
2660:
2659:
2633:
2624:
2618:
2617:
2608:(2–3): 275–282.
2597:
2591:
2590:
2580:
2563:(5): 1467–1473.
2548:
2542:
2541:
2521:
2515:
2514:
2504:
2494:
2462:
2456:
2455:
2432:J. Plant Physiol
2427:
2418:
2417:
2407:
2390:(1402): 1351–9.
2375:
2369:
2368:
2358:
2326:
2320:
2319:
2291:
2285:
2284:
2248:
2242:
2241:
2231:
2199:
2193:
2188:
2160:
2154:
2149:
2139:
2107:
2098:
2097:
2061:
2055:
2054:
2044:
2008:
1997:
1996:
1986:
1976:
1952:
1946:
1945:
1916:
1910:
1909:
1899:
1889:
1865:
1859:
1858:
1829:
1823:
1822:
1796:
1776:
1770:
1769:
1735:
1715:
1709:
1708:
1698:
1680:
1656:
1650:
1649:
1639:
1607:
1601:
1600:
1590:
1558:
1552:
1551:
1541:
1509:
1503:
1502:
1466:
1460:
1459:
1435:
1429:
1428:
1402:
1393:
1387:
1386:
1350:
1344:
1337:
1331:
1330:
1286:
1280:
1277:
1271:
1270:
1260:
1250:
1218:
1203:
1183:
1182:
1176:
1156:Oxygen evolution
867:Water photolysis
861:oxygen evolution
681:visible spectrum
600:that generate a
547:oxidative stress
502:organic solvents
482:sodium carbonate
126:
64:
54:
45:
39:
38:
21:
2779:
2778:
2774:
2773:
2772:
2770:
2769:
2768:
2739:
2738:
2732:
2711:
2684:
2668:
2663:
2631:
2625:
2621:
2598:
2594:
2549:
2545:
2538:
2522:
2518:
2463:
2459:
2428:
2421:
2376:
2372:
2327:
2323:
2302:(Pt 4): 421–6.
2292:
2288:
2249:
2245:
2200:
2196:
2161:
2157:
2108:
2101:
2062:
2058:
2009:
2000:
1953:
1949:
1917:
1913:
1866:
1862:
1830:
1826:
1777:
1773:
1716:
1712:
1657:
1653:
1608:
1604:
1559:
1555:
1510:
1506:
1467:
1463:
1436:
1432:
1400:
1394:
1390:
1351:
1347:
1338:
1334:
1287:
1283:
1278:
1274:
1219:
1206:
1201:Perseus Project
1177:
1173:
1169:
1136:André Jagendorf
1127:
1111:plasma membrane
1076:
1065:
1023:
969:
944:
938:
930:
926:
922:
898:
892:
881:
875:
869:
845:
815:signal peptides
803:
779:phosphorylation
750:
723:
710:
704:
695:
689:
640:antenna complex
636:
630:
559:
514:
491:
487:
474:
432:
403:
376:
339:
311:stroma lamellae
292:
270:comes from the
264:
237:thylakoid lumen
198:
197:
189:
182:
175:
168:
161:
151:
142:
135:Thylakoid space
127:
121:
111:
102:
93:
83:
76:
52:
43:
35:
32:Granum, Alberta
28:
23:
22:
18:Thylakoid lumen
15:
12:
11:
5:
2777:
2767:
2766:
2761:
2756:
2754:Photosynthesis
2751:
2737:
2736:
2730:
2715:
2709:
2688:
2682:
2667:
2664:
2662:
2661:
2619:
2592:
2543:
2536:
2516:
2477:(1): 170–177.
2457:
2419:
2370:
2321:
2286:
2243:
2194:
2155:
2099:
2072:(12): 963–77.
2056:
1998:
1967:(7): 4238–42.
1947:
1911:
1860:
1824:
1771:
1710:
1651:
1602:
1553:
1504:
1461:
1450:(3): 177–178.
1430:
1411:(4): 281–288.
1388:
1361:(6): 691–703.
1345:
1332:
1297:(6): 495–505.
