39:
656:
von der
Malsburg, K; Müller, JM; Bohnert, M; Oeljeklaus, S; Kwiatkowska, P; Becker, T; Loniewska-Lwowska, A; Wiese, S; Rao, S; Milenkovic, D; Hutu, DP; Zerbes, RM; Schulze-Specking, A; Meyer, HE; Martinou, JC; Rospert, S; Rehling, P; Meisinger, C; Veenhuis, M; Warscheid, B; van der Klei, IJ; Pfanner,
239:
For typical liver mitochondria, the area of the inner membrane is about 5 times as large as the outer membrane due to cristae. This ratio is variable and mitochondria from cells that have a greater demand for ATP, such as muscle cells, contain even more cristae. Cristae membranes are studded on the
260:
Cristae and the inner boundary membranes are separated by junctions. The end of cristae are partially closed by transmembrane protein complexes that bind head to head and link opposing crista membranes in a bottleneck-like fashion. For example, deletion of the junction protein
224:
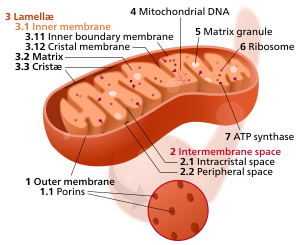
The inner membrane creates two compartments. The region between the inner and outer membrane, called the intermembrane space, is largely continuous with the cytosol, while the more sequestered space inside the inner membrane is called the matrix.
217:, separated by crista junctions from the inner boundary membrane juxtaposed to the outer membrane. Cristae significantly increase the total membrane surface area compared to a smooth inner membrane and thereby the available working space for
337:
from the cytosolic environment. This compartmentalization is a necessary feature for metabolism. The inner mitochondrial membrane is both an electrical insulator and chemical barrier. Sophisticated
265:
leads to a reduced inner membrane potential and impaired growth and to dramatically aberrant inner membrane structures which form concentric stacks instead of the typical invaginations.
748:
Comte J, Maïsterrena B, Gautheron DC (January 1976). "Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes".
699:
Rabl, R; Soubannier, V; Scholz, R; Vogel, F; Mendl, N; Vasiljev-Neumeyer, A; Körner, C; Jagasia, R; Keil, T; Baumeister, W; Cyrklaff, M; Neupert, W; Reichert, AS (15 June 2009).
814:
333:, carbon dioxide, and water only. It is much less permeable to ions and small molecules than the outer membrane, creating compartments by separating the
306:
mitochondria, phosphatidylcholine makes up 38.4% of the IMM, phosphatidylethanolamine makes up 24.0%, phosphatidylinositol 16.2%, cardiolipin 16.1%,
213:
The structure of the inner mitochondrial membrane is extensively folded and compartmentalized. The numerous invaginations of the membrane are called
904:
377:
1385:
821:
1380:
1471:
1053:
1139:
431:
948:
897:
550:
1239:
1174:
382:
1184:
1009:
461:
545:
1510:
1308:
943:
441:
426:
1677:
1505:
1169:
890:
466:
451:
372:
784:
1500:
1286:
1244:
446:
1090:
922:
480:
436:
318:
50:
1144:
847:
701:"Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g"
1318:
1313:
1117:
983:
513:
485:
1697:
1043:
218:
288:
makes up the majority of the inner mitochondrial membrane at 37.0% of the phospholipid composition.
1234:
1112:
363:
285:
1281:
1068:
658:
494:
421:
387:
278:
1276:
933:
397:
95:
1702:
1650:
1390:
1266:
1102:
1080:
555:
499:
471:
317:
In the inner mitochondrial membrane, the protein-to-lipid ratio is 80:20, in contrast to the
1491:
1397:
1254:
1203:
1149:
407:
334:
297:
198:
136:
580:
Mannella CA (2006). "Structure and dynamics of the mitochondrial inner membrane cristae".
