22:
9530:
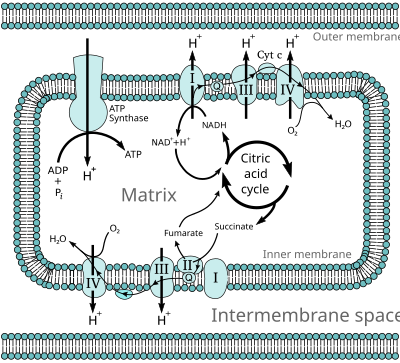
1948:". In this model, the various complexes exist as organized sets of interacting enzymes. These associations might allow channeling of substrates between the various enzyme complexes, increasing the rate and efficiency of electron transfer. Within such mammalian supercomplexes, some components would be present in higher amounts than others, with some data suggesting a ratio between complexes I/II/III/IV and the ATP synthase of approximately 1:1:3:7:4. However, the debate over this supercomplex hypothesis is not completely resolved, as some data do not appear to fit with this model.
484:
10763:
2571:, which can be shifted by altering the proton-motive force. In the absence of a proton-motive force, the ATP synthase reaction will run from right to left, hydrolyzing ATP and pumping protons out of the matrix across the membrane. However, when the proton-motive force is high, the reaction is forced to run in the opposite direction; it proceeds from left to right, allowing protons to flow down their concentration gradient and turning ADP into ATP. Indeed, in the closely related
10793:
10781:
1389:
10873:
10857:
10841:
10829:
10817:
10805:
649:; in other words, they will release a large amount of energy upon oxidation. However, the cell does not release this energy all at once, as this would be an uncontrollable reaction. Instead, the electrons are removed from NADH and passed to oxygen through a series of enzymes that each release a small amount of the energy. This set of enzymes, consisting of complexes I through IV, is called the electron transport chain and is found in the
1675:
1102:
886:
2379:, there are two different types of ubiquinol oxidase using oxygen as an electron acceptor. Under highly aerobic conditions, the cell uses an oxidase with a low affinity for oxygen that can transport two protons per electron. However, if levels of oxygen fall, they switch to an oxidase that transfers only one proton per electron, but has a high affinity for oxygen.
2330:
succinate/fumarate pair is unusual, as its midpoint potential is close to zero. Succinate can therefore be oxidized to fumarate if a strong oxidizing agent such as oxygen is available, or fumarate can be reduced to succinate using a strong reducing agent such as formate. These alternative reactions are catalyzed by
2983:
level can alter ATP production rates. Under anoxic conditions, ATP-synthase will commit 'cellular treason' and run in reverse, forcing protons from the matrix back into the inner membrane space, using up ATP in the process. The proton motive force and ATP production can be maintained by intracellular
2594:
portion contains six proteins of two different kinds (three α subunits and three β subunits), whereas the "stalk" consists of one protein: the γ subunit, with the tip of the stalk extending into the ball of α and β subunits. Both the α and β subunits bind nucleotides, but only the β subunits catalyze
1970:
possess a large variety of electron-transfer enzymes. These use an equally wide set of chemicals as substrates. In common with eukaryotes, prokaryotic electron transport uses the energy released from the oxidation of a substrate to pump ions across a membrane and generate an electrochemical gradient.
1655:
As coenzyme Q is reduced to ubiquinol on the inner side of the membrane and oxidized to ubiquinone on the other, a net transfer of protons across the membrane occurs, adding to the proton gradient. The rather complex two-step mechanism by which this occurs is important, as it increases the efficiency
503:
The electron transport chain carries both protons and electrons, passing electrons from donors to acceptors, and transporting protons across a membrane. These processes use both soluble and protein-bound transfer molecules. In mitochondria, electrons are transferred within the intermembrane space by
413:
ATP synthase releases this stored energy by completing the circuit and allowing protons to flow down the electrochemical gradient, back to the N-side of the membrane. The electrochemical gradient drives the rotation of part of the enzyme's structure and couples this motion to the synthesis of ATP.
1084:
in complex I that cause the protein to bind protons on the N-side of the membrane and release them on the P-side of the membrane. Finally, the electrons are transferred from the chain of iron–sulfur clusters to a ubiquinone molecule in the membrane. Reduction of ubiquinone also contributes to the
3009:
inhibits ATP synthase, protons cannot pass back into the mitochondrion. As a result, the proton pumps are unable to operate, as the gradient becomes too strong for them to overcome. NADH is then no longer oxidized and the citric acid cycle ceases to operate because the concentration of NAD falls
2931:
in complex III, as a highly reactive ubisemiquinone free radical is formed as an intermediate in the Q cycle. This unstable species can lead to electron "leakage" when electrons transfer directly to oxygen, forming superoxide. As the production of reactive oxygen species by these proton-pumping
2641:
and involves the active site of a β subunit cycling between three states. In the "open" state, ADP and phosphate enter the active site (shown in brown in the diagram). The protein then closes up around the molecules and binds them loosely – the "loose" state (shown in red). The enzyme then
680:
across the membrane. The energy stored in this potential is then used by ATP synthase to produce ATP. Oxidative phosphorylation in the eukaryotic mitochondrion is the best-understood example of this process. The mitochondrion is present in almost all eukaryotes, with the exception of anaerobic
2363:
Prokaryotes control their use of these electron donors and acceptors by varying which enzymes are produced, in response to environmental conditions. This flexibility is possible because different oxidases and reductases use the same ubiquinone pool. This allows many combinations of enzymes to
2329:
can grow with reducing agents such as formate, hydrogen, or lactate as electron donors, and nitrate, DMSO, or oxygen as acceptors. The larger the difference in midpoint potential between an oxidizing and reducing agent, the more energy is released when they react. Out of these compounds, the
1898:
have alternative NADH oxidases, which oxidize NADH in the cytosol rather than in the mitochondrial matrix, and pass these electrons to the ubiquinone pool. These enzymes do not transport protons, and, therefore, reduce ubiquinone without altering the electrochemical gradient across the inner
596:. Metal ion cofactors undergo redox reactions without binding or releasing protons, so in the electron transport chain they serve solely to transport electrons through proteins. Electrons move quite long distances through proteins by hopping along chains of these cofactors. This occurs by
2397:, is the final enzyme in the oxidative phosphorylation pathway. This enzyme is found in all forms of life and functions in the same way in both prokaryotes and eukaryotes. The enzyme uses the energy stored in a proton gradient across a membrane to drive the synthesis of ATP from ADP and
2926:
The cytochrome c oxidase complex is highly efficient at reducing oxygen to water, and it releases very few partly reduced intermediates; however small amounts of superoxide anion and peroxide are produced by the electron transport chain. Particularly important is the reduction of
3013:
Many site-specific inhibitors of the electron transport chain have contributed to the present knowledge of mitochondrial respiration. Synthesis of ATP is also dependent on the electron transport chain, so all site-specific inhibitors also inhibit ATP formation. The fish poison
387:
process, which requires an input of energy. Both the electron transport chain and the ATP synthase are embedded in a membrane, and energy is transferred from the electron transport chain to the ATP synthase by movements of protons across this membrane, in a process called
2626:
3280:
2932:
complexes is greatest at high membrane potentials, it has been proposed that mitochondria regulate their activity to maintain the membrane potential within a narrow range that balances ATP production against oxidant generation. For instance, oxidants can activate
2881:
1712:
oxygen is reduced to water in this step. Both the direct pumping of protons and the consumption of matrix protons in the reduction of oxygen contribute to the proton gradient. The reaction catalyzed is the oxidation of cytochrome c and the reduction of oxygen:
1943:
The original model for how the respiratory chain complexes are organized was that they diffuse freely and independently in the mitochondrial membrane. However, recent data suggest that the complexes might form higher-order structures called supercomplexes or
1980:
The main difference between eukaryotic and prokaryotic oxidative phosphorylation is that bacteria and archaea use many different substances to donate or accept electrons. This allows prokaryotes to grow under a wide variety of environmental conditions. In
2740:
because it is a strong oxidizing agent. The reduction of oxygen does involve potentially harmful intermediates. Although the transfer of four electrons and four protons reduces oxygen to water, which is harmless, transfer of one or two electrons produces
916:(kDa). The structure is known in detail only from a bacterium; in most organisms the complex resembles a boot with a large "ball" poking out from the membrane into the mitochondrion. The genes that encode the individual proteins are contained in both the
1595:
1871:
1124:, is a second entry point to the electron transport chain. It is unusual because it is the only enzyme that is part of both the citric acid cycle and the electron transport chain. Complex II consists of four protein subunits and contains a bound
2717:
So we can conclude that when NADH is oxidized, about 42% of energy is conserved in the form of three ATPs and the remaining (58%) energy is lost as heat (unless the chemical energy of ATP under physiological conditions was underestimated).
3288:
For another twenty years, the mechanism by which ATP is generated remained mysterious, with scientists searching for an elusive "high-energy intermediate" that would link oxidation and phosphorylation reactions. This puzzle was solved by
1693:, is the final protein complex in the electron transport chain. The mammalian enzyme has an extremely complicated structure and contains 13 subunits, two heme groups, as well as multiple metal ion cofactors – in all, three atoms of
2545:
1224:, an enzyme similar to complex II, fumarate reductase (menaquinol:fumarate oxidoreductase, or QFR), operates in reverse to oxidize ubiquinol and reduce fumarate. This allows the worm to survive in the anaerobic environment of the
1047:
7461:
Tsubaki M (January 1993). "Fourier-transform infrared study of cyanide binding to the Fea3-CuB binuclear site of bovine heart cytochrome c oxidase: implication of the redox-linked conformational change at the binuclear site".
3029:
Carbon monoxide, cyanide, hydrogen sulphide and azide effectively inhibit cytochrome oxidase. Carbon monoxide reacts with the reduced form of the cytochrome while cyanide and azide react with the oxidised form. An antibiotic,
3004:
that inhibit oxidative phosphorylation. Although any one of these toxins inhibits only one enzyme in the electron transport chain, inhibition of any step in this process will halt the rest of the process. For example, if
2351:
oxidize nitrite to nitrate, donating the electrons to oxygen. The small amount of energy released in this reaction is enough to pump protons and generate ATP, but not enough to produce NADH or NADPH directly for use in
6777:"Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model"
2622:. This movement of the tip of the γ subunit within the ball of α and β subunits provides the energy for the active sites in the β subunits to undergo a cycle of movements that produces and then releases ATP.
1342:
1197:
2663:
synthase, a form of the enzyme that contains additional proteins with little similarity in sequence to other bacterial and eukaryotic ATP synthase subunits. It is possible that, in some species, the A
394:. A current of protons is driven from the negative N-side of the membrane to the positive P-side through the proton-pumping enzymes of the electron transport chain. The movement of protons creates an
1931:, and infection by pathogens, as well as other factors that inhibit the full electron transport chain. Alternative pathways might, therefore, enhance an organism's resistance to injury, by reducing
1140:
and reduces ubiquinone. As this reaction releases less energy than the oxidation of NADH, complex II does not transport protons across the membrane and does not contribute to the proton gradient.
7390:
Joshi S, Huang YG (August 1991). "ATP synthase complex from bovine heart mitochondria: the oligomycin sensitivity conferring protein is essential for dicyclohexyl carbodiimide-sensitive ATPase".
1077:. The electrons are then transferred through a series of iron–sulfur clusters: the second kind of prosthetic group present in the complex. There are both and iron–sulfur clusters in complex I.
6355:
Van
Walraven HS, Strotmann H, Schwarz O, Rumberg B (February 1996). "The H+/ATP coupling ratio of the ATP synthase from thiol-modulated chloroplasts and two cyanobacterial strains is four".
453:
produces only 2 ATP molecules, but somewhere between 30 and 36 ATPs are produced by the oxidative phosphorylation of the 10 NADH and 2 succinate molecules made by converting one molecule of
2757:
4414:
Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, et al. (January 2003). "Architecture of succinate dehydrogenase and reactive oxygen species generation".
1065:
The start of the reaction, and indeed of the entire electron chain, is the binding of a NADH molecule to complex I and the donation of two electrons. The electrons enter complex I via a
7587:
Dervartanian DV, Veeger C (November 1964). "STUDIES ON SUCCINATE DEHYDROGENASE. I. SPECTRAL PROPERTIES OF THE PURIFIED ENZYME AND FORMATION OF ENZYME-COMPETITIVE INHIBITOR COMPLEXES".
1927:
yields than the full pathway. The advantages produced by a shortened pathway are not entirely clear. However, the alternative oxidase is produced in response to stresses such as cold,
1708:
This enzyme mediates the final reaction in the electron transport chain and transfers electrons to oxygen and hydrogen (protons), while pumping protons across the membrane. The final
5125:
Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, et al. (May 1996). "The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A".
1617:
donor to a cytochrome c acceptor at a time, the reaction mechanism of complex III is more elaborate than those of the other respiratory complexes, and occurs in two steps called the
1443:
group. The iron atoms inside complex III's heme groups alternate between a reduced ferrous (+2) and oxidized ferric (+3) state as the electrons are transferred through the protein.
2642:
changes shape again and forces these molecules together, with the active site in the resulting "tight" state (shown in pink) binding the newly produced ATP molecule with very high
584:
and cytochromes. There are several types of iron–sulfur cluster. The simplest kind found in the electron transfer chain consists of two iron atoms joined by two atoms of inorganic
2405:). Estimates of the number of protons required to synthesize one ATP have ranged from three to four, with some suggesting cells can vary this ratio, to suit different conditions.
1462:
588:; these are called clusters. The second kind, called , contains a cube of four iron atoms and four sulfur atoms. Each iron atom in these clusters is coordinated by an additional
9885:
569:
oxidized on the other, ubiquinone will couple these reactions and shuttle protons across the membrane. Some bacterial electron transport chains use different quinones, such as
367:. This means one cannot occur without the other. The chain of redox reactions driving the flow of electrons through the electron transport chain, from electron donors such as
1080:
As the electrons pass through this complex, four protons are pumped from the matrix into the intermembrane space. Exactly how this occurs is unclear, but it seems to involve
8442:
8430:
5453:
Ito Y, Saisho D, Nakazono M, Tsutsumi N, Hirai A (December 1997). "Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature".
1989:
of a chemical measures how much energy is released when it is oxidized or reduced, with reducing agents having negative potentials and oxidizing agents positive potentials.
8454:
2988:
can freely diffuse across the mitochondrial outer-membrane and acidify the inter-membrane space, hence directly contributing to the proton motive force and ATP production.
1721:
8516:
893:. The abbreviations are discussed in the text. In all diagrams of respiratory complexes in this article, the matrix is at the bottom, with the intermembrane space above.
4702:"Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification of a new 2-methyl branched chain acyl-CoA dehydrogenase"
257:. In eukaryotes, five main protein complexes are involved, whereas in prokaryotes many different enzymes are present, using a variety of electron donors and acceptors.
286:
across this membrane. This store of energy is tapped when protons flow back across the membrane and down the potential energy gradient, through a large enzyme called
9312:
8093:"Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase"
4939:
Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, et al. (July 1998). "Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex".
2649:
In some bacteria and archaea, ATP synthesis is driven by the movement of sodium ions through the cell membrane, rather than the movement of protons. Archaea such as
1132:
group that does not participate in electron transfer to coenzyme Q, but is believed to be important in decreasing production of reactive oxygen species. It oxidizes
6915:
Müller V (February 2004). "An exceptional variability in the motor of archael A1A0 ATPases: from multimeric to monomeric rotors comprising 6-13 ion binding sites".
230:, which is used throughout the cell whenever energy is needed. During oxidative phosphorylation, electrons are transferred from the electron donors to a series of
3313:, by his development in 1973 of the "binding change" mechanism, followed by his radical proposal of rotational catalysis in 1982. More recent work has included
8067:
6868:"Delta mu Na+ drives the synthesis of ATP via an delta mu Na(+)-translocating F1F0-ATP synthase in membrane vesicles of the archaeon Methanosarcina mazei Gö1"
2413:
7226:
Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, et al. (January 2002). "Superoxide activates mitochondrial uncoupling proteins".
7191:
Kadenbach B, Ramzan R, Wen L, Vogt S (March 2010). "New extension of the
Mitchell Theory for oxidative phosphorylation in mitochondria of living organisms".
4268:
Baranova EA, Holt PJ, Sazanov LA (February 2007). "Projection structure of the membrane domain of
Escherichia coli respiratory complex I at 8 A resolution".
2578:
ATP synthase is a massive protein complex with a mushroom-like shape. The mammalian enzyme complex contains 16 subunits and has a mass of approximately 600
7070:
Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J (2007). "Free radicals and antioxidants in normal physiological functions and human disease".
3275:
4000:
Boxma B, de Graaf RM, van der Staay GW, van Alen TA, Ricard G, Gabaldón T, et al. (March 2005). "An anaerobic mitochondrion that produces hydrogen".
951:
1228:, carrying out anaerobic oxidative phosphorylation with fumarate as the electron acceptor. Another unconventional function of complex II is seen in the
3358:
in the 1930s but was ultimately discontinued due to its dangerous side effects. However, illicit use of the drug for this purpose continues today. See
1985:, for example, oxidative phosphorylation can be driven by a large number of pairs of reducing agents and oxidizing agents, which are listed below. The
1660:
were used to directly reduce two molecules of cytochrome c, the efficiency would be halved, with only one proton transferred per cytochrome c reduced.
4541:
Painter HJ, Morrisey JM, Mather MW, Vaidya AB (March 2007). "Specific role of mitochondrial electron transport in blood-stage
Plasmodium falciparum".
465:
yields about 14 ATPs. These ATP yields are theoretical maximum values; in practice, some protons leak across the membrane, lowering the yield of ATP.
3258:
were known to be involved. However, in the early 1940s, the link between the oxidation of sugars and the generation of ATP was firmly established by
1644:
is bound and again passes its first electron to a cytochrome c acceptor. The second electron is passed to the bound ubisemiquinone, reducing it to QH
1454:, a heme protein loosely associated with the mitochondrion. Unlike coenzyme Q, which carries two electrons, cytochrome c carries only one electron.