1281:
1272:
1204:
1170:
1168:
1165:
1164:
1163:
1161:Photosynthesis
1158:
1153:
1148:
1143:
1138:
1133:
1126:
1123:
1119:phycobilisomes
1107:outer membrane
1095:photosynthesis
1075:
1072:
1063:
1022:
1021:ATP generation
1019:
1015:
1014:
999:
987:
982:, protons and
968:
965:
940:Main article:
937:
934:
933:
932:
928:
924:
920:
917:
909:
908:
905:
894:Main article:
891:
888:
879:
871:Main article:
868:
865:
844:
841:
802:
799:
749:
746:
722:
721:Lumen proteins
719:
706:Main article:
703:
700:
691:Main article:
688:
685:
646:and accessory
632:Main article:
629:
626:
612:reaction. The
590:
589:
584:
579:
558:
555:
513:
510:
489:
485:
478:centrifugation
473:
470:
431:
428:
402:
399:
395:photosynthesis
375:
372:
338:
335:
291:
288:
263:
260:
235:surrounding a
229:photosynthesis
200:
199:
194:Starch granule
187:Plastoglobulus
153:
152:
143:
113:
112:
109:Inner membrane
103:
94:
91:Outer membrane
65:
57:
56:
48:
47:
26:
9:
6:
4:
3:
2:
2776:
2765:
2762:
2760:
2759:Plant anatomy
2757:
2755:
2752:
2750:
2747:
2746:
2744:
2733:
2727:
2723:
2722:
2716:
2712:
2706:
2702:
2697:
2696:
2689:
2685:
2679:
2675:
2670:
2669:
2657:
2653:
2649:
2645:
2642:(2): 225–30.
2641:
2637:
2630:
2623:
2615:
2611:
2607:
2603:
2596:
2588:
2584:
2579:
2574:
2570:
2566:
2562:
2558:
2554:
2547:
2539:
2533:
2529:
2528:
2520:
2512:
2508:
2503:
2498:
2493:
2488:
2484:
2480:
2476:
2472:
2468:
2461:
2453:
2449:
2445:
2441:
2438:(3): 333–47.
2437:
2433:
2426:
2424:
2415:
2411:
2406:
2401:
2397:
2393:
2389:
2385:
2381:
2374:
2366:
2362:
2357:
2352:
2348:
2344:
2341:(1): 159–75.
2340:
2336:
2332:
2325:
2317:
2313:
2309:
2305:
2301:
2297:
2290:
2282:
2278:
2274:
2270:
2266:
2262:
2259:(3): 217–23.
2258:
2254:
2247:
2239:
2235:
2230:
2225:
2221:
2217:
2214:(3): 319–41.
2213:
2209:
2205:
2198:
2192:
2186:
2182:
2178:
2174:
2170:
2166:
2159:
2153:
2147:
2143:
2138:
2133:
2129:
2125:
2122:(2): 478–99.
2121:
2117:
2113:
2106:
2104:
2095:
2091:
2087:
2083:
2079:
2075:
2071:
2067:
2060:
2052:
2048:
2043:
2038:
2034:
2030:
2027:(1): 211–36.
2026:
2022:
2018:
2014:
2007:
2005:
2003:
1994:
1990:
1985:
1980:
1975:
1970:
1966:
1962:
1958:
1951:
1943:
1939:
1935:
1931:
1928:(2): 265–77.
1927:
1923:
1915:
1907:
1903:
1898:
1893:
1888:
1883:
1880:(7): 4243–8.
1879:
1875:
1871:
1864:
1856:
1852:
1848:
1844:
1841:(2): 119–28.
1840:
1836:
1828:
1820:
1816:
1812:
1808:
1804:
1800:
1795:
1790:
1787:(6): 065806.
1786:
1782:
1775:
1767:
1763:
1759:
1755:
1751:
1747:
1743:
1739:
1734:
1729:
1726:(3): 031102.
1725:
1721:
1720:Phys Rev Lett
1714:
1706:
1702:
1697:
1692:
1688:
1684:
1679:
1674:
1670:
1666:
1662:
1655:
1647:
1643:
1638:
1633:
1629:
1625:
1622:(2): 285–96.
1621:
1617:
1613:
1606:
1598:
1594:
1589:
1584:
1580:
1576:
1572:
1568:
1564:
1557:
1549:
1545:
1540:
1535:
1531:
1527:
1524:(9): 2580–6.