8:
392:
289:
202:
70:
1421:
1189:
1063:
725:
700:
560:
527:
307:
659:"Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis"
273:
The inner membrane of mitochondria is similar in lipid composition to the membrane of
1411:
1214:
995:
765:
761:
730:
681:
638:
597:
540:
311:
281:
of the origin of mitochondria as prokaryotes internalized by a eukaryotic host cell.
155:
1224:
1017:
757:
720:
712:
673:
628:
589:
1129:
1022:
677:
633:
616:
593:
456:
338:
60:
518:
1691:
913:
876:
302:
28:
1546:
1521:
1179:
1058:
971:
734:
685:
642:
601:
402:
341:
exist to allow specific molecules to cross this barrier. There are several
249:
248:, the site of proton-gradient driven ATP synthesis. Cristae affect overall
194:
174:
19:
769:
716:
882:
293:
617:"MINOS is plus: a Mitofilin complex for mitochondrial membrane contacts"
1298:
960:
655:
1229:
504:
38:
145:
1375:
1249:
412:
342:
274:
241:
167:
1535:
1530:
1450:
350:
234:
214:
1640:
1635:
1605:
1600:
1480:
1460:
1455:
1445:
1440:
1435:
1430:
346:
330:
1660:
1655:
1645:
1630:
1625:
1620:
1615:
1610:
1595:
1590:
1585:
1580:
1575:
1570:
1565:
1560:
1555:
1351:
1338:
582:
Biochimica et
Biophysica Acta (BBA) - Molecular Cell Research
345:
systems embedded in the inner membrane, allowing exchange of
262:
783:
Lomize, Andrel; Lomize, Mikhail; Pogozheva, Irina (2013).
747:
698:
240:
matrix side with small round protein complexes known as
782:
879:
Orientations of
Proteins in Membranes (OPM) database
657:
N; Chacinska, A; van der Laan, M (18 October 2011).
815:"Mitochondria: Structure and Role in Respiration"
1689:
378:Electron-transferring-flavoprotein dehydrogenase
898:
579:
1386:Mitochondrial permeability transition pore
912:
905:
891:
877:Mitochondrial inner membrane (97 proteins)
785:"Membrane Protein Lipid Composition Atlas"
741:
356:
329:The inner membrane is freely permeable to
277:. This phenomenon can be explained by the
820:. Nature Publishing Group. Archived from
724:
632:
1381:Mitochondrial membrane transport protein
845:
808:
806:
614:
846:Caprette, David R. (12 December 1996).
573:
1690:
1140:Cholesterol side-chain cleavage enzyme
812:
432:Cholesterol side-chain cleavage enzyme
886:
803:
789:Orientations of Proteins in Membranes
44:Components of a typical mitochondrion
255:
1054:Coenzyme Q – cytochrome c reductase
839:
551:Mitochondrial trifunctional protein
13:
1240:Oxoglutarate dehydrogenase complex
1175:Glycerol-3-phosphate dehydrogenase
383:Electron-transferring flavoprotein
14:
1714:
1185:Carnitine palmitoyltransferase II
870:
462:Translocase of the inner membrane
1309:Carbamoyl phosphate synthetase I
949:Long-chain-fatty-acid—CoA ligase
944:Carnitine palmitoyltransferase I
615:Herrmann, JM (18 October 2011).
442:Carnitine O-palmitoyltransferase
427:ATP-binding cassette transporter
37:
1170:Glutamate aspartate transporter
467:Glutamate aspartate transporter
452:Carnitine O-octanoyltransferase
373:NADH dehydrogenase (ubiquinone)
324:
1287:Pyruvate dehydrogenase complex
1245:Succinyl coenzyme A synthetase
776:
692:
649:
608:
353:and the mitochondrial matrix.