8855:
5790:"Relationship between lateral diffusion, collision frequency, and electron transfer of mitochondrial inner membrane oxidation-reduction components"
1250:
1380:
dehydrogenases. In plants, ETF-Q oxidoreductase is also important in the metabolic responses that allow survival in extended periods of darkness.
8509:
3914:
Page CC, Moser CC, Chen X, Dutton PL (November 1999). "Natural engineering principles of electron tunnelling in biological oxidation-reduction".
2708:
When one NADH is oxidized through the electron transfer chain, three ATPs are produced, which is equivalent to 7.3 kcal/mol x 3 = 21.9 kcal/mol.
2618:(the γ subunit stalk) within the α and β subunits. The α and β subunits are prevented from rotating themselves by the side-arm, which acts as a
8403:
8199:
1265:
and a cluster, but, unlike the other respiratory complexes, it attaches to the surface of the membrane and does not cross the lipid bilayer.
1238:. Here, the reversed action of complex II as an oxidase is important in regenerating ubiquinol, which the parasite uses in an unusual form of
8789:
8381:
9028:
5745:"The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes"
3390:
1894:
Many eukaryotic organisms have electron transport chains that differ from the much-studied mammalian enzymes described above. For example,
4887:
7338:"Acidosis Maintains the Function of Brain Mitochondria in Hypoxia-Tolerant Triplefin Fish: A Strategy to Survive Acute Hypoxic Exposure?"
3220:
can uncouple respiration from ATP synthesis. This rapid respiration produces heat, and is particularly important as a way of maintaining
7540:"Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I)"
2701:
The potential difference between these two redox pairs is 1.14 volt, which is equivalent to -52 kcal/mol or -2600 kJ per 6 mol of O
1640:. The first two substrates are released, but this ubisemiquinone intermediate remains bound. In the second step, a second molecule of QH
8502:
3301:
in 1978. Subsequent research concentrated on purifying and characterizing the enzymes involved, with major contributions being made by
2614:
interactions that propel the ring of c subunits past the proton channel. This rotating ring in turn drives the rotation of the central
4984:"The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex"
2646:. Finally, the active site cycles back to the open state, releasing ATP and binding more ADP and phosphate, ready for the next cycle.
6451:"Effects of carbon source on expression of F0 genes and on the stoichiometry of the c subunit in the F1F0 ATPase of Escherichia coli"
3116:
10777:
Single lines: pathways common to most lifeforms. Double lines: pathways not in humans (occurs in e.g. plants, fungi, prokaryotes).
8824:
8819:
6997:
6004:"Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors"
5383:
Moore AL, Siedow JN (August 1991). "The regulation and nature of the cyanide-resistant alternative oxidase of plant mitochondria".
3862:
429:
operate mainly on ΔpH. However, they also require a small membrane potential for the kinetics of ATP synthesis. In the case of the
8138:"A new concept for energy coupling in oxidative phosphorylation based on a molecular explanation of the oxygen exchange reactions"
4643:"Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool"
2360:
to produce enough proton-motive force to run part of the electron transport chain in reverse, causing complex I to generate NADH.
2010:
1986:
708:
9500:
8760:
5344:"Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role"
3195:
1273:
1148:
1625:, which is then oxidized, with one electron being passed to the second substrate, cytochrome c. The two protons released from QH
8482:
8439:
8427:
7981:
Mitchell P (July 1961). "Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism".
7667:
Borecký J, Vercesi AE (2005). "Plant uncoupling mitochondrial protein and alternative oxidase: energy metabolism and stress".
5774:
8487:
8477:
8451:
7514:
5016:
2599:
portion and back into the membrane is a long rod-like subunit that anchors the α and β subunits into the base of the enzyme.
25:
Oxidative phosphorylation is made up of two closely connected components: the electron transport chain and chemiosmosis. The
8472:
7825:"Esterification of inorganic phosphate coupled to electron transport between dihydrodiphosphopyridine nucleotide and oxygen"
6527:
4779:
3504:
8892:
6300:
3879:
Johnson DC, Dean DR, Smith AD, Johnson MK (2005). "Structure, function, and formation of biological iron-sulfur clusters".
4798:"The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation"
10906:
6806:
3270:
proved that the coenzyme NADH linked metabolic pathways such as the citric acid cycle and the synthesis of ATP. The term
1400:
1393:
765:
650:
8122:
8642:
8561:
8395:
4731:
3038:, an antidote used against chemical weapons, are the two important inhibitors of the site between cytochrome B and C1.
2876:{\displaystyle {\ce {O2->{\underset {Superoxide}{O2^{\underline {\bullet }}}}->{\underset {Peroxide}{O2^{2-}}}}}}
2168:
2078:
1258:
7854:
4502:"Role of complex II in anaerobic respiration of the parasite mitochondria from Ascaris suum and Plasmodium falciparum"
8365:
8346:
8327:
8308:
8282:
8260:
8241:
7807:
2068:
2064:
928:
728:
724:
547:, so it diffuses freely within the membrane. When Q accepts two electrons and two protons, it becomes reduced to the
5303:"Branched mitochondrial electron transport in the Animalia: presence of alternative oxidase in several animal phyla"
3965:
Leys D, Scrutton NS (December 2004). "Electrical circuitry in biology: emerging principles from protein structure".
3605:"Mitochondrial proton conductance and H+/O ratio are independent of electron transport rate in isolated hepatocytes"
8056:
5090:
Calhoun MW, Thomas JW, Gennis RB (August 1994). "The cytochrome oxidase superfamily of redox-driven proton pumps".
2331:
2282:
2187:
1113:
1106:
192:
carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative
9827:
9023:
3297:
in 1961. At first, this proposal was highly controversial, but it was slowly accepted and
Mitchell was awarded a
1962:
In contrast to the general similarity in structure and function of the electron transport chains in eukaryotes,
8799:
8629:
8617:
8566:
7437:
6260:"Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain"
5254:"Purification and characterization of a 43-kDa rotenone-insensitive NADH dehydrogenase from plant mitochondria"
4747:"A new iron-sulfur flavoprotein of the respiratory chain. A component of the fatty acid beta oxidation pathway"
2097:
9100:
6688:
Capaldi RA, Aggeler R (March 2002). "Mechanism of the F(1)F(0)-type ATP synthase, a biological rotary motor".
1590:{\displaystyle {\ce {QH2{}+ 2 Cyt\, c_{ox}{}+ 2H+_{matrix}-> Q{}+ 2 Cyt\, c_{red}{}+ 4H+_{intermembrane}}}}
10901:
9426:
8638:
2586:
and contains a ring of c subunits and the proton channel. The stalk and the ball-shaped headpiece is called F
6406:
Yoshida M, Muneyuki E, Hisabori T (September 2001). "ATP synthase--a marvellous rotary engine of the cell".
2575:, the hydrolysis reaction is used to acidify cellular compartments, by pumping protons and hydrolysing ATP.
425:
bacteria the electrical energy even has to compensate for a counteracting inverse pH difference. Inversely,
363:-releasing chemical reactions to drive energy-requiring reactions. The two sets of reactions are said to be
10533:
9529:
8625:
8466:
5348:
Brazilian
Journal of Medical and Biological Research = Revista Brasileira de Pesquisas Medicas e Biologicas
4594:"Reactions of electron-transfer flavoprotein and electron-transfer flavoprotein: ubiquinone oxidoreductase"
3241:
2920:
646:
265:
9673:
9662:
7870:"50 years of biological research--from oxidative phosphorylation to energy requiring transport regulation"
6119:"Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255"
2364:
function together, linked by the common ubiquinol intermediate. These respiratory chains therefore have a
1629:
pass into the intermembrane space. The third substrate is Q, which accepts the second electron from the QH
565:(Q) form. As a result, if two enzymes are arranged so that Q is reduced on one side of the membrane and QH
9493:
7497:
Heytler PG (1979). "Uncouplers of oxidative phosphorylation". In Sidney
Fleischer, Lester Packer (eds.).
4078:
Hirst J (June 2005). "Energy transduction by respiratory complex I--an evaluation of current knowledge".
3251:
3156:
Prevents the transfer of electrons from complex I to ubiquinone by blocking the ubiquinone-binding site.
2671:
form of the enzyme is a specialized sodium-driven ATP synthase, but this might not be true in all cases.
1125:
747:
445:
The amount of energy released by oxidative phosphorylation is high, compared with the amount produced by
193:
6215:
Iuchi S, Lin EC (July 1993). "Adaptation of
Escherichia coli to redox environments by gene expression".
253:, these proteins are located in the cell's outer membrane. These linked sets of proteins are called the
10707:
10582:
10109:
8933:
4065:
Medical CHEMISTRY Compendium. By Anders
Overgaard Pedersen and Henning Nielsen. Aarhus University. 2008
3834:"Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management"
3309:
on the ATP synthase. A critical step towards solving the mechanism of the ATP synthase was provided by
2341:
Some prokaryotes use redox pairs that have only a small difference in midpoint potential. For example,
2139:
2083:
1866:{\displaystyle {\ce {4Cyt\,c_{red}{}+O2{}+8H+_{matrix}->4Cyt\,c_{ox}{}+2H2O{}+4H+_{intermembrane}}}}
260:
The energy transferred by electrons flowing through this electron transport chain is used to transport
6117:
Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, et al. (March 2006).
5217:
Rasmusson AG, Soole KL, Elthon TE (2004). "Alternative NAD(P)H dehydrogenases of plant mitochondria".
3228:
animals, although these proteins may also have a more general function in cells' responses to stress.
3138:
proton pumping from ATP synthesis because it carries protons across the inner mitochondrial membrane.
1923:
The electron transport pathways produced by these alternative NADH and ubiquinone oxidases have lower
1257:, is a third entry point to the electron transport chain. It is an enzyme that accepts electrons from
21:
10612:
10293:
9842:
9565:
9297:
9219:
337:. The enzymes carrying out this metabolic pathway are also the target of many drugs and poisons that
7017:
4866:
3789:
Mitchell P (December 1979). "Keilin's respiratory chain concept and its chemiosmotic consequences".
520:
group in its structure. Cytochrome c is also found in some bacteria, where it is located within the
10270:
9980:
9767:
9317:
9038:
8979:
8885:
8721:
8681:
8556:
1261:
in the mitochondrial matrix, and uses these electrons to reduce ubiquinone. This enzyme contains a
677:
657:
is also oxidized by the electron transport chain, but feeds into the pathway at a different point.
613:
395:
254:
26:
5176:
Yoshikawa S, Muramoto K, Shinzawa-Itoh K, Aoyama H, Tsukihara T, Shimokata K, et al. (2006).
10861:
10849:
10785:
10507:
10252:
9411:
9160:
9155:
9141:
9116:
8746:
8662:
8573:
3359:
3355:
3237:
2900:
1928:
1220:
1085:
generation of a proton gradient, as two protons are taken up from the matrix as it is reduced to
738:
639:
581:
478:
434:
310:
3382:
1920:, and possibly some animals. This enzyme transfers electrons directly from ubiquinol to oxygen.
10735:
9952:
9750:
9486:
9446:
9380:
9266:
9261:
8966:
8705:
8621:
8195:
7140:
Finkel T, Holbrook NJ (November 2000). "Oxidants, oxidative stress and the biology of ageing".
7012:
5847:
Nealson KH (January 1999). "Post-Viking microbiology: new approaches, new data, new insights".
5606:"A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria"
4861:
4846:
2357:
2291:
1924:
446:
177:
8654:
7909:
Belitser VA, Tsibakova ET (1939). "About phosphorilation mechanism coupled with respiration".
7105:
Raha S, Robinson BH (October 2000). "Mitochondria, oxygen free radicals, disease and ageing".
1423:, with each subunit complex containing 11 protein subunits, an iron–sulfur cluster and three
512:. This carries only electrons, and these are transferred by the reduction and oxidation of an
10833:
10363:
10137:
9999:
9451:
9416:
9372:
9136:
8995:
8599:
5230:
4917:
4392:
4217:
Efremov RG, Baradaran R, Sazanov LA (May 2010). "The architecture of respiratory complex I".
3892:
3314:
2643:
2120:
2021:
1234:
1081:
1070:
743:
683:
295:
3421:
Mitchell P, Moyle J (January 1967). "Chemiosmotic hypothesis of oxidative phosphorylation".
10809:
10358:
9634:
9619:
9390:
9095:
9071:
9033:
8770:
8700:
8588:
8384:
8149:
7990:
7939:
7290:
7235:
7149:
6364:
6130:
5856:
5801:
5560:
5501:
5134:
5046:
4948:
4654:
4550:
4423:
4226:
4173:
4009:
3923:
3798:
3702:
3430:
3263:
3213:
2956:
2568:
2087:
1957:
1686:
1679:
1669:
921:
815:
197:
483:
8:
10911:
10652:
10412:
10386:
10102:
9335:
8954:
8878:
8850:
6928:
4465:
Horsefield R, Iwata S, Byrne B (April 2004). "Complex II from a structural perspective".
3530:
3294:
3267:
2540:{\displaystyle {\ce {ADP + P_i + 4H+_{intermembrane}<=> ATP + H2O + 4H+_{matrix}}}}
1904:
673:
421:
equivalent: In mitochondria, the largest part of energy is provided by the potential; in
399:
322:
8494:
8153:
7994:
7943:
7886:
7869:
7294:
7239:
7153:
7046:
6725:"Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series"
6596:
Philosophical
Transactions of the Royal Society of London. Series B, Biological Sciences
6368:
6134:
5860:
5805:
5564:
5505:
5490:"The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells"
5138:
5050:
4952:
4658:
4554:
4427:
4230:
4177:
4162:"Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus"
4013:
3927:
3833:
3802:
3706:
3434:
3411:
Voet, D.; Voet, J. G. (2004). "Biochemistry", 3rd ed., p. 804, Wiley.ISBN 0-471-19350-X.
10865:
10775:. Click any text (name of pathway or metabolites) to link to the corresponding article.
10169:
10130:
10076:
9939:
9458:
9131:
9076:
9063:
8741:
8230:
8014:
7963:
7799:
7768:
7692:
7644:
7623:
7569:
7364:
7337:
7259:
7173:
7038:
6940:
6845:
6749:
6724:
6670:
6616:
6591:
6431:
6388:
6240:
6228:
6192:
6175:
6151:
6118:
6102:
6085:
5880:
5635:
5586:
5418:
Vanlerberghe GC, McIntosh L (June 1997). "ALTERNATIVE OXIDASE: From Gene to Function".
5283:
5158:
5072:
4822:
4797:
4677:
4642:
4618:
4593:
4574:
4447:
4250:
4199:
4033:
3947:
3771:
3728:
3625:
3604:
3454:
3217:
3134:
that disrupt the proton gradient by carrying protons across a membrane. This ionophore
2933:
2335:
2305:
2268:
2059:
1977:
is understood in most detail, while archaeal systems are at present poorly understood.
897:
890:
719:
672:
across the inner membrane of the mitochondrion. This causes protons to build up in the
597:
407:
283:
270:
8172:
8137:
8109:
7841:
7118:
6892:
6867:
6793:
6701:
6567:
6542:
6475:
6450:
6287:
6061:
6044:
6020:
6003:
5976:
5951:
5924:
5899:
5824:
5789:
5720:
5703:
5679:
5654:
5573:
5548:
5466:
5396:
5059:
5034:
5003:
4904:
Crofts AR (2004). "The cytochrome bc1 complex: function in the context of structure".
4875:
4766:
4718:
4518:
4501:
10768:
10716:
10684:
9965:
9580:
9330:
9126:
8970:
8928:
8923:
8529:
8421:
8361:
8342:
8323:
8304:
8278:
8256:
8237:
8177:
8114:
8037:
8006:
7955:
7891:
7846:
7803:
7760:
7684:
7649:
7604:
7600:
7561:
7520:
7510:
7506:
7479:
7443:
7433:
7407:
7403:
7369:
7318:
7313:
7278:
7251:
7208:
7165:
7122:
7087:
7030:
6978:
6932:
6897:
6837:
6798:
6754:
6705:
6662:
6621:
6572:
6519:
6480:
6466:
6423:
6380:
6376:
6337:
6333:
6292:
6278:
6232:
6197:
6156:
6066:
6025:
5981:
5929:
5872:
5829:
5788:
Gupte S, Wu ES, Hoechli L, Hoechli M, Jacobson K, Sowers AE, et al. (May 1984).
5766:
5725:
5684:
5627:
5578:
5529:
5524:
5489:
5470:
5435:
5431:
5400:
5365:
5324:
5275:
5234:
5199:
5150:
5107:
5103:
5064:
5008:
4964:
4921:
4879:
4827:
4771:
4723:
4682:
4623:
4566:
4523:
4482:
4439:
4396:
4361:
4344:
Hirst J (December 2009). "Towards the molecular mechanism of respiratory complex I".
4326:
4285:
4242:
4191:
4139:
4095:
4025:
3982:
3939:
3896:
3854:
3814:
3763:
3720:
3715:
3690:
3671:
3666:
3649:
3630:
3585:
3550:
3496:
3446:
3290:
3120:
2737:
2633:. ATP is shown in red, ADP and phosphate in pink and the rotating γ subunit in black.
2244:
2225:
2206:
1709:
1439:. A cytochrome is a kind of electron-transferring protein that contains at least one
1042:{\displaystyle {\ce {NADH + Q + 5H+_{matrix}-> NAD+ + QH2 + 4H+_{intermembrane}}}}
631:
521:
372:
318:
208:
158:
34:
8870:
7930:
Slater EC (November 1953). "Mechanism of phosphorylation in the respiratory chain".
7772:
7696:
7573:
7042:
6944:
6883:
6849:
6674:
6392:
6244:
6142:
5884:
5639:
5590:
5360:
5343:
5162:
5076:
4796:
Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (September 2005).
4451:
4379:
Cecchini G (2003). "Function and structure of complex II of the respiratory chain".