1523:
1519:
1515:
1508:
1500:
1496:
1492:
1488:
1484:
1480:
1476:
1472:
1465:
1457:
1453:
1449:
1445:
1441:
1434:
1426:
1422:
1418:
1414:
1410:
1406:
1399:
1392:
1384:
1380:
1376:
1372:
1368:
1364:
1360:
1356:
1349:
1342:
1336:
1328:
1324:
1320:
1316:
1312:
1308:
1304:
1300:
1296:
1292:
1285:
1276:
1268:
1264:
1259:
1254:
1249:
1244:
1240:
1236:
1232:
1228:
1224:
1217:
1215:
1213:
1211:
1209:
1202:
1198:
1197:
1192:
1191:Scott, Robert
1188:
1184:
1175:
1171:
1162:
1159:
1157:
1154:
1152:
1151:Endosymbiosis
1149:
1147:
1144:
1142:
1139:
1137:
1134:
1132:
1129:
1128:
1122:
1120:
1114:
1112:
1108:
1104:
1100:
1096:
1092:
1091:Cyanobacteria
1085:
1084:Synechocystis
1080:
1071:
1069:
1061:
1057:
1053:
1049:
1044:
1040:
1036:
1032:
1028:
1018:
1012:
1008:
1004:
1003:plastoquinone
1000:
996:
992:
991:plastoquinone
988:
986:in the lumen.
985:
981:
977:
974:
973:
972:
964:
962:
958:
954:
950:
943:
918:
914:
913:
912:
906:
903:
902:
901:
897:
887:
885:
874:
864:
862:
858:
849:
840:
838:
833:
829:
825:
820:
816:
807:
798:
796:
792:
788:
784:
780:
776:
772:
771:transcription
768:
767:stoichiometry
763:
759:
758:endosymbiotic
755:
745:
743:
738:
736:
731:
728:
718:
716:
709:
699:
694:
684:
682:
678:
674:
670:
666:
662:
657:
653:
649:
645:
641:
635:
625:
623:
619:
615:
611:
607:
603:
599:
594:
588:
585:
583:
580:
578:
574:
571:
568:
567:
566:
564:
554:
552:
548:
544:
540:
535:
530:
527:
518:
509:
507:
503:
499:
495:
483:
479:
469:
467:
466:
461:
460:Chlamydomonas
457:
452:
450:
445:
441:
437:
427:
425:
420:
416:
412:
408:
398:
396:
392:
388:
384:
383:
371:
369:
364:
363:galactolipids
360:
359:phospholipids
356:
352:
348:
344:
334:
332:
323:
319:
312:
308:
304:
296:
287:
285:
281:
277:
273:
269:
259:
257:
252:
250:
246:
242:
238:
234:
230:
226:
222:
221:cyanobacteria
218:
214:
206:
196:
195:
192:
188:
185:
181:
178:
174:
171:
167:
164:
160:
157:
150:
147:
144:
140:
136:
133:
130:
129:
128:
125:
120:
117:
110:
107:
104:
101:
98:
95:
92:
89:
86:
85:
84:
82:
79:
75:
72:
68:
63:
58:
55:
49:
46:
40:
37:
33:
19:
2720:
2694:
2673:
2639:
2635:
2622:
2605:
2601:
2595:
2560:
2556:
2546:
2526:
2519:
2474:
2470:
2460:
2435:
2431:
2387:
2383:
2373:
2338:
2334:
2324:
2299:
2295:
2289:
2256:
2252:
2246:
2211:
2207:
2197:
2171:(3): 432–6.
2168:
2164:
2158:
2119:
2115:
2069:
2065:
2059:
2024:
2020:
2013:von Heijne G
1964:
1960:
1950:
1925:
1921:
1914:
1877:
1873:
1863:
1838:
1834:
1827:
1784:
1780:
1774:
1723:
1719:
1713:
1668:
1664:
1654:
1619:
1615:
1605:
1570:
1566:
1556:
1521:
1517:
1507:
1477:(3): 241–7.