268:
1:
566:
447:Carnitine O-acetyltransferase
197:membrane which separates the
1091:Dihydroorotate dehydrogenase
762:10.1016/0005-2736(76)90353-9
678:10.1016/j.devcel.2011.08.026
634:10.1016/j.devcel.2011.09.013
594:10.1016/j.bbamcr.2006.04.006
481:Dihydroorotate dehydrogenase
437:Protein tyrosine phosphatase
208:
187:inner mitochondrial membrane
7:
1145:Steroid 11-beta-hydroxylase
848:"Structure of Mitochondria"
705:The Journal of Cell Biology
534:
284:In pig heart mitochondria,
10:
1719:
1319:N-Acetylglutamate synthase
1314:Ornithine transcarbamylase
1118:Glycerol phosphate shuttle
984:monoamine neurotransmitter
514:Protoporphyrinogen oxidase
486:Thymidylate synthase (FAD)
252:function of mitochondria.
232:
228:
1673:
1544:
1519:
1489:
1469:
1419:
1410:
1371:
1364:
1347:
1327:
1295:
1263:
1211:
1202:
1158:
1126:
1099:
1077:
1044:oxidative phosphorylation
1040:
1031:
1008:
980:
957:
930:
921:
219:oxidative phosphorylation
36:
27:
18:
1235:Isocitrate dehydrogenase
1113:Malate-aspartate shuttle
852:Experimental Biosciences
791:. University of Michigan
364:Electron transport chain
286:phosphatidylethanolamine
1282:Glutamate dehydrogenase
1069:Succinate dehydrogenase
813:Krauss, Stefan (2001).
495:HtrA serine peptidase 2
388:Succinate dehydrogenase
357:IMM-associated proteins
279:endosymbiont hypothesis
120:Inner boundary membrane
1678:mitochondrial diseases
1277:Aspartate transaminase
934:fatty acid degradation
750:Biochim. Biophys. Acta
398:Cytochrome bc1 complex
292:makes up about 26.5%,
1391:Mitochondrial carrier
1267:anaplerotic reactions
1103:mitochondrial shuttle
1081:pyrimidine metabolism
717:10.1083/jcb.200811099
556:Mitochondrial shuttle
500:Adrenodoxin reductase
472:Pyrimidine metabolism
1398:Translocator protein
1255:Malate dehydrogenase
1150:Aldosterone synthase
408:Cytochrome c oxidase
298:phosphatidylinositol
199:mitochondrial matrix
1010:Intermembrane space
422:ATP–ADP translocase
393:Alternative oxidase
290:Phosphatidylcholine
203:intermembrane space
71:Intermembrane space
1365:Other/to be sorted
1330:alcohol metabolism
1190:Uncoupling protein
1064:NADH dehydrogenase
827:on 21 October 2012
666:Developmental Cell
621:Developmental Cell
561:Transport proteins
528:Uncoupling protein
321:, which is 50:50.
308:phosphatidylserine
80:Intracristal space
1685:
1684:
1669:
1668:
1412:Mitochondrial DNA
1406:
1405:
1360:
1359:
1215:citric acid cycle
1198:
1197:
1004:
1003:
996:Monoamine oxidase
854:. Rice University
541:Citric acid cycle
505:Heme biosynthesis
312:phosphatidic acid
256:Cristae junctions
183:
182:
156:Mitochondrial DNA
1710:
1698:Membrane biology
1549:
1524:
1494:
1474:
1424:
1417:
1416:
1369:
1368:
1332:
1302:
1270:
1225:Citrate synthase
1218:
1209:
1208:
1163:
1133:
1106:
1084:
1047:
1038:
1037:
1018:Adenylate kinase
989:
965:
937:
928:
927:
907:
900:
893:
884:
883:
864:
863:
861:
859:
843:
837:
836:
834:
832:
826:
819:
810:
801:
800:
798:
796:
780:
774:
773:
745:
739:
738:
728:
696:
690:
689:
663:
653:
647:
646:
636:
612:
606:
605:
588:(5–6): 542–548.