3775:
3546:
3477:"Crucial role of the membrane potential for ATP synthesis by F(1)F(o) ATP synthases"
2602:
As protons cross the membrane through the channel in the base of ATP synthase, the F
912:
with the mammalian complex I having 46 subunits and a molecular mass of about 1,000
10877:
10227:
10176:
10123:
10095:
9593:
9552:
9353:
8692:
8546:
8167:
8157:
8104:
8018:
7998:
7967:
7947:
7881:
7836:
7795:
7786:
Lipmann F (1941). "Metabolic generation and utilization of phosphate bond energy".
7752:
7723:
7676:
7639:
7631:
7596:
7589:
Biochimica et Biophysica Acta (BBA) - Specialized Section on Enzymological Subjects
7551:
7502:
7471:
7399:
7359:
7349:
7308:
7298:
7263:
7243:
7200:
7177:
7157:
7114:
7079:
7022:
6970:
6924:
6887:
6879:
6829:
6788:
6744:
6736:
6697:
6652:
6611:
6603:
6562:
6554:
6511:
6470:
6462:
6435:
6415:
6372:
6329:
6282:
6274:
6224:
6187:
6146:
6138:
6097:
6056:
6015:
5971:
5967:
5963:
5919:
5915:
5911:
5864:
5819:
5809:
5756:
5744:
5715:
5674:
5666:
5617:
5568:
5519:
5509:
5462:
5427:
5392:
5355:
5314:
5265:
5226:
5189:
5142:
5099:
5054:
4998:
4956:
4913:
4871:
4817:
4809:
4761:
4713:
4672:
4662:
4613:
4605:
4578:
4558:
4513:
4474:
4431:
4388:
4353:
4316:
4277:
4254:
4234:
4203:
4181:
4129:
4087:
4037:
4017:
3974:
3951:
3931:
3888:
3844:
3806:
3759:
3755:
3732:
3710:
3661:
3620:
3612:
3577:
3542:
3488:
3458:
3438:
3255:
3221:
3070:
2727:
2272:
1973:
1932:
1066:
913:
540:
338:
275:
189:
102:
55:
5287:
5146:
4983:
3568:
Rich PR (December 2003). "The molecular machinery of Keilin's respiratory chain".
3262:, confirming the central role of ATP in energy transfer that had been proposed by
2679:
The transport of electrons from redox pair NAD/ NADH to the final redox pair 1/2 O
33:
is the site of oxidative phosphorylation. The NADH and succinate generated in the
10721:
9986:
9926:
9810:
9735:
9539:
9307:
9189:
8458:
8446:
8434:
8399:
7204:
7083:
6499:
5194:
5177:
4746:
4321:
4304:
4134:
4117:
3476:
3335:
3081:
Inhibit the electron transport chain by binding more strongly than oxygen to the
3062:
2985:
2564:
1225:
299:
246:
231:
7743:
Kalckar HM (November 1974). "Origins of the concept oxidative phosphorylation".
7635:
7336:
Devaux JB, Hedges CP, Birch N, Herbert N, Renshaw GM, Hickey AJ (January 2019).
6259:
4960:
3849:
10916:
10821:
10797:
10589:
10519:
10183:
10067:
10054:
9690:
9649:
9384:
9302:
9292:
9241:
9229:
9202:
8142:
Proceedings of the National Academy of Sciences of the United States of America
7283:
Proceedings of the National Academy of Sciences of the United States of America
5794:
Proceedings of the National Academy of Sciences of the United States of America
5494:
Proceedings of the National Academy of Sciences of the United States of America
5319:
5302:
4647:
Proceedings of the National Academy of Sciences of the United States of America
3531:"Structures and proton-pumping strategies of mitochondrial respiratory enzymes"
3318:
3302:
3259:
2365:
2295:
2263:
2106:
1361:
1216:
927:
The reaction that is catalyzed by this enzyme is the two electron oxidation of
635:
458:
430:
235:
212:
162:
30:
8408:
7680:
7026:
6776:
5868:
5670:
5655:"Supercomplexes in the respiratory chains of yeast and mammalian mitochondria"
4281:
3978:
664:, the enzymes in this electron transport system use the energy released from O
402:. It has two components: a difference in proton concentration (a H gradient, Δ
309:
Although oxidative phosphorylation is a vital part of metabolism, it produces
10895:
10379:
10325:
10302:
10261:
10153:
10144:
10025:
9780:
9256:
8943:
8092:
7354:
6740:
5270:
5253:
4478:
3310:
3306:
3247:
2651:
2611:
2590:
and is the site of ATP synthesis. The ball-shaped complex at the end of the F
2371:
In addition to this metabolic diversity, prokaryotes also possess a range of
2342:
2314:
2196:
2173:
1420:
1133:
1073:(FMN). The addition of electrons to FMN converts it to its reduced form, FMNH
908:, is the first protein in the electron transport chain. Complex I is a giant
688:
654:
418:
354:
8392:
8162:
8057:"David Keilin's Respiratory Chain Concept and Its Chemiosmotic Consequences"
8041:
7447:
5514:
4701:
4667:
4435:
4186:
4161:
687:
that instead reduce protons to hydrogen in a remnant mitochondrion called a
10693:
10661:
10568:
10563:
10547:
10419:
10243:
10197:
10050:
9705:
9236:
9206:
9011:
8983:
8765:
8751:
8673:
8118:
8010:
7959:
7850:
7824:
7728:
7711:
7688:
7653:
7608:
7565:
7556:
7539:
7373:
7322:
7303:
7255:
7212:
7169:
7126:
7091:
7034:
6936:
6758:
6709:
6666:
6657:
6640:
6625:
6607:
6576:
6558:
6523:
6427:
6160:
6070:
5933:
5876:
5814:
5770:
5761:
5729:
5688:
5631:
5622:
5605:
5582:
5533:
5439:
5328:
5238:
5203:
5068:
4925:
4883:
4831:
4813:
4686:
4570:
4527:
4486:
4443:
4400:
4365:
4330:
4289:
4246:
4195:
4143:
4099:
4029:
3986:
3943:
3900:
3858:
3810:
3767:
3589:
3554:
3500:
3330:
3165:
3023:
2630:
2388:
2375: – different enzymes that catalyze the same reaction. For example, in
2310:
2192:
2125:
2110:
1637:
1451:
1262:
1137:
932:
917:
840:
820:
798:
774:
617:
577:
549:
532:
509:
390:
350:
291:
287:
227:
185:
42:
8181:
7895:
7764:
7483:
7411:
6982:
6901:
6841:
6802:
6484:
6384:
6341:
6296:
6236:
6201:
6029:
5985:
5833:
5474:
5404:
5369:
5279:
5154:
5111:
5012:
4968:
4727:
4627:
3724:
3675:
3634:
3450:
2984:
acidosis. Cytosolic protons that have accumulated with ATP hydrolysis and
2711:
The conservation of the energy can be calculated by the following formula
1446:
The reaction catalyzed by complex III is the oxidation of one molecule of
10772:
10638:
10575:
10526:
10220:
10204:
10190:
10088:
10083:
10012:
9900:
9870:
9795:
9362:
9224:
7524:
6592:"Structural model of F1-ATPase and the implications for rotary catalysis"
6515:
6498:
Nelson N, Perzov N, Cohen A, Hagai K, Padler V, Nelson H (January 2000).
4775:
3818:
3526:
3492:
3298:
3225:
3181:
3035:
3031:
3019:
2970:
2964:
2940:
2731:
2347:
2129:
2102:
2040:
2026:
1945:
1634:
1436:
1396:. After each step, Q (in the upper part of the figure) leaves the enzyme.
426:
422:
7475:
6543:"Structure of the mitochondrial ATP synthase by electron cryomicroscopy"
4562:
4413:
4238:
4021:
3246:
The field of oxidative phosphorylation began with the report in 1906 by
2606:
proton-driven motor rotates. Rotation might be caused by changes in the
561:
releases two electrons and two protons, it becomes oxidized back to the
306:
of a part of the enzyme. The ATP synthase is a rotary mechanical motor.
10744:
10675:
10626:
10605:
10596:
10540:
10495:
10459:
10372:
10311:
10236:
10211:
10160:
10041:
9720:
9606:
9509:
9463:
9185:
9106:
9087:
9019:
8962:
8905:
8901:
8609:
8525:
7911:
7756:
6974:
6833:
4357:
4091:
3581:
3099:
3094:
3006:
2928:
2907:
radical, are very harmful to cells, as they oxidize proteins and cause
2742:
2607:
2579:
2155:
1428:
1424:
1388:
1377:
1369:
1365:
1239:
627:
589:
544:
492:
488:
462:
450:
384:
334:
314:
250:
181:
7624:"The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP"
6258:
Calhoun MW, Oden KL, Gennis RB, de Mattos MJ, Neijssel OM (May 1993).
6045:"Succinate dehydrogenase and fumarate reductase from Escherichia coli"
4609:
3616:
383:
process – it releases energy, whereas the synthesis of ATP is an
302:
reaction. The reaction is driven by the proton flow, which forces the
10645:
10630:
10621:
10499:
10489:
10477:
10464:
10450:
10439:
10435:
10404:
10399:
10392:
10351:
9913:
9358:
9165:
8909:
8551:
8270:
8002:
7951:
7161:
6500:"The cellular biology of proton-motive force generation by V-ATPases"
6419:
4116:
Lenaz G, Fato R, Genova ML, Bergamini C, Bianchi C, Biondi A (2006).
3442:
3203:
3199:
3186:
3148:
3135:
3131:
2952:
2948:
2398:
2353:
1698:
1447:
1086:
661:
623:
570:
380:
242:
7247:
6116:
5175:
5035:"Protonmotive pathways and mechanisms in the cytochrome bc1 complex"
3089:
center in cytochrome c oxidase, preventing the reduction of oxygen.
2967:, which detoxify the reactive species, limiting damage to the cell.
2939:
To counteract these reactive oxygen species, cells contain numerous
2828:
2780:
238:
ending in oxygen, whose reaction releases half of the total energy.
10668:
10472:
10346:
10339:
10116:
8583:
6961:
Davies KJ (1995). "Oxidative stress: the paradox of aerobic life".
6354:
6320:
Boyer PD (1997). "The ATP synthase--a splendid molecular machine".
3746:
Crane FL (December 2001). "Biochemical functions of coenzyme Q10".
3161:
3143:
3107:
Inhibits ATP synthase by blocking the flow of protons through the F
3015:
2960:
2908:
2904:
2746:
2572:
2049:
1963:
593:
474:
303:
173:
6998:"Theories of biological aging: genes, proteins, and free radicals"
6043:
Cecchini G, Schröder I, Gunsalus RP, Maklashina E (January 2002).
6042:
3999:
3935:
1656:
of proton transfer. If, instead of the Q cycle, one molecule of QH
1245:
249:
within the inner membrane of the cell's mitochondria, whereas, in
10845:
10728:
10556:
10514:
10485:
10332:
10286:
9478:
9421:
9406:
4500:
Kita K, Hirawake H, Miyadera H, Amino H, Takeo S (January 2002).
4305:"The gross structure of the respiratory complex I: a Lego System"
3058:
2944:
2916:
2372:
2253:
2249:
2234:
2230:
2177:
2151:
2147:
2030:
1967:
1917:
1621:. In the first step, the enzyme binds three substrates, first, QH
1618:
1373:
1229:
940:
505:
454:
326:
204:
10762:
5124:
3176:
Competitive inhibitors of succinate dehydrogenase (complex II).
924:, as is the case for many enzymes present in the mitochondrion.
10318:
10279:
10060:
8532:
7225:
5704:"Respiratory chain supercomplexes of mitochondria and bacteria"
5178:"Proton pumping mechanism of bovine heart cytochrome c oxidase"
3212:
Not all inhibitors of oxidative phosphorylation are toxins. In
3086:
2976:
2619:
2211:
2045:
1913:
1902:
Another example of a divergent electron transport chain is the
1694:
909:
669:
585:
376:
360:
261:
166:
4844:
1648:
as it gains two protons from the mitochondrial matrix. This QH
1383:
8834:
8829:
7499:
Biomembranes Part F: Bioenergetics: Oxidative Phosphorylation
6820:
Dimroth P (1994). "Bacterial sodium ion-coupled energetics".
5420:
Annual Review of Plant Physiology and Plant Molecular Biology
3321:, with Walker and Boyer being awarded a Nobel Prize in 1997.
3305:
on the complexes of the electron-transport chain, as well as
3066:
3001:
2979:
is fundamental for oxidative phosphorylation, a shortage in O
2215:
2005:
1909:
1895:
1674:
1101:
880:
703:
536:
528:
330:
170:
108:
8091:
Pullman ME, Penefsky HS, Datta A, Racker E (November 1960).
7072:
The International Journal of Biochemistry & Cell Biology
5603:
4118:"Mitochondrial Complex I: structural and functional aspects"
2595:
the ATP synthesis reaction. Reaching along the side of the F
1337:{\displaystyle {\ce {ETF_{red}{}+ Q -> ETF_{ox}{}+ QH2}}}
1192:{\displaystyle {\ce {{Succinate}+ Q -> {Fumarate}+ QH2}}}
885:
10428:
8814:
8809:
8804:
8794:
8784:
8577:
7279:"Mitochondria as ATP consumers: cellular treason in anoxia"
5604:
Heinemeyer J, Braun HP, Boekema EJ, Kouril R (April 2007).
5549:"A critical appraisal of the mitochondrial coenzyme Q pool"
4795:
4540:
3082:
2997:
2615:
1702:
1613:
As only one of the electrons can be transferred from the QH
1440:
1129:
642:
517:
513:
438:
it drives the counter-rotation of subunits a and c of the F
368:
219:
215:
129:
79:
8090:
7069:
6448:
6257:
6176:"The nitrite oxidizing system of Nitrobacter winogradskyi"
1096:
939:(represented as Q in the equation below), a lipid-soluble
695:
Typical respiratory enzymes and substrates in eukaryotes.
58:
10700:
8524:
8275:
Power, Sex, Suicide: Mitochondria and the Meaning of Life
7501:. Methods in Enzymology. Vol. 55. pp. 462–472.
6540:
6449:
Schemidt RA, Qu J, Williams JR, Brusilow WS (June 1998).
5452:
4499:
2912:
2772:
2674:
2625:
2508:
1832:
1810:
1762:
1744:
1559:
1503:
1477:
1330:
1312:
1288:
1185:
1013:
468:
138:
120:
88:
70:
8382:
Animated diagrams illustrating oxidative phosphorylation
7335:
6722:
5032:
4845:
Berry EA, Guergova-Kuras M, Huang LS, Crofts AR (2000).
4115:
3317:
on the enzymes involved in oxidative phosphorylation by
1951:
1251:
Electron transfer flavoprotein-ubiquinone oxidoreductase
176:, thereby releasing chemical energy in order to produce
7276:
6541:
Rubinstein JL, Walker JE, Henderson R (December 2003).
6405:
4216:
3878:
3383:"oxidative Meaning in the Cambridge English Dictionary"
607:
403:
279:
9103:(amino acid→pyruvate, acetyl CoA, or TCA intermediate)
7193:
Biochimica et Biophysica Acta (BBA) - General Subjects
7190:
6497:
5897:
3650:"The structure, function and evolution of cytochromes"
3535:
Annual Review of Biophysics and Biomolecular Structure
2971:
Oxidative phosphorylation in hypoxic/anoxic conditions
2582:. The portion embedded within the membrane is called F
2473:
2368:, with easily interchangeable sets of enzyme systems.
1889:
222:. Oxidative phosphorylation uses these molecules and O
8900:
5787:
4591:
2760:
2416:
1724:
1465:
1276:
1151:
954:
245:, these redox reactions are catalyzed by a series of
123:
117:
111:
67:
61:
9520:
5417:
5300:
5216:
4699:
4464:
3010:
below the concentration that these enzymes can use.
1663:
645:. This coenzyme contains electrons that have a high
141:
135:
126:
91:
85:
73:
8358:
Biophysical and Structural Aspects of Bioenergetics
8253:
Into the Cool: Energy Flow, Thermodynamics and Life
6049:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
6008:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
5898:Schäfer G, Engelhard M, Müller V (September 1999).
5708:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
5487:
5385:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
5341:
5182:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
5089:
4847:"Structure and function of cytochrome bc complexes"
4592:Ramsay RR, Steenkamp DJ, Husain M (February 1987).
4506:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
4309:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
4267:
4122:
Biochimica et Biophysica Acta (BBA) - Bioenergetics
3913:
1360:In mammals, this metabolic pathway is important in
576:Within proteins, electrons are transferred between
105:
64:
8229:
7586:
7432:(2nd ed.). Kolkata, India: Books and Allied.
7392:Biochimica et Biophysica Acta (BBA) - Biomembranes
3474:
2875:
2539:
1865:
1589:
1336:
1191:
1041:
417:The two components of the proton-motive force are
7908:
7822:
7621:
7277:St-Pierre J, Brand MD, Boutilier RG (July 2000).
6774:
6723:Dimroth P, von Ballmoos C, Meier T (March 2006).
5033:Hunte C, Palsdottir H, Trumpower BL (June 2003).
4302:
4159:
2610:of amino acids in the ring of c subunits causing
2481:
2480:
2463:
2462:
457:to carbon dioxide and water, while each cycle of
294:. The ATP synthase uses the energy to transform
10893:
8856:Electron-transferring-flavoprotein dehydrogenase
8298:
8135:
6173:
5742:
5652:
4938:
4155:
4153:
1392:The two electron transfer steps in complex III:
1255:electron transferring-flavoprotein dehydrogenase
600:, which is rapid over distances of less than 1.4
8761:Complex III/Coenzyme Q - cytochrome c reductase
6775:Gresser MJ, Myers JA, Boyer PD (October 1982).
4700:Ikeda Y, Dabrowski C, Tanaka K (January 1983).
4640:
1246:Electron transfer flavoprotein-Q oxidoreductase
8277:(1st ed.). Oxford University Press, USA.