1474:
1470:
1464:
1447:
1443:
1433:
1408:
1404:
1391:
1358:
1354:
1348:
1335:
1294:
1290:
1284:
1275:
1230:
1226:
1194:
1174:
1141:Chemiosmosis
1115:
1089:
1083:
1060:ATP synthase
1056:ATP synthase
1027:mitochondria
1024:
1016:
970:
945:
942:chemiosmosis
936:Chemiosmosis
910:
899:
876:
854:
812:
762:cell nucleus
751:
739:
732:
727:plastocyanin
724:
711:
708:ATP synthase
702:ATP synthase
696:
668:
664:
660:
644:chlorophylls
637:
628:Photosystems
614:ATP synthase
595:
591:
587:ATP synthase
570:Photosystems
560:
531:
523:
504:solubilizes
475:
463:
455:
453:
433:
414:
410:
406:
404:
379:
377:
342:
340:
328:
321:
306:
283:
279:
275:
267:
265:
253:
244:
217:chloroplasts
212:
211:
190:
183:
176:
169:
162:
155:
154:
145:
134:
131:
124:You are here
123:
118:
115:
114:
105:
96:
87:
77:
70:
69:
66:
44:Cell biology
36:
1343:9 Apr. 2008
1291:J Plant Res
1099:respiration
824:Toc and Tic
787:translation
765:the proper
652:carotenoids
634:Photosystem
551:translation
492:) detaches
436:proplastids
256:quantasomes
243:(singular:
53:Chloroplast
2743:Categories
2335:Plant Cell
2208:Plant Cell
2116:Plant Cell
2021:Plant Cell
1794:1602.03215
1678:1509.00410
1567:Plant Cell
1518:Plant Cell
1167:References
1007:ferredoxin
976:Photolysis
873:photolysis
642:that uses
545:with 11%,
526:proteomics
498:detergents
449:etiolation
444:etioplasts
380:thylakoid
213:Thylakoids
1733:1410.2197
1062:, ADP + P
984:electrons
440:seedlings
430:Formation
355:nanometre
290:Structure
284:thylakoid
268:Thylakoid
266:The word
262:Etymology
122: ◄
119:Thylakoid
2764:Plastids
2656:21473741
2587:17304210
2452:16386331
2414:11127990
2365:16339851
2316:11498001
2281:10066592
2238:10715320
2185:16418230
2146:14729914
2094:15707834
2051:11826309
1993:11274447
1942:17355436
1906:11274448
1855:17346982
1819:28272522
1766:12021024
1758:25658989
1705:36462725
1646:23870120
1597:18952780
1548:16055630
1499:16603410
1327:27225926
1319:15538651
1267:31611387
1125:See also
1103:bacteria
843:Function
832:arginine
650:such as
534:proteome
512:Proteins
415:lamellae
409:(plural
337:Membrane
276:thylakos
180:Ribosome
2701:115–127
2578:1817639
2511:5220864
2479:Bibcode
2405:1692884
2356:1323491
2261:Bibcode
2074:Bibcode
1922:Plant J
1799:Bibcode
1738:Bibcode
1683:Bibcode
1637:3767119
1588:2590735
1539:1197436
1479:Bibcode
1456:3428918
1383:6076741
1375:3320041
1299:Bibcode
1258:6825288
1235:Bibcode
1199:at the
1181:θύλακος
1009:during
993:during
961:protons
951:to the
783:kinases
620:during
543:folding
393:during
280:θύλακος
249:stromal
2728:
2707:
2680:
2654:
2585:
2575:
2557:EMBO J
2534:
2509:
2502:285771
2499:
2450:
2412:
2402:
2363:
2353:
2314:
2279:
2236:
2229:139834
2226:
2183:
2144:
2137:341918
2134:
2092:
2049:
2042:150561
2039:
1991:
1981:
1940:
1904:
1894:
1853:
1817:
1764:
1756:
1703:
1644:
1634:
1595:
1585:
1546:
1536:
1497:
1454:
1425:360223
1423:
1381:
1373:
1325:
1317:
1265:
1255:
1052:stroma
980:oxygen
957:acidic
949:stroma
754:genome
553:(8%).