577:
339:ion transporters
127:Cristal membrane
113:
87:Peripheral space
41:
31:
22:
16:
15:
1718:
1717:
1713:
1712:
1711:
1709:
1708:
1707:
1688:
1687:
1686:
1681:
1665:
1545:
1540:
1520:
1515:
1490:
1485:
1470:
1465:
1420:
1402:
1356:
1343:
1328:
1323:
1296:
1291:
1264:
1259:
1212:
1194:
1159:
1154:
1130:steroidogenesis
1127:
1122:
1100:
1095:
1078:
1073:
1041:
1027:
1023:Creatine kinase
1000:
986:
981:
976:
958:
953:
931:
917:
911:
873:
868:
867:
857:
855:
844:
840:
830:
828:
824:
817:
811:
804:
794:
792:
781:
777:
746:
742:
697:
693:
661:
654:
650:
613:
609:
578:
574:
569:
546:Proton gradient
537:
532:
457:Cytochrome P450
359:
327:
271:
258:
245:
237:
231:
211:
179:
178:
170:
163:
158:
148:
139:
128:
121:
114:
108:
98:
88:
81:
73:
63:
53:
29:
20:
12:
11:
5:
1716:
1706:
1705:
1700:
1683:
1682:
1674:
1671:
1670:
1667:
1666:
1664:
1663:
1658:
1653:
1648:
1643:
1638:
1633:
1628:
1623:
1618:
1613:
1608:
1603:
1598:
1593:
1588:
1583:
1578:
1573:
1568:
1563:
1558:
1552:
1550:
1542:
1541:
1539:
1538:
1533:
1527:
1525:
1517:
1516:
1514:
1513:
1508:
1503:
1497:
1495:
1487:
1486:
1484:
1483:
1477:
1475:
1467:
1466:
1464:
1463:
1458:
1453:
1448:
1443:
1438:
1433:
1427:
1425:
1414:
1408:
1407:
1404:
1403:
1401:
1400:
1395:
1394:
1393:
1388:
1378:
1372:
1366:
1362:
1361:
1358:
1357:
1355:
1354:
1348:
1345:
1344:
1342:
1341:
1335:
1333:
1325:
1324:
1322:
1321:
1316:
1311:
1305:
1303:
1293:
1292:
1290:
1289:
1284:
1279:
1273:
1271:
1261:
1260:
1258:
1257:
1252:
1247:
1242:
1237:
1232:
1227:
1221:
1219:
1206:
1200:
1199:
1196:
1195:
1193:
1192:
1187:
1182:
1177:
1172:
1166:
1164:
1156:
1155:
1153:
1152:
1147:
1142:
1136:
1134:
1124:
1123:
1121:
1120:
1115:
1109:
1107:
1097:
1096:
1094:
1093:
1087:
1085:
1075:
1074:
1072:
1071:
1066:
1061:
1056:
1050:
1048:
1035:
1033:Inner membrane
1029:
1028:
1026:
1025:
1020:
1014:
1012:
1006:
1005:
1002:
1001:
999:
998:
992:
990:
978:
977:
975:
974:
968:
966:
955:
954:
952:
951:
946:
940:
938:
925:
923:Outer membrane
919:
918:
910:
909:
902:
895:
887:
881:
880:
872:
871:External links
869:
866:
865:
838:
802:
775:
740:
711:(6): 1047–63.
691:
672:(4): 694–707.
648:
627:(4): 599–600.