8250:
7666:
7139:
6687:
6641:"The rotary machine in the cell, ATP synthase"
6001:
5849:Origins of Life and Evolution of the Biosphere
4744:
1971:In the bacteria, oxidative phosphorylation in
9494:
8886:
8510:
8255:(1st ed.). University of Chicago Press.
8136:Boyer PD, Cross RL, Momsen W (October 1973).
7427:
5949:
4150:
3475:Dimroth P, Kaim G, Matthey U (January 2000).
3470:
3468:
3420:
3266:in 1941. Later, in 1949, Morris Friedkin and
943:that is found in the mitochondrion membrane:
527:Within the inner mitochondrial membrane, the
211:, producing carbon dioxide and the energetic
8360:(1st ed.). Royal Society of Chemistry.
8322:(1st ed.). Cambridge University Press.
7537:
7104:
6865:
6589:
5952:"The respiratory chains of Escherichia coli"
5488:Maxwell DP, Wang Y, McIntosh L (July 1999).
4641:Zhang J, Frerman FE, Kim JJ (October 2006).
4111:
4109:
3964:
3831:
3748:Journal of the American College of Nutrition
3654:Progress in Biophysics and Molecular Biology
1938:
1128:(FAD) cofactor, iron–sulfur clusters, and a
535:(Q) carries both electrons and protons by a
410:, with the N-side having a negative charge.
7709:
6716:
6638:
6083:
5945:
5943:
5382:
4061:
4059:
4057:
4055:
4053:
4051:
4049:
4047:
3602:
3524:
2382:
1384:Q-cytochrome c oxidoreductase (complex III)
203:The energy stored in the chemical bonds of
132:
114:
82:
76:
9501:
9487:
8893:
8879:
8517:
8503:
8404:University of Illinois at Urbana–Champaign
7389:
6770:
6768:
5904:Microbiology and Molecular Biology Reviews
3465:
3250:of a vital role for phosphate in cellular
2721:
2637:This ATP synthesis reaction is called the
881:NADH-coenzyme Q oxidoreductase (complex I)
495:form (Q) to the reduced ubiquinol form (QH
8227:
8171:
8161:
8108:
7885:
7840:
7727:
7643:
7555:
7363:
7353:
7312:
7302:
7016:
6956:
6954:
6917:Journal of Bioenergetics and Biomembranes
6891:
6861:
6859:
6792:
6748:
6656:
6615:
6566:
6474:
6286:
6191:
6150:
6101:
6060:
6019:
5975:
5923:
5823:
5813:
5760:
5719:
5678:
5621:
5572:
5540:
5523:
5513:
5359:
5318:
5269:
5193:
5058:
5002:
4981:
4865:
4821:
4765:
4717:
4676:
4666:
4617:
4517:
4320:
4185:
4133:
4106:
3848:
3714:
3665:
3624:
3520:
3518:
3516:
3514:
2915:. This cellular damage may contribute to
2903:and their reaction products, such as the
2859:
2520:
2436:
1846:
1821:
1799:
1795:
1773:
1733:
1729:
1570:
1548:
1544:
1514:
1492:
1488:
1022:
971:
359:Oxidative phosphorylation works by using
151:electron transport-linked phosphorylation
8355:
8054:
7980:
7622:Ricquier D, Bouillaud F (January 2000).
7385:
7383:
6315:
6313:
6214:
6174:Yamanaka T, Fukumori Y (December 1988).
5997:
5995:
5950:Ingledew WJ, Poole RK (September 1984).
5940:
5701:
5342:Sluse FE, Jarmuszkiewicz W (June 1998).
5301:McDonald A, Vanlerberghe G (June 2004).
5231:10.1146/annurev.arplant.55.031903.141720
4918:10.1146/annurev.physiol.66.032102.150251
4393:10.1146/annurev.biochem.72.121801.161700
4378:
4303:Friedrich T, Böttcher B (January 2004).
4073:
4071:
4044:
3893:10.1146/annurev.biochem.74.082803.133518
3788:
2749:anions, which are dangerously reactive.
2624:
1673:
1387:
1376:, as it accepts electrons from multiple
1100:
884:
482:
298:(ADP) into adenosine triphosphate, in a
20:
8415:
8125:from the original on 29 September 2007.
7867:
7823:Friedkin M, Lehninger AL (April 1949).
7785:
7742:
7538:Lambert AJ, Brand MD (September 2004).
7496:
7460:
6819:
6809:from the original on 29 September 2007.
6765:
6530:from the original on 30 September 2007.
5846:
5743:Schägger H, Pfeiffer K (October 2001).
5251:
4745:Ruzicka FJ, Beinert H (December 1977).
4734:from the original on 29 September 2007.
4160:Sazanov LA, Hinchliffe P (March 2006).
3872:
3825:
3647:
3507:from the original on 30 September 2007.
2456:
1097:Succinate-Q oxidoreductase (complex II)
37:are oxidized, releasing the energy of O
10894:
10742:
10733:
10726:
10705:
10698:
10691:
10682:
10673:
10666:
10659:
10650:
10643:
10636:
10619:
10610:
10603:
10594:
10587:
10580:
10573:
10545:
10538:
10531:
10524:
10512:
10505:
10483:
10470:
10457:
10433:
10426:
10417:
10410:
10384:
10377:
10356:
10337:
10323:
10309:
10300:
10291:
10284:
10277:
10259:
10250:
10241:
10234:
10225:
10218:
10209:
10202:
10195:
10174:
10167:
10158:
10151:
10142:
10135:
10128:
10121:
10114:
10107:
10100:
10093:
9976:
9961:
9948:
9935:
9922:
9909:
9896:
9881:
9851:
9838:
9823:
9806:
9791:
9776:
9763:
9746:
9731:
9716:
9670:
9659:
9646:
9631:
9616:
9603:
9590:
9577:
9562:
9549:
9536:
8317:
7929:
7857:from the original on 16 December 2008.
7712:"The alcoholic ferment of yeast-juice"
6995:
6960:
6951:
6914:
6856:
6408:Nature Reviews. Molecular Cell Biology
6123:Applied and Environmental Microbiology
4903:
3603:Porter RK, Brand MD (September 1995).
3511:
2714:Efficiency = (21.9 x 100%) / 52 = 42%
2675:Oxidative phosphorylation - energetics
1993:Respiratory enzymes and substrates in
1450:and the reduction of two molecules of
1253:(ETF-Q oxidoreductase), also known as
871:
469:Electron and proton transfer molecules
10714:
10561:
10554:
10448:
10397:
10370:
10344:
10268:
10188:
10181:
10081:
10074:
10065:
10048:
10039:
10021:
10008:
9995:
9866:
9701:
9686:
9482:
8874:
8498:
8336:
7423:
7421:
7380:
6319:
6310:
5992:
5653:Schägger H, Pfeiffer K (April 2000).
5546:
4467:Current Protein & Peptide Science
4343:
4210:
4077:
4068:
3967:Current Opinion in Structural Biology
3745:
1952:Prokaryotic electron transport chains
516:atom that the protein holds within a
325:, damaging cells and contributing to
9003:Electron acceptors other than oxygen
8375:
8269:
8232:Lehninger Principles of Biochemistry
8031:
7098:
6086:"Energy conservation in Nitrobacter"
3688:
3567:
3393:from the original on 24 January 2018
3053:Effect on oxidative phosphorylation
2751:
2736:Molecular oxygen is a good terminal
2407:
2356:. This problem is solved by using a
1715:
1456:
1267:
1142:
945:
608:Eukaryotic electron transport chains
8483:Coenzyme Q - cytochrome c reductase
8196:"The Nobel Prize in Chemistry 1997"
8097:The Journal of Biological Chemistry
7887:10.1146/annurev.bi.60.070191.000245
7829:The Journal of Biological Chemistry
7745:Molecular and Cellular Biochemistry
7544:The Journal of Biological Chemistry
6781:The Journal of Biological Chemistry
6645:The Journal of Biological Chemistry
6590:Leslie AG, Walker JE (April 2000).
6504:The Journal of Experimental Biology
5749:The Journal of Biological Chemistry
5610:The Journal of Biological Chemistry
5258:The Journal of Biological Chemistry
4991:The Journal of Biological Chemistry
4754:The Journal of Biological Chemistry
4706:The Journal of Biological Chemistry
3832:Søballe B, Poole RK (August 1999).
3481:The Journal of Experimental Biology
3405:
3216:, regulated proton channels called
1890:Alternative reductases and oxidases
651:inner membrane of the mitochondrion
398:across the membrane, is called the
13:
9508:
8747:Complex II/Succinate dehydrogenase
8643:Pyruvate dehydrogenase phosphatase
8216:
8034:Peter Mitchell and the Vital Force
7800:10.4159/harvard.9780674366701.c141
7418:
6929:10.1023/B:JOBB.0000019603.68282.04
6639:Noji H, Yoshida M (January 2001).
6229:10.1111/j.1365-2958.1993.tb01664.x
6193:10.1111/j.1574-6968.1988.tb02746.x
6103:10.1111/j.1574-6968.1990.tb03989.x
6002:Unden G, Bongaerts J (July 1997).
5252:Menz RI, Day DA (September 1996).
3691:"Why do c-type cytochromes exist?"
2955:, and antioxidant enzymes such as
2079:Glycerol-3-phosphate dehydrogenase
1652:is then released from the enzyme.
1633:and is reduced to Q, which is the
1259:electron-transferring flavoprotein
14:
10928:
8299:Nicholls DG, Ferguson SJ (2002).
4876:10.1146/annurev.biochem.69.1.1005
1664:Cytochrome c oxidase (complex IV)
10871:
10855:
10839:
10827:
10815:
10803:
10791:
10779:
10761:
9528:
8465:Interactive molecular models at
8303:(1st ed.). Academic Press.
8188:
8129:
8084:
8048:
8025:
7974:
7923:
7902:
7861:
7816:
7779:
7736:
7716:Proceedings of the Royal Society
6467:10.1128/jb.180.12.3205-3208.1998
6334:10.1146/annurev.biochem.66.1.717
6306:from the original on 2007-09-27.
6279:10.1128/jb.175.10.3020-3025.1993
5777:from the original on 2007-09-29.
5432:10.1146/annurev.arplant.48.1.703
5022:from the original on 2007-09-27.
4893:from the original on 2015-12-28.
4785:from the original on 2007-09-27.
4080:Biochemical Society Transactions
3868:from the original on 2008-05-29.
3570:Biochemical Society Transactions
2936:that reduce membrane potential.
2919:and is proposed as one cause of
1215:In some eukaryotes, such as the
592:, usually by the sulfur atom of
101:
54:
9024:Substrate-level phosphorylation
8771:Complex IV/Cytochrome c oxidase
8236:(4th ed.). W. H. Freeman.
8221:
8202:from the original on 2017-03-25
8073:from the original on 2007-09-27
7703:
7660:
7615:
7580:
7531:
7490:
7454:
7329:
7270:
7219:
7184:
7133:
7063:
6989:
6908:
6884:10.1128/jb.176.9.2543-2550.1994
6866:Becher B, Müller V (May 1994).
6813:
6681:
6632:
6583:
6534:
6491:
6442:
6399:
6348:
6251:
6208:
6167:
6143:10.1128/AEM.72.3.2050-2063.2006
6110:
6077:
6036:
5891:
5840:
5781:
5736:
5695:
5646:
5597:
5481:
5446:
5411:
5376:
5361:10.1590/S0100-879X1998000600003
5335:
5294:
5245:
5210:
5169:
5118:
5083:
5026:
4975:
4932:
4897:
4838:
4789:
4738:
4693:
4634:
4585:
4534:
4493:
4458:
4407:
4372:
4337:
4296:
4261:
3993:
3958:
3907:
3782:
3739:
3611:. 310 ( Pt 2) (Pt 2): 379–382.
3547:10.1146/annurev.biophys.30.1.23
3354:DNP was extensively used as an
3348:
2943:systems, including antioxidant
1419:. In mammals, this enzyme is a
626:biochemical processes, such as
344:
321:, which lead to propagation of
207:is released by the cell in the
8618:Pyruvate dehydrogenase complex
8251:Schneider ED, Sagan D (2006).
7107:Trends in Biochemical Sciences
6690:Trends in Biochemical Sciences
5968:10.1128/mmbr.48.3.222-271.1984
5916:10.1128/MMBR.63.3.570-620.1999
5900:"Bioenergetics of the Archaea"
5219:Annual Review of Plant Biology
5092:Trends in Biochemical Sciences
3843:. 145 ( Pt 8) (8): 1817–1830.
3760:10.1080/07315724.2001.10719063
3682:
3641:
3596:
3561:
3414:
3375:
2483:
2458:
1789:
1530:
1299:
1164:
987:
898:NADH-coenzyme Q oxidoreductase
379:and hydrogen (protons), is an
1:
9427:Reverse cholesterol transport
8639:Pyruvate dehydrogenase kinase
8339:Introduction to Bioenergetics
8110:10.1016/S0021-9258(20)81361-1
7874:Annual Review of Biochemistry
7842:10.1016/S0021-9258(18)56879-4
7119:10.1016/S0968-0004(00)01674-1
6963:Biochemical Society Symposium
6794:10.1016/S0021-9258(18)33672-X
6702:10.1016/S0968-0004(01)02051-5
6322:Annual Review of Biochemistry
6062:10.1016/S0005-2728(01)00238-9
6021:10.1016/S0005-2728(97)00034-0
5721:10.1016/S0005-2728(02)00271-2
5702:Schägger H (September 2002).
5574:10.1016/S0014-5793(01)03172-6
5467:10.1016/S0378-1119(97)00502-7
5397:10.1016/S0005-2728(05)80197-5
5147:10.1126/science.272.5265.1136
5060:10.1016/S0014-5793(03)00391-0
5004:10.1016/S0021-9258(19)38410-8
4854:Annual Review of Biochemistry
4767:10.1016/S0021-9258(19)75238-7
4719:10.1016/S0021-9258(18)33160-0
4519:10.1016/S0005-2728(01)00237-7
4381:Annual Review of Biochemistry
3881:Annual Review of Biochemistry
3369:
3360:2,4-Dinitrophenol#Dieting aid
3026:inhibit NADH and coenzyme Q.
2996:There are several well-known
2991:
1401:Q-cytochrome c oxidoreductase
1394:Q-cytochrome c oxidoreductase
573:, in addition to ubiquinone.
9079:(protein→peptide→amino acid)
8742:Complex I/NADH dehydrogenase
8467:Universidade Fernando Pessoa
7630:. 345 Pt 2 (Pt 2): 161–179.
7601:10.1016/0926-6569(64)90182-8
7507:10.1016/0076-6879(79)55060-5
7404:10.1016/0005-2736(91)90051-9
7205:10.1016/j.bbagen.2009.04.019
7084:10.1016/j.biocel.2006.07.001
6377:10.1016/0014-5793(95)01536-1
5195:10.1016/j.bbabio.2006.06.004
5104:10.1016/0968-0004(94)90071-X
4322:10.1016/j.bbabio.2003.10.002
4270:Journal of Molecular Biology
4135:10.1016/j.bbabio.2006.05.007
3716:10.1016/0014-5793(83)80289-0
3667:10.1016/0079-6107(85)90004-5
3293:with the publication of the
3242:History of molecular biology
3185:
3169:
3147:
3124:
3098:
3074:
2318:
2299:
2276:
2264:Dimethyl sulfoxide reductase
2257:
2238:
2219:
2200:
2181:
2162:
2133:
2114:
2091:
2072:
2053:
2034:
266:inner mitochondrial membrane
7:
8393:On-line biophysics lectures
8292:
7710:Harden A, Young WJ (1906).
6996:Rattan SI (December 2006).
4961:10.1126/science.281.5373.64
4906:Annual Review of Physiology
3850:10.1099/13500872-145-8-1817
3324:
2889:
2553:
1879:
1603:
1350:
1205:
1126:flavin adenine dinucleotide
1055:
282:gradient and the resulting
10:
10933:
10907:Integral membrane proteins
9464:Phospagen system (ATP-PCr)
8934:Primary nutritional groups
8562:Oxoglutarate dehydrogenase
8228:Nelson DL, Cox MM (2004).
6084:Freitag A, Bock E (1990).
5320:10.1080/1521-6540400000876
4982:Trumpower BL (July 1990).
3235:
3231:
2725:
2393:ATP synthase, also called
2386:
1955:
1667:
1114:Succinate-Q oxidoreductase
1107:Succinate-Q oxidoreductase
856:
836:
814:
611:
531:-soluble electron carrier
508:electron transfer protein
472:
348:
184:, this takes place inside
9517:
9439:
9399:
9371:
9352:
9326:
9298:Anoxygenic photosynthesis
9288:
9281:
9252:
9220:Pentose phosphate pathway
9215:
9198:
9181:
9174:
9154:
9115:
9086:
9062:
9053:
9010:
8994:
8975:Oxidative phosphorylation
8953:
8942:
8916:
8843:
8734:
8727:oxidative phosphorylation
8718:
8690:
8671:
8652:
8607:
8598:
8539:
8320:Biological Thermodynamics
7681:10.1007/s10540-005-2889-2
7636:10.1042/0264-6021:3450161
7027:10.1080/10715760600911303
6180:FEMS Microbiology Reviews
6090:FEMS Microbiology Letters
5547:Lenaz G (December 2001).
4282:10.1016/j.jmb.2006.11.026
3979:10.1016/j.sbi.2004.10.002
3689:Wood PM (December 1983).
3272:oxidative phosphorylation
3198:, thereby inhibiting the
2143:-amino acid dehydrogenase
1939:Organization of complexes
1069:attached to the complex,
50:Oxidative phosphorylation
9318:Entner-Doudoroff pathway
8980:electron transport chain
8967:Pyruvate decarboxylation
8722:electron transport chain
8682:Methylmalonyl-CoA mutase
8557:Isocitrate dehydrogenase
8388:Concepts in Biochemistry
8356:Wikstrom M, ed. (2005).
7428:Satyanarayana U (2002).