407:granum
331:stroma
245:granum
166:Stroma
74:Granum
2632:(PDF)
1984:31209
1897:31210
1815:S2CID
1789:arXiv
1762:S2CID
1728:arXiv
1701:S2CID
1673:arXiv
1421:S2CID
1401:(PDF)
1379:S2CID
1323:S2CID
953:lumen
775:light
610:redox
606:NADPH
438:when
411:grana
382:lumen
374:Lumen
274:word
272:Greek
241:grana
139:lumen
2726:ISBN
2705:ISBN
2678:ISBN
2652:PMID
2606:1319
2583:PMID
2532:ISBN
2507:PMID
2448:PMID
2410:PMID
2361:PMID
2312:PMID
2277:PMID
2234:PMID
2181:PMID
2142:PMID
2090:PMID
2047:PMID
1989:PMID
1938:PMID
1902:PMID
1851:PMID
1754:PMID
1642:PMID
1616:Cell
1593:PMID
1544:PMID
1495:PMID
1452:PMID
1371:PMID
1315:PMID
1263:PMID
1097:and
927:O (O
795:mRNA
677:P680
673:P700
654:and
575:and
500:and
378:The
361:and
341:The
219:and
2644:doi
2640:436
2610:doi
2573:PMC
2565:doi
2497:PMC
2487:doi
2440:doi
2436:163
2400:PMC
2392:doi
2388:355
2351:PMC
2343:doi
2304:doi
2269:doi
2224:PMC
2216:doi
2173:doi
2132:PMC
2124:doi
2082:doi
2037:PMC
2029:doi
1979:PMC
1969:doi
1930:doi
1892:PMC
1882:doi
1843:doi
1807:doi
1746:doi
1724:114
1691:doi
1632:PMC
1624:doi
1620:154
1583:PMC
1575:doi
1534:PMC
1526:doi
1487:doi
1413:doi
1363:doi
1307:doi
1295:117
1253:PMC
1243:doi
1231:116
1005:by
916:it.
742:Tat
618:ATP
484:(Na
278:or
227:of
146:3.2
132:3.1
106:2.3
97:2.2
88:2.1
2745::
2703:.
2650:.
2638:.
2634:.
2604:.
2581:.
2571:.
2561:26
2559:.
2555:.
2505:.
2495:.
2485:.
2475:55
2473:.
2469:.
2446:.
2434:.
2422:^
2408:.
2398:.
2386:.
2382:.
2359:.
2349:.
2339:18
2337:.
2333:.
2310:.
2300:29
2298:.
2275:.
2267:.
2255:.
2232:.
2222:.
2212:12
2210:.
2206:.
2189:–
2179:.
2169:47
2167:.
2150:-
2140:.
2130:.
2120:16
2118:.
2114:.
2102:^
2088:.
2080:.
2070:42
2068:.
2045:.
2035:.
2025:14
2023:.
2019:.
2001:^
1987:.
1977:.
1965:98
1963:.
1959:.
1936:.
1926:50
1924:.
1900:.
1890:.
1878:98
1876:.
1872:.
1849:.
1839:45
1837:.
1813:.
1805:.
1797:.
1785:93
1783:.
1760:.
1752:.
1744:.
1736:.
1722:.
1699:.
1689:.
1681:.
1669:94
1667:.
1663:.
1640:.
1630:.
1618:.
1614:.
1591:.
1581:.
1571:20
1569:.
1565:.
1542:.
1532:.
1522:17
1520:.
1516:.
1493:.
1485:.
1473:.
1448:24
1446:.
1442:.
1419:.
1409:15
1407:.
1403:.
1377:.
1369:.
1359:19
1357:.
1321:.
1313:.
1305:.
1293:.
1261:.
1251:.
1241:.
1229:.
1225:.
1207:^
1193:;
1189:;
1185:.
1109:,
624:.
577:II
508:.
488:CO
468:.
325:).
2734:.
2713:.
2686:.
2658:.
2646::
2616:.
2612::
2589:.
2567::
2540:.
2513:.
2489::
2481::
2454:.
2442::
2416:.
2394::
2367:.
2345::
2318:.
2306::
2283:.
2271::
2263::
2257:1
2240:.
2218::
2187:.
2175::
2148:.
2126::
2096:.
2084::
2076::
2053:.
2031::
1995:.
1971::
1944:.
1932::
1908:.
1884::
1857:.
1845::
1821:.
1809::
1801::
1791::
1768:.
1748::
1740::
1730::
1707:.
1693::
1685::
1675::
1648:.
1626::
1599:.
1577::
1550:.
1528::
1501:.
1489::
1481::
1475:9
1458:.
1427:.
1415::
1385:.
1365::
1329:.
1309::
1301::
1269:.
1245::
1237::
1086:)
1064:i
929:2
925:2
921:2
880:2
835:(
669:a
665:a
661:a
573:I
490:3
486:2
191:9
184:8
177:7
170:6
163:5
156:4
141:)
137:(
116:3
78:2
71:1
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.