607:
571:
570:
568:
565:
564:
563:
558:
553:
548:
543:
536:
533:
531:
530:
524:
523:
522:
521:
519:Ferrochelatase
516:
508:
507:
502:
497:
491:
490:
489:
488:
483:
475:
474:
469:
464:
459:
454:
449:
444:
439:
434:
429:
424:
418:
417:
416:
415:
410:
405:
400:
395:
390:
385:
380:
375:
367:
366:
360:
358:
355:
326:
323:
319:outer membrane
270:
267:
257:
254:
243:
233:Main article:
230:
227:
210:
207:
181:
180:
162:Matrix granule
150:
149:
140:
131:
130:
129:
122:
106:Inner membrane
90:
89:
82:
65:
64:
51:Outer membrane
42:
34:
33:
25:
24:
9:
6:
4:
3:
2:
1715:
1704:
1701:
1699:
1696:
1695:
1693:
1680:
1679:
1672:
1662:
1659:
1657:
1654:
1652:
1649:
1647:
1644:
1642:
1639:
1637:
1634:
1632:
1629:
1627:
1624:
1622:
1619:
1617:
1614:
1612:
1609:
1607:
1604:
1602:
1599:
1597:
1594:
1592:
1589:
1587:
1584:
1582:
1579:
1577:
1574:
1572:
1569:
1567:
1564:
1562:
1559:
1557:
1554:
1553:
1551:
1548:
1543:
1537:
1534:
1532:
1529:
1528:
1526:
1523:
1518:
1512:
1509:
1507:
1504:
1502:
1499:
1498:
1496:
1493:
1488:
1482:
1479:
1478:
1476:
1473:
1468:
1462:
1459:
1457:
1454:
1452:
1449:
1447:
1444:
1442:
1439:
1437:
1434:
1432:
1429:
1428:
1426:
1423:
1418:
1415:
1413:
1409:
1399:
1396:
1392:
1389:
1387:
1384:
1383:
1382:
1379:
1377:
1374:
1373:
1370:
1367:
1363:
1353:
1350:
1349:
1346:
1340:
1337:
1336:
1334:
1331:
1326:
1320:
1317:
1315:
1312:
1310:
1307:
1306:
1304:
1301:
1300:
1294:
1288:
1285:
1283:
1280:
1278:
1275:
1274:
1272:
1269:
1268:
1262:
1256:
1253:
1251:
1248:
1246:
1243:
1241:
1238:
1236:
1233:
1231:
1228:
1226:
1223:
1222:
1220:
1217:
1216:
1210:
1207:
1205:
1201:
1191:
1188:
1186:
1183:
1181:
1178:
1176:
1173:
1171:
1168:
1167:
1165:
1162:
1157:
1151:
1148:
1146:
1143:
1141:
1138:
1137:
1135:
1132:
1131:
1125:
1119:
1116:
1114:
1111:
1110:
1108:
1105:
1104:
1098:
1092:
1089:
1088:
1086:
1083:
1082:
1076:
1070:
1067:
1065:
1062:
1060:
1057:
1055:
1052:
1051:
1049:
1046:
1045:
1039:
1036:
1034:
1030:
1024:
1021:
1019:
1016:
1015:
1013:
1011:
1007:
997:
994:
993:
991:
988:
985:
979:
973:
970:
969:
967:
964:
962:
956:
950:
947:
945:
942:
941:
939:
936:
935:
929:
926:
924:
920:
915:
914:Mitochondrial
908:
903:
901:
896:
894:
889:
888:
885:
878:
875:
874:
853:
849:
842:
823:
816:
809:
807:
790:
786:
779:
771:
767:
763:
759:
756:(2): 271–84.
755:
751:
744:
736:
732:
727:
722:
718:
714:
710:
706:
702:
695:
687:
683:
679:
675:
671:
667:
660:
652:
644:
640:
635:
630:
626:
622:
618:
611:
603:
599:
595:
591:
587:
583:
576:
572:
562:
559:
557:
554:
552:
549:
547:
544:
542:
539:
538:
529:
526:
525:
520:
517:
515:
512:
511:
510:
509:
506:
503:
501:
498:
496:
493:
492:
487:
484:
482:
479:
478:
477:
476:
473:
470:
468:
465:
463:
460:
458:
455:
453:
450:
448:
445:
443:
440:
438:
435:
433:
430:
428:
425:
423:
420:
419:
414:
411:
409:
406:
404:
401:
399:
396:
394:
391:
389:
386:
384:
381:
379:
376:
374:
371:
370:
369:
368:
365:
362:
361:
354:
352:
348:
344:
340:
336:
332:
322:
320:
315:
313:
309:
305:
304:
303:S. cerevisiae
299:
295:
291:
287:
282:
280:
276:
266:
264:
253:
251:
247:
236:
226:
222:
220:
216:
206:
204:
200:
196:
195:mitochondrial
192:
188:
177:
176:
173:
169:
166:
161:
157:
154:
147:
144:
141:
138:
135:
132:
126:
123:
119:
116:
115:
112:
107:
104:
101:
100:
99:
97:
94:
86:
83:
79:
76:
75:
74:
72:
69:
62:
59:
56:
55:
54:
52:
49:
45:
40:
35:
32:
30:mitochondrion
26:
23:
17:
1703:Mitochondria
1675:
1522:ATP synthase
1329:
1297:
1265:
1213:
1180:ATP synthase
1160:
1128:
1101:
1079:
1059:Cytochrome c
1042:
1032:
982:
972:Kynureninase
959:
932:
856:. Retrieved
851:
841:
829:. Retrieved
822:the original
793:. Retrieved
788:
778:
753:
749:
743:
708:
704:
694:
669:
665:
651:
624:
620:
610:
585:
581:
575:
403:Cytochrome c
349:between the
328:
325:Permeability
316:
301:
283:
272:
259:
250:chemiosmotic
238:
223:
212:
190:
186:
184:
175:ATP synthase
171:
164:
159:
152:
151:
142:
133:
124:
117:
111:You are here
110:
105:
102:
92:
91:
84:
77:
67:
66:
57:
47:
46:
43:
21:Cell biology
1472:Complex III
296:25.4%, and
294:cardiolipin
269:Composition
1692:Categories
1492:Complex IV
1299:urea cycle
987:metabolism
963:metabolism
961:tryptophan
567:References
310:3.8%, and
1676:see also
1422:Complex I
1230:Aconitase
300:4.5%. In
246:particles
209:Structure
201:from the
193:) is the
109: ◄
1376:Frataxin
1250:Fumarase
916:proteins
795:10 April
735:19528297
686:21944719
643:22014515
602:16730811
535:See also
413:F-ATPase
343:antiport
275:bacteria
168:Ribosome
1536:MT-ATP8
1531:MT-ATP6
1451:MT-ND4L
858:9 April
831:9 April
770:1247555
726:2711607
351:cytosol
235:Cristae
229:Cristae
215:cristae
96:Lamella
1641:MT-TS2
1636:MT-TS1
1606:MT-TL2
1601:MT-TL1
1511:MT-CO3
1506:MT-CO2
1501:MT-CO1
1481:MT-CYB
1461:MT-ND6
1456:MT-ND5
1446:MT-ND4
1441:MT-ND3
1436:MT-ND2
1431:MT-ND1
1204:Matrix
768:
733:
723:
684:
641:
600:
347:anions
335:matrix
331:oxygen
314:1.5%.
146:Cristæ
137:Matrix
1661:MT-TY
1656:MT-TW
1651:MT-TV
1646:MT-TT
1631:MT-TR
1626:MT-TQ
1621:MT-TP
1616:MT-TN
1611:MT-TM
1596:MT-TK
1591:MT-TI
1586:MT-TH
1581:MT-TG
1576:MT-TF
1571:MT-TE
1566:MT-TD
1561:MT-TC
1556:MT-TA
1352:PMPCB
1339:ALDH2
1161:other
825:(PDF)
818:(PDF)
662:(PDF)
61:Porin
1547:tRNA
860:2014
833:2014
797:2014
766:PMID
731:PMID
682:PMID
639:PMID
598:PMID
586:1763
263:IMMT
185:The
125:3.12
118:3.11
758:doi
754:419
721:PMC
713:doi
709:185
674:doi
629:doi
590:doi
191:IMM
143:3.3
134:3.2
103:3.1
85:2.2
78:2.1
58:1.1
1694::
850:.
805:^
787:.
764:.
752:.
729:.
719:.
707:.
703:.
680:.
670:21
668:.
664:.
637:.
625:21
623:.
619:.
596:.
584:.
221:.
205:.
906:e
899:t
892:v
862:.
835:.
799:.
772:.
760::
737:.
715::
688:.
676::
645:.
631::
604:.
592::
244:1
242:F
189:(
172:7
165:6
160:5
153:4
93:3
68:2
48:1
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.