7355:10.3389/fphys.2018.01941
6741:10.1038/sj.embor.7400646
5271:10.1074/jbc.271.38.23117
4479:10.2174/1389203043486847
3387:dictionary.cambridge.org
3341:
3194:Binds to the Qi site of
2639:binding change mechanism
2573:vacuolar type H+-ATPases
2383:ATP synthase (complex V)
678:electrochemical gradient
614:Electron transport chain
396:electrochemical gradient
255:electron transport chain
27:electron transport chain
10786:carbohydrate metabolism
9412:Sphingolipid metabolism
9313:DeLey-Doudoroff pathway
9161:carbohydrate catabolism
9156:Carbohydrate metabolism
9142:Purine nucleotide cycle
8663:Glutamate dehydrogenase
8574:Succinate dehydrogenase
8567:Succinyl CoA synthetase
8478:succinate dehydrogenase
8424:molecule of the month:
8341:(1st ed.). Anmol.
8163:10.1073/pnas.70.10.2837
7628:The Biochemical Journal
7342:Frontiers in Physiology
6872:Journal of Bacteriology
6822:Antonie van Leeuwenhoek
6455:Journal of Bacteriology
6267:Journal of Bacteriology
5956:Microbiological Reviews
5869:10.1023/A:1006515817767
5671:10.1093/emboj/19.8.1777
5515:10.1073/pnas.96.14.8271
4668:10.1073/pnas.0604567103
4598:The Biochemical Journal
4436:10.1126/science.1079605
4346:The Biochemical Journal
4187:10.1126/science.1123809
3609:The Biochemical Journal
3356:anti-obesity medication
3238:History of biochemistry
3018:, the barbiturate drug
2901:reactive oxygen species
2722:Reactive oxygen species
2687:O can be summarized as
2332:succinate dehydrogenase
2188:Succinate dehydrogenase
1929:reactive oxygen species
1122:succinate dehydrogenase
739:Succinate dehydrogenase
479:Cofactor (biochemistry)
442:motor of ATP synthase.
435:Propionigenium modestum
311:reactive oxygen species
9381:Fatty acid degradation
9101:Amino acid degradation
8706:Aspartate transaminase
7729:10.1098/rspb.1906.0029
7557:10.1074/jbc.M406576200
7304:10.1073/pnas.140093597
6658:10.1074/jbc.R000021200
6608:10.1098/rstb.2000.0588
6217:Molecular Microbiology
5815:10.1073/pnas.81.9.2606
5762:10.1074/jbc.M106474200
5623:10.1074/jbc.M610545200
4814:10.1105/tpc.105.035162
3811:10.1126/science.388618
3196:cytochrome c reductase
2877:
2634:
2541:
2358:nitrite oxidoreductase
1867:
1683:
1591:
1405:cytochrome c reductase
1397:
1338:
1193:
1110:
1082:conformational changes
1043:
894:
638:, produce the reduced
500:
447:anaerobic fermentation
406:) and a difference in
268:, in a process called
178:adenosine triphosphate
46:
10834:amino acid metabolism
9417:Eicosanoid metabolism
9373:Fatty acid metabolism
9137:Pyrimidine metabolism
8996:Anaerobic respiration
7005:Free Radical Research
3362:for more information.
3254:, but initially only
3236:Further information:
3125:Poisons, weight-loss
3036:British anti-Lewisite
3022:, and the antibiotic
2878:
2726:Further information:
2628:
2542:
2387:Further information:
2169:Glucose dehydrogenase
2121:Lactate dehydrogenase
2022:Formate dehydrogenase
1956:Further information:
1868:
1677:
1668:Further information:
1592:
1391:
1339:
1235:Plasmodium falciparum
1194:
1104:
1071:flavin mononucleotide
1044:
891:NADH-Q oxidoreductase
888:
684:Trichomonas vaginalis
612:Further information:
486:
473:Further information:
349:Further information:
296:adenosine diphosphate
24:
10902:Cellular respiration
10810:cellular respiration
9391:Fatty acid synthesis
9096:Amino acid synthesis
8701:Pyruvate carboxylase
8589:Malate dehydrogenase
8488:cytochrome c oxidase
8452:Cytochrome c oxidase
8416:Structural resources
8198:. Nobel Foundation.
8066:. Nobel Foundation.
6559:10.1093/emboj/cdg608
6516:10.1242/jeb.203.1.89
3493:10.1242/jeb.203.1.51
3264:Fritz Albert Lipmann
3214:brown adipose tissue
3063:Carbon monoxide
2957:superoxide dismutase
2758:
2414:
1958:Microbial metabolism
1908:, which is found in
1722:
1687:Cytochrome c oxidase
1680:cytochrome c oxidase
1670:cytochrome c oxidase
1463:
1274:
1149:
952:
922:mitochondrial genome
582:iron–sulfur clusters
290:in a process called
284:electrical potential
198:anaerobic glycolysis
9982:Transcription &
8955:Aerobic respiration
8851:Alternative oxidase
8655:α-ketoglutaric acid
8154:1973PNAS...70.2837B
8055:Mitchell P (1978).
7995:1961Natur.191..144M
7944:1953Natur.172..975S
7868:Kalckar HM (1991).
7550:(38): 39414–39420.
7476:10.1021/bi00052a022
7295:2000PNAS...97.8670S
7240:2002Natur.415...96E
7154:2000Natur.408..239F
6787:(20): 12030–12038.
6369:1996FEBSL.379..309V
6135:2006ApEnM..72.2050S
5861:1999OLEB...29...73N
5806:1984PNAS...81.2606G
5755:(41): 37861–37867.
5616:(16): 12240–12248.
5565:2001FEBSL.509..151L
5506:1999PNAS...96.8271M
5264:(38): 23117–23120.
5188:(9–10): 1110–1116.
5139:1996Sci...272.1136T
5133:(5265): 1136–1144.
5051:2003FEBSL.545...39H
4997:(20): 11409–11412.
4953:1998Sci...281...64I
4659:2006PNAS..10316212Z
4653:(44): 16212–16217.
4563:10.1038/nature05572
4555:2007Natur.446...88P
4428:2003Sci...299..700Y
4239:10.1038/nature09066
4231:2010Natur.465..441E
4178:2006Sci...311.1430S
4172:(5766): 1430–1436.
4128:(9–10): 1406–1420.
4022:10.1038/nature03343
4014:2005Natur.434...74B
3928:1999Natur.402...47P
3803:1979Sci...206.1148M
3797:(4423): 1148–1159.
3707:1983FEBSL.164..223W
3648:Mathews FS (1985).
3576:(Pt 6): 1095–1105.
3435:1967Natur.213..137M
3295:chemiosmotic theory
3268:Albert L. Lehninger
3218:uncoupling proteins
2934:uncoupling proteins
2866:
2841:
2817:
2793:
2774:
2535:
2510:
2469:
2451:
2002:Respiratory enzyme
1998:
1905:alternative oxidase
1861:
1834:
1812:
1788:
1764:
1746:
1585:
1561:
1529:
1505:
1479:
1332:
1314:
1290:
1187:
1037:
1015:
986:
700:Respiratory enzyme
696:
676:, and generates an
674:intermembrane space
400:proton-motive force
10769:metabolic pathways
9459:Ethanol metabolism
9407:Steroid metabolism
9132:Nucleotide salvage
9064:Protein metabolism
8473:NADH dehydrogenase
8457:2020-07-24 at the
8445:2020-07-24 at the
8433:2020-07-24 at the
8398:2009-05-02 at the
7757:10.1007/BF01874172
7669:Bioscience Reports
6975:10.1042/bss0610001
6834:10.1007/BF00872221
4358:10.1042/BJ20091382
4092:10.1042/BST0330525
3582:10.1042/bst0311095
3315:structural studies
3276:Volodymyr Belitser
2873:
2870:
2846:
2821:
2815:
2798:
2762:
2655:also contain the A
2635:
2537:
2521:
2498:
2488:
2437:
2336:fumarate reductase
2306:Fumarate reductase
2060:NADH dehydrogenase
2011:Midpoint potential
1992:
1987:midpoint potential
1912:, as well as some
1863:
1847:
1822:
1800:
1774:
1752:
1734:
1684:
1587:
1571:
1549:
1515:
1493:
1467:
1398:
1368:and catabolism of
1334:
1320:
1302:
1278:
1189:
1175:
1111:
1039:
1023:
1003:
972:
902:NADH dehydrogenase
895:
873:Conditions: pH = 7
720:NADH dehydrogenase
709:Midpoint potential
694:
647:transfer potential
598:quantum tunnelling
539:cycle. This small
501:
408:electric potential
373:electron acceptors
341:their activities.
271:electron transport
232:electron acceptors
196:processes such as
155:terminal oxidation
47:
10887:
10886:
10757:
10756:
10717:Neurotransmitters
10096:Nucleotide sugars
9829:Direct / C4 / CAM
9476:
9475:
9472:
9471:
9435:
9434:
9348:
9347:
9344:
9343:
9331:Xylose metabolism
9277:
9276:
9150:
9149:
9127:Purine metabolism
9072:Protein synthesis
9049:
9048:
8971:Citric acid cycle
8929:Metabolic network
8924:Metabolic pathway
8868:
8867:
8864:
8863:
8714:
8713:
8530:Citric acid cycle
8376:General resources
8337:Rajan SS (2003).
8318:Haynie D (2001).
8148:(10): 2837–2839.
8103:(11): 3322–3329.
7989:(4784): 144–148.
7938:(4387): 975–978.
7516:978-0-12-181955-2
7289:(15): 8670–8674.
7148:(6809): 239–247.
7011:(12): 1230–1238.
6602:(1396): 465–471.
6553:(23): 6182–6192.
6461:(12): 3205–3208.
6273:(10): 3020–3025.
5500:(14): 8271–8276.
4760:(23): 8440–8445.
4610:10.1042/bj2410883
4422:(5607): 700–704.
4225:(7297): 441–445.
4086:(Pt 3): 525–529.
3617:10.1042/bj3100379
3429:(5072): 137–139.
3291:Peter D. Mitchell
3210:
3209:
3121:2,4-Dinitrophenol
2897:
2896:
2869:
2849:
2845:
2842:
2833:
2820:
2808:
2801:
2797:
2794:
2785:
2765:
2738:electron acceptor
2561:
2560:
2528:
2524:
2513:
2501:
2494:
2490:
2444:
2440:
2429:
2426:
2420:
2345:bacteria such as
2323:
2322:
2245:Nitrite reductase
2226:Nitrate reductase
2207:Ubiquinol oxidase
2158:
2142:
1887:
1886:
1854:
1850:
1837:
1825:
1807:
1803:
1798:
1781:
1777:
1755:
1741:
1737:
1732:
1710:electron acceptor
1611:
1610:
1578:
1574:
1556:
1552:
1547:
1535:
1522:
1518:
1500:
1496:
1491:
1470:
1403:is also known as
1358:
1357:
1323:
1309:
1305:
1298:
1285:
1281:
1213:
1212:
1178:
1170:
1163:
1156:
1063:
1062:
1030:
1026:
1006:
993:
979:
975:
964:
958:
878:
877:
681:protozoa such as
632:citric acid cycle
543:molecule is very
522:periplasmic space
419:thermodynamically
319:hydrogen peroxide
278:in the form of a
274:. This generates
247:protein complexes
209:citric acid cycle
190:aerobic organisms
159:metabolic pathway
35:citric acid cycle
16:Metabolic pathway
10924:
10881:
10878:lipid metabolism
10875:
10874:
10869:
10859:
10858:
10853:
10843:
10842:
10837:
10831:
10830:
10825:
10819:
10818:
10813:
10807:
10806:
10801:
10795:
10794:
10789:
10783:
10782:
10765:
10764:
10747:
10740:
10731:
10724:
10722:thyroid hormones
10712:
10703:
10696:
10689:
10680:
10671:
10664:
10657:
10648:
10641:
10634:
10617:
10608:
10601:
10592:
10585:
10578:
10571:
10559:
10552:
10543:
10536:
10529:
10522:
10510:
10503:
10481:
10468:
10455:
10446:
10431:
10424:
10415:
10408:
10395:
10382:
10375:
10368:
10354:
10342:
10335:
10321:
10307:
10298:
10289:
10282:
10275:
10266:
10257:
10248:
10239:
10232:
10223:
10216:
10207:
10200:
10193:
10186:
10179:
10172:
10165:
10156:
10149:
10140:
10133:
10126:
10119:
10112:
10105:
10098:
10091:
10079:
10072:
10063:
10046:
10034:
10030:
10019:
10015:
10006:
10002:
9993:
9989:
9983:
9974:
9970:
9959:
9955:
9946:
9942:
9933:
9929:
9920:
9916:
9907:
9903:
9894:
9890:
9879:
9875:
9864:
9860:
9849:
9845:
9836:
9832:
9821:
9817:
9804:
9800:
9789:
9785:
9774:
9770:
9761:
9757:
9744:
9740:
9729:
9725:
9714:
9710:
9699:
9695:
9681:
9678:
9668:
9665:
9657:
9654:
9644:
9641:
9629:
9626:
9614:
9611:
9601:
9598:
9588:
9585:
9575:
9572:
9560:
9557:
9547:
9544:
9532:
9521:
9503:
9496:
9489:
9480:
9479:
9447:Metal metabolism
9369:
9368:
9354:Lipid metabolism
9286:
9285:
9179:
9178:
9172:
9171:
9084:
9083:
9060:
9059:
8986:
8951:
8950:
8895:
8888:
8881:
8872:
8871:
8732:
8731:
8693:oxaloacetic acid
8605:
8604:
8547:Citrate synthase
8519:
8512:
8505:
8496:
8495:
8371:
8352:
8333:
8314:
8288:
8266:
8247:
8235:
8211:
8210:
8208:
8207:
8192:
8186:
8185:
8175:
8165:
8133:
8127:
8126:
8112:
8088:
8082:
8081:
8079:
8078:
8072:
8061:
8052:
8046:
8045:
8029:
8023:
8022:
8003:10.1038/191144a0
7978:
7972:
7971:
7952:10.1038/172975a0
7927:
7921:
7920:
7906:
7900:
7899:
7889:
7865:
7859:
7858:
7844:
7820:
7814:
7813:
7783:
7777:
7776:
7740:
7734:
7733:
7731:
7707:
7701:
7700:
7675:(3–4): 271–286.
7664:
7658:
7657:
7647:
7619:
7613:
7612:
7584:
7578:
7577:
7559:
7535:
7529:
7528:
7494:
7488:
7487:
7458:
7452:
7451:
7425:
7416:
7415:
7387:
7378:
7377:
7367:
7357:
7333:
7327:
7326:
7316:
7306:
7274:
7268:
7267:
7223:
7217:
7216:
7188:
7182:
7181:
7162:10.1038/35041687
7137:
7131:
7130:
7102:
7096:
7095:
7067:
7061:
7060:
7058:
7057:
7051:
7045:. Archived from
7020:
7002:
6993:
6987:
6986:
6958:
6949:
6948:
6912:
6906:
6905:
6895:
6878:(9): 2543–2550.
6863:
6854:
6853:
6817:
6811:
6810:
6796:
6772:
6763:
6762:
6752:
6720:
6714:
6713:
6685:
6679:
6678:
6660:
6651:(3): 1665–1668.
6636:
6630:
6629:
6619:
6587:
6581:
6580:
6570:
6547:The EMBO Journal
6538:
6532:
6531:
6495:
6489:
6488:
6478:
6446:
6440:
6439:
6420:10.1038/35089509
6403:
6397:
6396:
6352:
6346:
6345:
6317:
6308:
6307:
6305:
6290:
6264:
6255:
6249:
6248:
6212:
6206:
6205:
6195:
6171:
6165:
6164:
6154:
6129:(3): 2050–2063.
6114:
6108:
6107:
6105:
6081:
6075:
6074:
6064:
6055:(1–2): 140–157.
6040:
6034:
6033:
6023:
5999:
5990:
5989:
5979:
5947:
5938:
5937:
5927:
5895:
5889:
5888:
5844:
5838:
5837:
5827:
5817:
5800:(9): 2606–2610.
5785:
5779:
5778:
5764:
5740:
5734:
5733:
5723:
5714:(1–3): 154–159.
5699:
5693:
5692:
5682:
5665:(8): 1777–1783.
5659:The EMBO Journal
5650:
5644:
5643:
5625:
5601:
5595:
5594:
5576:
5544:
5538:
5537:
5527:
5517:
5485:
5479:
5478:
5450:
5444:
5443:
5415:
5409:
5408:
5380:
5374:
5373:
5363:
5339:
5333:
5332:
5322:
5298:
5292:
5291:
5273:
5249:
5243:
5242:
5214:
5208:
5207:
5197:
5173:
5167:
5166:
5122:
5116:
5115:
5087:
5081:
5080:
5062:
5030:
5024:
5023:
5021:
5006:
4988:
4979:
4973:
4972:
4936:
4930:
4929:
4901:
4895:
4894:
4892:
4869:
4851:
4842:
4836:
4835:
4825:
4808:(9): 2587–2600.
4793:
4787:
4786:
4784:
4769:
4751:
4742:
4736:
4735:
4721:
4712:(2): 1066–1076.
4697:
4691:
4690:
4680:
4670:
4638:
4632:
4631:
4621:
4589:
4583:
4582:
4538:
4532:
4531:
4521:
4512:(1–2): 123–139.
4497:
4491:
4490:
4462:
4456:
4455:
4411:
4405:
4404:
4376:
4370:
4369:
4341:
4335:
4334:
4324:
4300:
4294:
4293:
4265:
4259:
4258:
4214:
4208:
4207:
4189:
4157:
4148:
4147:
4137:
4113:
4104:
4103:
4075:
4066:
4063:
4042:
4041:
3997:
3991:
3990:
3962:
3956:
3955:
3911:
3905:
3904:
3876:
3870:
3869:
3867:
3852:
3838:
3829:
3823:
3822:
3786:
3780:
3779:
3743:
3737:
3736:
3718:
3686:
3680:
3679:
3669:
3645:
3639:
3638:
3628:
3600:
3594:
3593:
3565:
3559:
3558:
3522:
3509:
3508:
3472:
3463:
3462:
3443:10.1038/213137a0
3418:
3412:
3409:
3403:
3402:
3400:
3398:
3379:
3363:
3352:
3284:
3256:sugar phosphates
3222:body temperature
3071:Hydrogen sulfide
3041:
3040:
2891:
2882:
2880:
2879:
2874:
2872:
2871:
2867:
2865:
2864:
2854:
2847:
2843:
2840:
2839:
2838:
2831:
2824:
2822:
2818:
2816:
2806:
2799:
2795:
2792:
2791:
2790:
2783:
2776:
2773:
2770:
2763:
2752:
2728:Oxidative stress
2555:
2546:
2544:
2543:
2538:
2536:
2534:
2529:
2526:
2522:
2511:
2509:
2506:
2499:
2492:
2491:
2489:
2487:
2486:
2479:
2471:
2470:
2468:
2461:
2453:
2450:
2445:
2442:
2438:
2427:
2424:
2418:
2408:
2338:, respectively.
2325:As shown above,
2287:-oxide reductase
2156:
2140:
2098:Pyruvate oxidase
1999:
1991:
1974:Escherichia coli
1933:oxidative stress
1881:
1872:
1870:
1869:
1864:
1862:
1860:
1855:
1852:
1848:
1839:
1835:
1833:
1830:
1823:
1814:
1811:
1808:
1805:
1801:
1796:
1787:
1782:
1779:
1775:
1766:
1763:
1760:
1753:
1748:
1745:
1742:
1739:
1735:
1730:
1716:
1689:, also known as
1605:
1596:
1594:
1593:
1588:
1586:
1584:
1579:
1576:
1572:
1563:
1560:
1557:
1554:
1550:
1545:
1537:
1533:
1528:
1523:
1520:
1516:
1507:
1504:
1501:
1498:
1494:
1489:
1481:
1478:
1475:
1468:
1457:
1352:
1343:
1341:
1340:
1335:
1333:
1331:
1328:
1321:
1316:
1313:
1310:
1307:
1303:
1296:
1292:
1289:
1286:
1283:
1279:
1268:
1207:
1198:
1196:
1195:
1190:
1188:
1186:
1183:
1176:
1171:
1168:
1161:
1157:
1154:
1143:
1116:, also known as
1067:prosthetic group
1057:
1048:
1046:
1045:
1040:
1038:
1036:
1031:
1028:
1024:
1014:
1011:
1004:
999:
998:
991:
985:
980:
977:
973:
962:
956:
946:
900:, also known as
874:
697:
693:
668:by NADH to pump
603:
276:potential energy
148:
147:
144:
143:
140:
137:
134:
131:
128:
125:
122:
119:
116:
113:
110:
107:
98:
97:
94:
93:
90:
87:
84:
81:
78:
75:
72:
69:
66:
63:
60:
10932:
10931:
10927:
10926:
10925:
10923:
10922:
10921:
10892:
10891:
10888:
10883:
10882:
10872:
10870:
10856:
10854:
10840:
10838:
10828:
10826:
10816:
10814:
10804:
10802:
10792:
10790:
10780:
10778:
10776:
10773:metro-style map
10766:
10758:
10753:
10752:
10751:
10750:
10749:
10748:
10743:
10741:
10737:
10734:
10732:
10727:
10725:
10719:
10715:
10713:
10709:
10708:Polyunsaturated
10706:
10704:
10699:
10697:
10692:
10690:
10686:
10683:
10681:
10677:
10674:
10672:
10667:
10665:
10660:
10658:
10654:
10651:
10649:
10644:
10642:
10637:
10635:
10624:
10620:
10618:
10614:
10611:
10609:
10604:
10602:
10598:
10595:
10593:
10588:
10586:
10581:
10579:
10574:
10572:
10566:
10562:
10560:
10555:
10553:
10549:
10546:
10544:
10539:
10537:
10532:
10530:
10525:
10523:
10517:
10513:
10511:
10506:
10504:
10493:
10484:
10482:
10475:
10471:
10469:
10462:
10458:
10456:
10452:
10449:
10447:
10443:
10434:
10432:
10427:
10425:
10421:
10418:
10416:
10411:
10409:
10402:
10398:
10396:
10388:
10385:
10383:
10378:
10376:
10371:
10369:
10365:
10362:
10357:
10355:
10349:
10345:
10343:
10338:
10336:
10330:
10327:
10324:
10322:
10316:
10313:
10310:
10308:
10304:
10301:
10299:
10295:
10292:
10290:
10285:
10283:
10278:
10276:
10272:
10269:
10267:
10263:
10260:
10258:
10254:
10251:
10249:
10245:
10242:
10240:
10235:
10233:
10229:
10226:
10224:
10219:
10217:
10213:
10210:
10208:
10203:
10201:
10196:
10194:
10189:
10187:
10182:
10180:
10175:
10173:
10168:
10166:
10162:
10159:
10157:
10152:
10150:
10146:
10143:
10141:
10136:
10134:
10129:
10127:
10122:
10120:
10115:
10113:
10108:
10106:
10101:
10099:
10094:
10092:
10086:
10082:
10080:
10075:
10073:
10069:
10066:
10064:
10056:
10049:
10047:
10043:
10040:
10037:
10035:
10028:
10026:
10022:
10020:
10013:
10009:
10007:
10000:
9996:
9994:
9987:
9985:
9981:
9977:
9975:
9968:
9966:
9962:
9960:
9953:
9949:
9947:
9940:
9936:
9934:
9928:Steroidogenesis
9927:
9923:
9921:
9914:
9910:
9908:
9901:
9897:
9895:
9888:
9886:
9882:
9880:
9873:
9871:
9867:
9865:
9859:phosphorylation
9858:
9856:
9852:
9850:
9843:
9839:
9837:
9830:
9828:
9824:
9822:
9815:
9813:
9811:
9807:
9805:
9798:
9796:
9792:
9790:
9783:
9781:
9777:
9775:
9768:
9764:
9762:
9755:
9753:
9751:
9747:
9745:
9738:
9736:
9732:
9730:
9723:
9721:
9717:
9715:
9708:
9706:
9702:
9700:
9693:
9691:
9687:
9684:
9682:
9676:
9674:
9671:
9669:
9663:
9660:
9658:
9652:
9650:
9647:
9645:
9639:
9637:
9635:
9632:
9630:
9624:
9622:
9620:
9617:
9615:
9609:
9607:
9604:
9602:
9596:
9594:
9591:
9589:
9583:
9581:
9578:
9576:
9570:
9568:
9566:
9563:
9561:
9555:
9553:
9550:
9548:
9542:
9540:
9537:
9533:
9513:
9507:
9477:
9468:
9452:Iron metabolism
9431:
9395:
9356:
9340:
9322:
9308:Carbon fixation
9273:
9248:
9211:
9194:
9190:Gluconeogenesis
9163:
9158:
9146:
9118:
9111:
9082:
9055:
9045:
9006:
8990:
8978:
8945:
8938:
8912:
8899:
8869:
8860:
8839:
8781:
8755:
8725:
8720:
8710:
8686:
8667:
8648:
8594:
8535:
8523:
8459:Wayback Machine
8447:Wayback Machine
8435:Wayback Machine
8418:
8402:Antony Crofts,
8400:Wayback Machine
8378:
8368:
8349:
8330:
8311:
8301:Bioenergetics 3
8295:
8285:
8263:
8244:
8224:
8219:
8217:Further reading
8214:
8205:
8203:
8194:
8193:
8189:
8134:
8130:
8089:
8085:
8076:
8074:
8070:
8059:
8053:
8049:
8030:
8026:
7979:
7975:
7928:
7924:
7907:
7903:
7866:
7862:
7821:
7817:
7810:
7784:
7780:
7741:
7737:
7708:
7704:
7665:
7661:
7620:
7616:
7585:
7581:
7536:
7532:
7517:
7495:
7491:
7459:
7455:
7440:
7426:
7419:
7388:
7381:
7334:
7330:
7275:
7271:
7248:10.1038/415096a
7234:(6867): 96–99.
7224:
7220:
7189:
7185:
7138:
7134:
7113:(10): 502–508.
7103:
7099:
7068:
7064:
7055:
7053:
7049:
7018:10.1.1.476.9259
7000:
6994:
6990:
6959:
6952:
6913:
6909:
6864:
6857:
6818:
6814:
6773:
6766:
6721:
6717:
6686:
6682:
6637:
6633:
6588:
6584:
6539:
6535:
6510:(Pt 1): 89–95.
6496:
6492:
6447:
6443:
6404:
6400:
6353:
6349:
6318:
6311:
6303:
6262:
6256:
6252:
6213:
6209:
6172:
6168:
6115:
6111:
6096:(1–3): 157–62.
6082:
6078:
6041:
6037:
6000:
5993:
5948:
5941:
5896:
5892:
5845:
5841:
5786:
5782:
5741:
5737:
5700:
5696:
5651:
5647:
5602:
5598:
5545:
5541:
5486:
5482:
5451:
5447:
5416:
5412:
5381:
5377:
5340:
5336:
5299:
5295:
5250:
5246:
5215:
5211:
5174:
5170:
5123:
5119:
5088:
5084:
5031:
5027:
5019:
4986:
4980:
4976:
4947:(5373): 64–71.
4937:
4933:
4902:
4898:
4890:
4867:10.1.1.319.5709
4849:
4843:
4839:
4794:
4790:
4782:
4749:
4743:
4739:
4698:
4694:
4639:
4635:
4590:
4586:
4549:(7131): 88–91.
4539:
4535:
4498:
4494:
4463:
4459:
4412:
4408:
4377:
4373:
4342:
4338:
4301:
4297:
4266:
4262:
4215:
4211:
4158:
4151:
4114:
4107:
4076:
4069:
4064:
4045:
4008:(7029): 74–79.
3998:
3994:
3963:
3959:
3922:(6757): 47–52.
3912:
3908:
3877:
3873:
3865:
3836:
3830:
3826:
3787:
3783:
3744:
3740:
3687:
3683:
3646:
3642:
3601:
3597:
3566:
3562:
3523:
3512:
3487:(Pt 1): 51–59.
3473:
3466:
3419:
3415:
3410:
3406:
3396:
3394:
3381:
3380:
3376:
3372:
3367:
3366:
3353:
3349:
3344:
3336:TIM/TOM Complex
3327:
3278:
3244:
3234:
3128:Inner membrane
3119:
3110:
3069:
3065:
3061:
3050:Site of action
2994:
2986:lactic acidosis
2982:
2973:
2860:
2855:
2850:
2844:
2834:
2830:
2829:
2823:
2807:
2802:
2796:
2786:
2782:
2781:
2775:
2771:
2766:
2761:
2759:
2756:
2755:
2734:
2724:
2704:
2697:
2694:+ NADH + H → H
2693:
2686:
2682:
2677:
2670:
2666:
2662:
2658:
2605:
2598:
2593:
2589:
2585:
2567:reaction is an
2565:phosphorylation
2530:
2525:
2507:
2502:
2482:
2475:
2474:
2472:
2464:
2457:
2455:
2454:
2452:
2446:
2441:
2417:
2415:
2412:
2411:
2404:
2391:
2385:
2283:Trimethylamine
1960:
1954:
1941:
1892:
1856:
1851:
1838:
1831:
1826:
1813:
1809:
1804:
1783:
1778:
1765:
1761:
1756:
1747:
1743:
1738:
1725:
1723:
1720:
1719:
1672:
1666:
1659:
1651:
1647:
1643:
1632:
1628:
1624:
1616:
1580:
1575:
1562:
1558:
1553:
1536:
1524:
1519:
1506:
1502:
1497:
1480:
1476:
1471:
1466:
1464:
1461:
1460:
1434:
1412:
1386:
1329:
1324:
1315:
1311:
1306:
1291:
1287:
1282:
1277:
1275:
1272:
1271:
1248:
1226:large intestine
1184:
1179:
1167:
1153:
1152:
1150:
1147:
1146:
1099:
1092:
1076:
1032:
1027:
1012:
1007:
994:
990:
981:
976:
955:
953:
950:
949:
883:
872:
863:
849:
845:
829:
825:
807:
803:
794:
783:
779:
769:
757:
753:
667:
620:
610:
601:
568:
560:
556:
498:
481:
471:
441:
357:
347:
329:and, possibly,
300:phosphorylation
236:redox reactions
234:in a series of
225:
213:electron donors
104:
100:
57:
53:
40:
17:
12:
11:
5:
10930:
10920:
10919:
10914:
10909:
10904:
10885:
10884:
10822:cell signaling
10798:photosynthesis
10796:Violet nodes:
10784:Orange nodes:
10760:
10759:
10755:
10754:
10441:
10387:Aromatic amino
10294:Branched-chain
9844:Light reaction
9535:
9534:
9527:
9526:
9525:
9524:
9519:
9518:
9515:
9514:
9506:
9505:
9498:
9491:
9483:
9474:
9473:
9470:
9469:
9467:
9466:
9461:
9456:
9455:
9454:
9443:
9441:
9437:
9436:
9433:
9432:
9430:
9429:
9424:
9419:
9414:
9409:
9403:
9401:
9397:
9396:
9394:
9393:
9388:
9385:Beta oxidation
9377:
9375:
9366:
9350:
9349:
9346:
9345:
9342:
9341:
9339:
9338:
9333:
9327:
9324:
9323:
9321:
9320:
9315:
9310:
9305:
9303:Chemosynthesis
9300:
9295:
9293:Photosynthesis
9289:
9283:
9279:
9278:
9275:
9274:
9272:
9271:
9270:
9269:
9264:
9253:
9250:
9249:
9247:
9246:
9245:
9244:
9242:Leloir pathway
9234:
9233:
9232:
9230:Polyol pathway
9222:
9216:
9213:
9212:
9210:
9209:
9203:Glycogenolysis
9199:
9196:
9195:
9193:
9192:
9182:
9176:
9169:
9152:
9151:
9148:
9147:
9145:
9144:
9139:
9134:
9129:
9123:
9121:
9113:
9112:
9110:
9109:
9104:
9098:
9092:
9090:
9081:
9080:
9074:
9068:
9066:
9057:
9051:
9050:
9047:
9046:
9044:
9043:
9042:
9041:
9036:
9031:
9016:
9014:
9008:
9007:
9005:
9004:
9000:
8998:
8992:
8991:
8989:
8988:
8959:
8957:
8948:
8940:
8939:
8937:
8936:
8931:
8926:
8920:
8918:
8914:
8913:
8898:
8897:
8890:
8883:
8875:
8866:
8865:
8862:
8861:
8859:
8858:
8853:
8847:
8845:
8841:
8840:
8838:
8837:
8832:
8827:
8822:
8817:
8812:
8807:
8802:
8797:
8792:
8787:
8779:
8774:
8773:
8768:
8763:
8758:
8753:
8749:
8744:
8738:
8736:
8729:
8716:
8715:
8712:
8711:
8709:
8708:
8703:
8697:
8695:
8688:
8687:
8685:
8684:
8678:
8676:
8669:
8668:
8666:
8665:
8659:
8657:
8650:
8649:
8647:
8646:
8637:(regulated by
8634:
8633:
8614:
8612:
8602:
8596:
8595:
8593:
8592:
8586:
8581:
8570:
8569:
8564:
8559:
8554:
8549:
8543:
8541:
8537:
8536:
8522:
8521:
8514:
8507:
8499:
8493:
8492:
8491:
8490:
8485:
8480:
8475:
8463:
8462:
8461:
8449:
8437:
8417:
8414:
8413:
8412:
8411:Graham Johnson
8406:
8390:
8377:
8374:
8373:
8372:
8366:
8353:
8347:
8334:
8328:
8315:
8309:
8294:
8291:
8290:
8289:
8283:
8267:
8261:
8248:
8242:
8223:
8220:
8218:
8215:
8213:
8212:
8187:
8128:
8083:
8047:
8024:
7973:
7922:
7901:
7860:
7835:(2): 611–644.
7815:
7808:
7778:
7751:(1–2): 55–63.
7735:
7722:(77): 405–20.
7702:
7659:
7614:
7595:(2): 233–247.
7579:
7530:
7515:
7489:
7470:(1): 164–173.
7453:
7438:
7417:
7398:(2): 255–258.
7379:
7328:
7269:
7218:
7199:(3): 205–212.
7183:
7132:
7097:
7062:
6988:
6950:
6923:(1): 115–125.
6907:
6855:
6828:(4): 381–395.
6812:
6764:
6735:(3): 276–282.
6715:
6696:(3): 154–160.
6680:
6631:
6582:
6533:
6490:
6441:
6414:(9): 669–677.
6398:
6363:(3): 309–313.
6347:
6309:
6250:
6207:
6186:(4): 259–270.
6166:
6109:
6076:
6035:
6014:(3): 217–234.
5991:
5962:(3): 222–271.
5939:
5910:(3): 570–620.
5890:
5839:
5780:
5735:
5694:
5645:
5596:
5559:(2): 151–155.
5539:
5480:
5461:(2): 121–129.
5445:
5410:
5391:(2): 121–140.
5375:
5354:(6): 733–747.
5334:
5313:(6): 333–341.
5293:
5244:
5209:
5168:
5117:
5098:(8): 325–330.
5082:
5025:
4974:
4931:
4896:
4837:
4802:The Plant Cell
4788:
4737:
4692:
4633:
4604:(3): 883–892.
4584:
4533:
4492:
4473:(2): 107–118.
4457:
4406:
4371:
4352:(2): 327–339.
4336:
4295:
4276:(1): 140–154.
4260:
4209:
4149:
4105:
4067:
4043:
3992:
3973:(6): 642–647.
3957:
3906:
3871:
3824:
3781:
3754:(6): 591–598.
3738:
3701:(2): 223–226.
3681:
3640:
3595:
3560:
3510:
3464:
3413:
3404:
3373:
3371:
3368:
3365:
3364:
3346:
3345:
3343:
3340:
3339:
3338:
3333:
3326:
3323:
3319:John E. Walker
3303:David E. Green
3274:was coined by
3260:Herman Kalckar
3233:
3230:
3208:
3207:
3192:
3189:
3184:
3178:
3177:
3174:
3171:
3168:
3158:
3157:
3154:
3151:
3146:
3140:
3139:
3129:
3126:
3123:
3113:
3112:
3108:
3105:
3102:
3097:
3091:
3090:
3079:
3076:
3073:
3055:
3054:
3051:
3048:
3045:
2993:
2990:
2980:
2972:
2969:
2895:
2894:
2885:
2883:
2863:
2858:
2853:
2837:
2827:
2814:
2811:
2805:
2789:
2779:
2769:
2723:
2720:
2702:
2695:
2691:
2684:
2680:
2676:
2673:
2668:
2664:
2660:
2656:
2603:
2596:
2591:
2587:
2583:
2559:
2558:
2549:
2547:
2533:
2519:
2516:
2505:
2497:
2485:
2478:
2467:
2460:
2449:
2435:
2432:
2423:
2402:
2384:
2381:
2366:modular design
2321:
2320:
2317:
2308:
2302:
2301:
2298:
2289:
2279:
2278:
2275:
2266:
2260:
2259:
2256:
2247:
2241:
2240:
2237:
2228:
2222:
2221:
2218:
2209:
2203:
2202:
2199:
2190:
2184:
2183:
2180:
2171:
2165:
2164:
2161:
2145:
2136:
2135:
2132:
2123:
2117:
2116:
2113:
2107:Carbon dioxide
2100:
2094:
2093:
2090:
2081:
2075:
2074:
2071:
2062:
2056:
2055:
2052:
2043:
2037:
2036:
2033:
2024:
2018:
2017:
2008:
2003:
1953:
1950:
1940:
1937:
1891:
1888:
1885:
1884:
1875:
1873:
1859:
1845:
1842:
1829:
1820:
1817:
1794:
1791:
1786:
1772:
1769:
1759:
1751:
1728:
1665:
1662:
1657:
1649:
1645:
1641:
1635:ubisemiquinone
1630:
1626:
1622:
1614:
1609:
1608:
1599:
1597:
1583:
1569:
1566:
1543:
1540:
1532:
1527:
1513:
1510:
1487:
1484:
1474:
1432:
1410:
1385:
1382:
1362:beta oxidation
1356:
1355:
1346:
1344:
1327:
1319:
1301:
1295:
1247:
1244:
1242:biosynthesis.
1217:parasitic worm
1211:
1210:
1201:
1199:
1182:
1174:
1166:
1160:
1098:
1095:
1090:
1074:
1061:
1060:
1051:
1049:
1035:
1021:
1018:
1010:
1002:
997:
989:
984:
970:
967:
961:
882:
879:
876:
875:
869:
868:
865:
861:
858:
854:
853:
850:
847:
846:/ Cytochrome a
843:
838:
834:
833:
830:
827:
826:/ Cytochrome c
823:
818:
812:
811:
808:
805:
804:/ Cytochrome b
801:
796:
792:
788:
787:
784:
781:
780:/ Coenzyme Q10
777:
772:
767:
762:
761:
758:
755:
751:
741:
735:
734:
731:
722:
716:
715:
706:
701:
665:
636:beta oxidation
609:
606:
566:
558:
554:
496:
470:
467:
459:beta oxidation
439:
346:
343:
223:
38:
15:
9:
6:
4:
3:
2:
10929:
10918:
10915:
10913:
10910:
10908:
10905:
10903:
10900:
10899:
10897:
10890:
10879:
10876:Green nodes:
10867:
10863:
10860:Brown nodes:
10851:
10847:
10835:
10823:
10811:
10799:
10787:
10774:
10770:
10746:
10739:
10730:
10723:
10718:
10711:
10702:
10695:
10694:Sphingolipids
10688:
10687:sphingolipids
10679:
10670:
10663:
10662:Glycerolipids
10656:
10655:phospholipids
10647:
10640:
10632:
10628:
10623:
10616:
10607:
10600:
10591:
10584:
10577:
10570:
10569:proteoglycans
10565:
10564:Glycoproteins
10558:
10551:
10542:
10535:
10528:
10521:
10516:
10509:
10501:
10497:
10491:
10487:
10479:
10474:
10466:
10461:
10454:
10444:
10437:
10430:
10423:
10414:
10406:
10401:
10394:
10390:
10381:
10374:
10367:
10360:
10353:
10348:
10341:
10334:
10329:
10320:
10315:
10306:
10297:
10288:
10281:
10274:
10265:
10256:
10247:
10238:
10231:
10222:
10215:
10206:
10199:
10192:
10185:
10178:
10171:
10164:
10155:
10148:
10139:
10132:
10125:
10118:
10111:
10104:
10097:
10090:
10085:
10078:
10071:
10062:
10058:
10052:
10045:
10038:
10033:
10032:
10031:
10018:
10017:
10016:
10005:
10004:
10003:
9992:
9991:
9990:
9984:
9973:
9972:
9971:
9958:
9957:
9956:
9945:
9944:
9943:
9932:
9931:
9930:
9919:
9918:
9917:
9906:
9905:
9904:
9893:
9892:
9891:
9878:
9877:
9876:
9863:
9862:
9861:
9848:
9847:
9846:
9835:
9834:
9833:
9831:carbon intake
9820:
9819:
9818:
9803:
9802:
9801:
9788:
9787:
9786:
9773:
9772:
9771:
9760:
9759:
9758:
9743:
9742:
9741:
9728:
9727:
9726:
9713:
9712:
9711:
9698:
9697:
9696:
9685:
9680:
9679:
9667:
9666:
9656:
9655:
9643:
9642:
9628:
9627:
9613:
9612:
9600:
9599:
9587:
9586:
9574:
9573:
9559:
9558:
9546:
9545:
9531:
9523:
9522:
9516:
9511:
9504:
9499:
9497:
9492:
9490:
9485:
9484:
9481:
9465:
9462:
9460:
9457:
9453:
9450:
9449:
9448:
9445:
9444:
9442:
9438:
9428:
9425:
9423:
9420:
9418:
9415:
9413:
9410:
9408:
9405:
9404:
9402:
9398:
9392:
9389:
9386:
9382:
9379:
9378:
9376:
9374:
9370:
9367:
9364:
9360:
9355:
9351:
9337:
9336:Radiotrophism
9334:
9332:
9329:
9328:
9325:
9319:
9316:
9314:
9311:
9309:
9306:
9304:
9301:
9299:
9296:
9294:
9291:
9290:
9287:
9284:
9280:
9268:
9265:
9263:
9260:
9259:
9258:
9257:Glycosylation
9255:
9254:
9251:
9243:
9240:
9239:
9238:
9235:
9231:
9228:
9227:
9226:
9223:
9221:
9218:
9217:
9214:
9208:
9204:
9201:
9200:
9197:
9191:
9187:
9184:
9183:
9180:
9177:
9173:
9170:
9167:
9162:
9157:
9153:
9143:
9140:
9138:
9135:
9133:
9130:
9128:
9125:
9124:
9122:
9120:
9114:
9108:
9105:
9102:
9099:
9097:
9094:
9093:
9091:
9089:
9085:
9078:
9075:
9073:
9070:
9069:
9067:
9065:
9061:
9058:
9052:
9040:
9037:
9035:
9032:
9030:
9027:
9026:
9025:
9021:
9018:
9017:
9015:
9013:
9009:
9002:
9001:
8999:
8997:
8993:
8985:
8981:
8976:
8972:
8968:
8964:
8961:
8960:
8958:
8956:
8952:
8949:
8947:
8941:
8935:
8932:
8930:
8927:
8925:
8922:
8921:
8919:
8915:
8911:
8907:
8903:
8896:
8891:
8889:
8884:
8882:
8877:
8876:
8873:
8857:
8854:
8852:
8849:
8848:
8846:
8842:
8836:
8833:
8831:
8828:
8826:
8823:
8821:
8818:
8816:
8813:
8811:
8808:
8806:
8803:
8801:
8798:
8796:
8793:
8791:
8788:
8786:
8782:
8776:
8775:
8772:
8769:
8767:
8764:
8762:
8759:
8757:
8750:
8748:
8745:
8743:
8740:
8739:
8737:
8733:
8730:
8728:
8723:
8719:Mitochondrial
8717:
8707:
8704:
8702:
8699:
8698:
8696:
8694:
8689:
8683:
8680:
8679:
8677:
8675:
8670:
8664:
8661:
8660:
8658:
8656:
8651:
8644:
8640:
8636:
8635:
8631:
8627:
8623:
8619:
8616:
8615:
8613:
8611:
8606:
8603:
8601:
8597:
8590:
8587:
8585:
8582:
8579:
8575:
8572:
8571:
8568:
8565:
8563:
8560:
8558:
8555:
8553:
8550:
8548:
8545:
8544:
8542:
8538:
8534:
8531:
8527:
8520:
8515:
8513:
8508:
8506:
8501:
8500:
8497:
8489:
8486:
8484:
8481:
8479:
8476:
8474:
8471:
8470:
8468:
8464:
8460:
8456:
8453:
8450:
8448:
8444:
8441:
8438:
8436:
8432:
8429:
8426:
8425:
8423:
8420:
8419:
8410:
8407:
8405:
8401:
8397:
8394:
8391:
8389:
8386:
8383:
8380:
8379:
8369:
8367:0-85404-346-2
8363:
8359:
8354:
8350:
8348:81-261-1364-2
8344:
8340:
8335:
8331:
8329:0-521-79549-4
8325:
8321:
8316:
8312:
8310:0-12-518121-3
8306:
8302:
8297:
8296:
8286:
8284:0-19-920564-7
8280:
8276:
8272:
8268:
8264:
8262:0-226-73937-6
8258:
8254:
8249:
8245:
8243:0-7167-4339-6
8239:
8234:
8233:
8226:
8225:
8201:
8197:
8191:
8183:
8179:
8174:
8169:
8164:
8159:
8155:
8151:
8147:
8143:
8139:
8132:
8124:
8120:
8116:
8111:
8106:
8102:
8098:
8094:
8087:
8069:
8065:
8064:Nobel lecture
8058:
8051:
8043:
8039:
8035:
8032:Saier Jr MH.
8028:
8020:
8016:
8012:
8008:
8004:
8000:
7996:
7992:
7988:
7984:
7977:
7969:
7965:
7961:
7957:
7953:
7949:
7945:
7941:
7937:
7933:
7926:
7918:
7914:
7913:
7905:
7897:
7893:
7888:
7883:
7879:
7875:
7871:
7864:
7856:
7852:
7848:
7843:
7838:
7834:
7830:
7826:
7819:
7811:
7809:9780674366701
7805:
7801:
7797:
7793:
7789:
7782:
7774:
7770:
7766:
7762:
7758:
7754:
7750:
7746:
7739:
7730:
7725:
7721:
7717:
7713:
7706:
7698:
7694:
7690:
7686:
7682:
7678:
7674:
7670:
7663:
7655:
7651:
7646:
7641:
7637:
7633:
7629:
7625:
7618:
7610:
7606:
7602:
7598:
7594:
7590:
7583:
7575:
7571:
7567:
7563:
7558:
7553:
7549:
7545:
7541:
7534:
7526:
7522:
7518:
7512:
7508:
7504:
7500:
7493:
7485:
7481:
7477:
7473:
7469:
7465:
7457:
7449:
7445:
7441:
7435:
7431:
7424:
7422:
7413:
7409:
7405:
7401:
7397:
7393:
7386:
7384:
7375:
7371:
7366:
7361:
7356:
7351:
7347:
7343:
7339:
7332:
7324:
7320:
7315:
7310:
7305:
7300:
7296:
7292:
7288:
7284:
7280:
7273:
7265:
7261:
7257:
7253:
7249:
7245:
7241:
7237:
7233:
7229:
7222:
7214:
7210:
7206:
7202:
7198:
7194:
7187:
7179:
7175:
7171:
7167:
7163:
7159:
7155:
7151:
7147:
7143:
7136:
7128:
7124:
7120:
7116:
7112:
7108:
7101:
7093:
7089:
7085:
7081:
7077:
7073:
7066:
7052:on 2014-06-14
7048:
7044:
7040:
7036:
7032:
7028:
7024:
7019:
7014:
7010:
7006:
6999:
6992:
6984:
6980:
6976:
6972:
6968:
6964:
6957:
6955:
6946:
6942:
6938:
6934:
6930:
6926:
6922:
6918:
6911:
6903:
6899:
6894:
6889:
6885:
6881:
6877:
6873:
6869:
6862:
6860:
6851:
6847:
6843:
6839:
6835:
6831:
6827:
6823:
6816:
6808:
6804:
6800:
6795:
6790:
6786:
6782:
6778:
6771:
6769:
6760:
6756:
6751:
6746:
6742:
6738:
6734:
6730:
6726:
6719:
6711:
6707:
6703:
6699:
6695:
6691:
6684:
6676:
6672:
6668:
6664:
6659:
6654:
6650:
6646:
6642:
6635:
6627:
6623:
6618:
6613:
6609:
6605:
6601:
6597:
6593:
6586:
6578:
6574:
6569:
6564:
6560:
6556:
6552:
6548:
6544:
6537:
6529:
6525:
6521:
6517:
6513:
6509:
6505:
6501:
6494:
6486:
6482:
6477:
6472:
6468:
6464:
6460:
6456:
6452:
6445:
6437:
6433:
6429:
6425:
6421:
6417:
6413:
6409:
6402:
6394:
6390:
6386:
6382:
6378:
6374:
6370:
6366:
6362:
6358:
6351:
6343:
6339:
6335:
6331:
6327:
6323:
6316:
6314:
6302:
6298:
6294:
6289:
6284:
6280:
6276:
6272:
6268:
6261:
6254:
6246:
6242:
6238:
6234:
6230:
6226:
6222:
6218:
6211:
6203:
6199:
6194:
6189:
6185:
6181:
6177:
6170:
6162:
6158:
6153:
6148:
6144:
6140:
6136:
6132:
6128:
6124:
6120:
6113:
6104:
6099:
6095:
6091:
6087:
6080:
6072:
6068:
6063:
6058:
6054:
6050:
6046:
6039:
6031:
6027:
6022:
6017:
6013:
6009:
6005:
5998:
5996:
5987:
5983:
5978:
5973:
5969:
5965:
5961:
5957:
5953:
5946:
5944:
5935:
5931:
5926:
5921:
5917:
5913:
5909:
5905:
5901:
5894:
5886:
5882:
5878:
5874:
5870:
5866:
5862:
5858:
5854:
5850:
5843:
5835:
5831:
5826:
5821:
5816:
5811:
5807:
5803:
5799:
5795:
5791:
5784:
5776:
5772:
5768:
5763:
5758:
5754:
5750:
5746:
5739:
5731:
5727:
5722:
5717:
5713:
5709:
5705:
5698:
5690:
5686:
5681:
5676:
5672:
5668:
5664:
5660:
5656:
5649:
5641:
5637:
5633:
5629:
5624:
5619:
5615:
5611:
5607:
5600:
5592:
5588:
5584:
5580:
5575:
5570:
5566:
5562:
5558:
5554:
5550:
5543:
5535:
5531:
5526:
5521:
5516:
5511:
5507:
5503:
5499:
5495:
5491:
5484:
5476:
5472:
5468:
5464:
5460:
5456:
5449:
5441:
5437:
5433:
5429:
5425:
5421:
5414:
5406:
5402:
5398:
5394:
5390:
5386:
5379:
5371:
5367:
5362:
5357:
5353:
5349:
5345:
5338:
5330:
5326:
5321:
5316:
5312:
5308:
5304:
5297:
5289:
5285:
5281:
5277:
5272:
5267:
5263:
5259:
5255:
5248:
5240:
5236:
5232:
5228:
5224:
5220:
5213:
5205:
5201:
5196:
5191:
5187:
5183:
5179:
5172:
5164:
5160:
5156:
5152:
5148:
5144:
5140:
5136:
5132:
5128:
5121:
5113:
5109:
5105:
5101:
5097:
5093:
5086:
5078:
5074:
5070:
5066:
5061:
5056:
5052:
5048:
5044:
5040:
5036:
5029:
5018:
5014:
5010:
5005:
5000:
4996:
4992:
4985:
4978:
4970:
4966:
4962:
4958:
4954:
4950:
4946:
4942:
4935:
4927:
4923:
4919:
4915:
4911:
4907:
4900:
4889:
4885:
4881:
4877:
4873:
4868:
4863:
4860:: 1005–1075.
4859:
4855:
4848:
4841:
4833:
4829:
4824:
4819:
4815:
4811:
4807:
4803:
4799:
4792:
4781:
4777:
4773:
4768:
4763:
4759:
4755:
4748:
4741:
4733:
4729:
4725:
4720:
4715:
4711:
4707:
4703:
4696:
4688:
4684:
4679:
4674:
4669:
4664:
4660:
4656:
4652:
4648:
4644:
4637:
4629:
4625:
4620:
4615:
4611:
4607:
4603:
4599:
4595:
4588:
4580:
4576:
4572:
4568:
4564:
4560:
4556:
4552:
4548:
4544:
4537:
4529:
4525:
4520:
4515:
4511:
4507:
4503:
4496:
4488:
4484:
4480:
4476:
4472:
4468:
4461:
4453:
4449:
4445:
4441:
4437:
4433:
4429:
4425:
4421:
4417:
4410:
4402:
4398:
4394:
4390:
4386:
4382:
4375:
4367:
4363:
4359:
4355:
4351:
4347:
4340:
4332:
4328:
4323:
4318:
4314:
4310:
4306:
4299:
4291:
4287:
4283:
4279:
4275:
4271:
4264:
4256:
4252:
4248:
4244:
4240:
4236:
4232:
4228:
4224:
4220:
4213:
4205:
4201:
4197:
4193:
4188:
4183:
4179:
4175:
4171:
4167:
4163:
4156:
4154:
4145:
4141:
4136:
4131:
4127:
4123:
4119:
4112:
4110:
4101:
4097:
4093:
4089:
4085:
4081:
4074:
4072:
4062:
4060:
4058:
4056:
4054:
4052:
4050:
4048:
4039:
4035:
4031:
4027:
4023:
4019:
4015:
4011:
4007:
4003:
3996:
3988:
3984:
3980:
3976:
3972:
3968:
3961:
3953:
3949:
3945:
3941:
3937:
3936:10.1038/46972
3933:
3929:
3925:
3921:
3917:
3910:
3902:
3898:
3894:
3890:
3886:
3882:
3875:
3864:
3860:
3856:
3851:
3846:
3842:
3835:
3828:
3820:
3816:
3812:
3808:
3804:
3800:
3796:
3792:
3785:
3777:
3773:
3769:
3765:
3761:
3757:
3753:
3749:
3742:
3734:
3730:
3726:
3722:
3717:
3712:
3708:
3704:
3700:
3696:
3692:
3685:
3677:
3673:
3668:
3663:
3659:
3655:
3651:
3644:
3636:
3632:
3627:
3622:
3618:
3614:
3610:
3606:
3599:
3591:
3587:
3583:
3579:
3575:
3571:
3564:
3556:
3552:
3548:
3544:
3540:
3536:
3532:
3528:
3521:
3519:
3517:
3515:
3506:
3502:
3498:
3494:
3490:
3486:
3482:
3478:
3471:
3469:
3460:
3456:
3452:
3448:
3444:
3440:
3436:
3432:
3428:
3424:
3417:
3408:
3392:
3388:
3384:
3378:
3374:
3361:
3357:
3351:
3347:
3337:
3334:
3332:
3329:
3328:
3322:
3320:
3316:
3312:
3311:Paul D. Boyer
3308:
3307:Efraim Racker
3304:
3300:
3296:
3292:
3286:
3282:
3277:
3273:
3269:
3265:
3261:
3257:
3253:
3249:
3248:Arthur Harden
3243:
3239:
3229:
3227:
3223:
3219:
3215:
3205:
3201:
3197:
3193:
3190:
3188:
3183:
3180:
3179:
3175:
3172:
3167:
3163:
3160:
3159:
3155:
3152:
3150:
3145:
3142:
3141:
3137:
3133:
3130:
3127:
3122:
3118:
3115:
3114:
3106:
3103:
3101:
3096:
3093:
3092:
3088:
3084:
3080:
3077:
3072:
3068:
3064:
3060:
3057:
3056:
3052:
3049:
3046:
3043:
3042:
3039:
3037:
3033:
3027:
3025:
3021:
3017:
3011:
3008:
3003:
2999:
2989:
2987:
2978:
2968:
2966:
2962:
2958:
2954:
2950:
2946:
2942:
2937:
2935:
2930:
2924:
2922:
2918:
2914:
2910:
2906:
2902:
2893:
2886:
2884:
2861:
2856:
2851:
2835:
2825:
2812:
2809:
2803:
2787:
2777:
2767:
2754:
2753:
2750:
2748:
2744:
2739:
2733:
2729:
2719:
2715:
2712:
2709:
2706:
2699:
2688:
2672:
2654:
2653:
2652:Methanococcus
2647:
2645:
2640:
2632:
2629:Mechanism of
2627:
2623:
2621:
2617:
2613:
2612:electrostatic
2609:
2600:
2581:
2576:
2574:
2570:
2566:
2557:
2550:
2548:
2531:
2517:
2514:
2503:
2495:
2476:
2465:
2447:
2443:intermembrane
2433:
2430:
2421:
2410:
2409:
2406:
2400:
2396:
2390:
2380:
2378:
2374:
2369:
2367:
2361:
2359:
2355:
2350:
2349:
2344:
2339:
2337:
2333:
2328:
2316:
2312:
2309:
2307:
2304:
2303:
2297:
2293:
2290:
2288:
2286:
2281:
2280:
2274:
2270:
2267:
2265:
2262:
2261:
2255:
2251:
2248:
2246:
2243:
2242:
2236:
2232:
2229:
2227:
2224:
2223:
2217:
2213:
2210:
2208:
2205:
2204:
2198:
2194:
2191:
2189:
2186:
2185:
2179:
2175:
2172:
2170:
2167:
2166:
2160:
2153:
2149:
2146:
2144:
2138:
2137:
2131:
2127:
2124:
2122:
2119:
2118:
2112:
2108:
2104:
2101:
2099:
2096:
2095:
2089:
2085:
2082:
2080:
2077:
2076:
2070:
2066:
2063:
2061:
2058:
2057:
2051:
2047:
2044:
2042:
2039:
2038:
2032:
2028:
2025:
2023:
2020:
2019:
2016:
2012:
2009:
2007:
2004:
2001:
2000:
1996:
1990:
1988:
1984:
1978:
1976:
1975:
1969:
1965:
1959:
1949:
1947:
1936:
1934:
1930:
1926:
1921:
1919:
1915:
1911:
1907:
1906:
1900:
1897:
1883:
1876:
1874:
1857:
1853:intermembrane
1843:
1840:
1827:
1818:
1815:
1792:
1784:
1770:
1767:
1757:
1749:
1726:
1718:
1717:
1714:
1711:
1706:
1704:
1700:
1696:
1692:
1688:
1681:
1676:
1671:
1661:
1653:
1639:
1636:
1620:
1607:
1600:
1598:
1581:
1577:intermembrane
1567:
1564:
1541:
1538:
1525:
1511:
1508:
1485:
1482:
1472:
1459:
1458:
1455:
1453:
1449:
1444:
1442:
1438:
1430:
1426:
1422:
1418:
1414:
1409:cytochrome bc
1406:
1402:
1395:
1390:
1381:
1379:
1375:
1371:
1367:
1363:
1354:
1347:
1345:
1325:
1317:
1293:
1270:
1269:
1266:
1264:
1260:
1256:
1252:
1243:
1241:
1237:
1236:
1231:
1227:
1223:
1222:
1218:
1209:
1202:
1200:
1180:
1172:
1158:
1145:
1144:
1141:
1139:
1135:
1131:
1127:
1123:
1119:
1115:
1108:
1103:
1094:
1088:
1083:
1078:
1072:
1068:
1059:
1052:
1050:
1033:
1029:intermembrane
1019:
1016:
1008:
1000:
995:
982:
968:
965:
959:
948:
947:
944:
942:
938:
934:
930:
925:
923:
919:
915:
911:
907:
903:
899:
892:
889:Complex I or
887:
870:
866:
859:
855:
851:
842:
839:
835:
831:
822:
819:
817:
813:
809:
800:
797:
791:Cytochrome bc
790:
789:
785:
776:
773:
771:
766:Cytochrome bc
764:
763:
759:
749:
745:
742:
740:
737:
736:
732:
730:
726:
723:
721:
718:
717:
714:
710:
707:
705:
702:
699:
698:
692:
690:
689:hydrogenosome
686:
685:
679:
675:
671:
663:
658:
656:
652:
648:
644:
641:
637:
633:
629:
625:
619:
615:
605:
599:
595:
591:
587:
583:
579:
574:
572:
564:
552:
551:
546:
542:
538:
534:
530:
525:
523:
519:
515:
511:
507:
494:
490:
487:Reduction of
485:
480:
476:
466:
464:
460:
456:
452:
448:
443:
437:
436:
432:
431:fusobacterium
428:
424:
420:
415:
411:
409:
405:
401:
397:
393:
392:
386:
382:
378:
374:
370:
366:
362:
356:
355:Bioenergetics
352:
342:
340:
336:
332:
328:
324:
323:free radicals
320:
316:
312:
307:
305:
301:
297:
293:
289:
285:
281:
277:
273:
272:
267:
263:
258:
256:
252:
248:
244:
239:
237:
233:
229:
221:
217:
214:
210:
206:
201:
199:
195:
191:
188:. Almost all
187:
183:
179:
175:
172:
168:
164:
160:
156:
152:
146:
96:
51:
44:
41:to power the
36:
32:
28:
23:
19:
10889:
10844:Grey nodes:
10832:Blue nodes:
10820:Pink nodes:
10738:cannabinoids
10576:Chlorophylls
10527:Antioxidants
10280:Serine group
10191:Photosystems
10177:P-glycerates
10124:P-glycerates
10089:sialic acids
10084:Amino sugars
10036:
10024:
10023:
10011:
10010:
9998:
9997:
9979:
9978:
9964:
9963:
9951:
9950:
9938:
9937:
9925:
9924:
9912:
9911:
9899:
9898:
9884:
9883:
9869:
9868:
9855:
9854:
9853:
9841:
9840:
9826:
9825:
9809:
9808:
9794:
9793:
9779:
9778:
9769:Fermentation
9766:
9765:
9749:
9748:
9734:
9733:
9719:
9718:
9704:
9703:
9689:
9688:
9683:
9672:
9661:
9648:
9633:
9618:
9605:
9592:
9579:
9564:
9551:
9538:
9237:Galactolysis
9207:Glycogenesis
9012:Fermentation
8984:ATP synthase
8974:
8777:
8766:Cytochrome c
8726:
8674:succinyl-CoA
8440:Cytochrome c
8428:ATP synthase
8409:ATP Synthase
8387:
8385:Wiley and Co
8357:
8338:
8319:
8300:
8274:
8252:
8231:
8222:Introductory
8204:. Retrieved
8190:
8145:
8141:
8131:
8100:
8096:
8086:
8075:. Retrieved
8063:
8050:
8033:
8027:
7986:
7982:
7976:
7935:
7931:
7925:
7916:
7910:
7904:
7877:
7873:
7863:
7832:
7828:
7818:
7791:
7787:
7781:
7748:
7744:
7738:
7719:
7715:
7705:
7672:
7668:
7662:
7627:
7617:
7592:
7588:
7582:
7547:
7543:
7533:
7498:
7492:
7467:
7464:Biochemistry
7463:
7456:
7430:Biochemistry
7429:
7395:
7391:
7345:
7341:
7331:
7286:
7282:
7272:
7231:
7227:
7221:
7196:
7192:
7186:
7145:
7141:
7135:
7110:
7106:
7100:
7078:(1): 44–84.
7075:
7071:
7065:
7054:. Retrieved
7047:the original
7008:
7004:
6991:
6966:
6962:
6920:
6916:
6910:
6875:
6871:
6825:
6821:
6815:
6784:
6780:
6732:
6729:EMBO Reports
6728:
6718:
6693:
6689:
6683:
6648:
6644:
6634:
6599:
6595:
6585:
6550:
6546:
6536:
6507:
6503:
6493:
6458:
6454:
6444:
6411:
6407:
6401:
6360:
6357:FEBS Letters
6356:
6350:
6325:
6321:
6270:
6266:
6253:
6220:
6216:
6210:
6183:
6179:
6169:
6126:
6122:
6112:
6093:
6089:
6079:
6052:
6048:
6038:
6011:
6007:
5959:
5955:
5907:
5903:
5893:
5855:(1): 73–93.
5852:
5848:
5842:
5797:
5793:
5783:
5752:
5748:
5738:
5711:
5707:
5697:
5662:
5658:
5648:
5613:
5609:
5599:
5556:
5553:FEBS Letters
5552:
5542:
5497:
5493:
5483:
5458:
5454:
5448:
5423:
5419:
5413:
5388:
5384:
5378:
5351:
5347:
5337:
5310:
5306:
5296:
5261:
5257:
5247:
5222:
5218:
5212:
5185:
5181:
5171:
5130:
5126:
5120:
5095:
5091:
5085:
5045:(1): 39–46.
5042:
5039:FEBS Letters
5038:
5028:
4994:
4990:
4977:
4944:
4940:
4934:
4909:
4905:
4899:
4857:
4853:
4840:
4805:
4801:
4791:
4757:
4753:
4740:
4709:
4705:
4695:
4650:
4646:
4636:
4601:
4597:
4587:
4546:
4542:
4536:
4509:
4505:
4495:
4470:
4466:
4460:
4419:
4415:
4409:
4384:
4380:
4374:
4349:
4345:
4339:
4312:
4308:
4298:
4273:
4269:
4263:
4222:
4218:
4212:
4169:
4165:
4125:
4121:
4083:
4079:
4005:
4001:
3995:
3970:
3966:
3960:
3919:
3915:
3909:
3884:
3880:
3874:
3841:Microbiology
3840:
3827:
3794:
3790:
3784:
3751:
3747:
3741:
3698:
3695:FEBS Letters
3694:
3684:
3657:
3653:
3643:
3608:
3598:
3573:
3569:
3563:
3538:
3534:
3525:Schultz BE,
3484:
3480:
3426:
3422:
3416:
3407:
3395:. Retrieved
3386:
3377:
3350:
3331:Respirometry
3287:
3271:
3252:fermentation
3245:
3211:
3191:Complex III
3166:oxaloacetate
3028:
3024:piericidin A
3012:
2995:
2974:
2938:
2925:
2898:
2887:
2735:
2716:
2713:
2710:
2707:
2700:
2689:
2678:
2650:
2648:
2638:
2636:
2631:ATP synthase
2601:
2577:
2562:
2551:
2394:
2392:
2389:ATP synthase
2376:
2370:
2362:
2346:
2340:
2326:
2324:
2284:
2014:
1994:
1982:
1979:
1972:
1961:
1946:respirasomes
1942:
1922:
1903:
1901:
1893:
1877:
1707:
1690:
1685:
1678:Complex IV:
1654:
1638:free radical
1612:
1601:
1452:cytochrome c
1445:
1416:
1415:, or simply
1408:
1404:
1399:
1359:
1348:
1254:
1249:
1233:
1221:Ascaris suum
1219:
1214:
1203:
1121:
1117:
1112:
1105:Complex II:
1079:
1064:
1053:
936:
933:coenzyme Q10
926:
918:cell nucleus
905:
901:
896:
841:Cytochrome a
821:Cytochrome c
799:Cytochrome b
775:Coenzyme Q10
712:
682:
659:
621:
618:Chemiosmosis
575:
562:
548:
541:benzoquinone
533:coenzyme Q10
526:
510:cytochrome c
502:
444:
433:
427:chloroplasts
416:
412:
391:chemiosmosis
389:
364:
358:
351:Chemiosmosis
345:Chemiosmosis
308:
292:chemiosmosis
288:ATP synthase
269:
259:
240:
202:
194:fermentation
186:mitochondria
154:
150:
49:
48:
43:ATP synthase
18:
10868:metabolism.
10852:metabolism.
10808:Red nodes:
10745:Eicosanoids
10710:fatty acids
10639:Cholesterol
10627:carotenoids
10606:Polyketides
10541:Nucleotides
10496:tocopherols
10460:Calciferols
10373:Amino acids
10366:amino acids
10296:amino acids
10271:Respiratory
10014:Proteolysis
10001:Translation
9988:replication
9954:MEP pathway
9941:MVA pathway
9902:Lipogenesis
9874:deamination
9664:Peroxisomal
9556:respiration
9363:lipogenesis
9225:Fructolysis
9039:Lactic acid
8783:synthesis:
8600:Anaplerotic
7788:Adv Enzymol
6328:: 717–749.
6223:(1): 9–15.
5426:: 703–734.
4912:: 689–733.
3887:: 247–281.
3660:(1): 1–56.
3299:Nobel prize
3279: [
3226:hibernating
3182:Antimycin A
3173:Complex II
3078:Complex IV
3032:antimycin A
2965:peroxidases
2941:antioxidant
2732:Antioxidant
2580:kilodaltons
2569:equilibrium
2348:Nitrobacter
2159:-amino acid
2041:Hydrogenase
2027:Bicarbonate
1701:and one of
1437:cytochromes
1425:cytochromes
1417:complex III
1370:amino acids
1366:fatty acids
914:kilodaltons
857:Complex IV
837:Complex IV
580:cofactors,
571:menaquinone
545:hydrophobic
423:alkaliphile
264:across the
251:prokaryotes
228:produce ATP
10912:Metabolism
10896:Categories
10862:nucleotide
10646:Bile acids
10622:Terpenoids
10453:vitamin Bs
10436:Cobalamins
10364:glucogenic
10352:polyamines
10312:Homoserine
10184:Glyoxylate
10077:Inositol-P
9872:Amino acid
9812:feeders to
9640:elongation
9595:Glyoxylate
9584:acid cycle
9510:Metabolism
9186:Glycolysis
9119:metabolism
9117:Nucleotide
9107:Urea cycle
9088:Amino acid
9077:Catabolism
9020:Glycolysis
8963:Glycolysis
8946:metabolism
8906:catabolism
8902:Metabolism
8778:Coenzyme Q
8752:Coenzyme Q
8610:acetyl-CoA
8526:Metabolism
8206:2007-07-21
8077:2007-07-21
7919:: 516–534.
7912:Biokhimiya
7794:: 99–162.
7439:8187134801
7056:2017-10-27
5307:IUBMB Life
4387:: 77–109.
4315:(1): 1–9.
3370:References
3153:Complex I
3132:Ionophores
3104:Complex V
3100:Antibiotic
3095:Oligomycin
3044:Compounds
3007:oligomycin
2992:Inhibitors
2929:coenzyme Q
2819:Superoxide
2743:superoxide
2608:ionization
2343:nitrifying
2006:Redox pair
1899:membrane.
1691:complex IV
1435:and two b
1429:cytochrome
1378:acetyl-CoA
1240:pyrimidine
1118:complex II
937:ubiquinone
816:Complex IV
704:Redox pair
662:eukaryotes
628:glycolysis
590:amino acid
563:ubiquinone
557:); when QH
504:the water-
493:ubiquinone
489:coenzyme Q
463:fatty acid
451:Glycolysis
385:endergonic
335:senescence
315:superoxide
243:eukaryotes
182:eukaryotes
180:(ATP). In
10631:vitamin A
10615:backbones
10613:Terpenoid
10508:Cofactors
10500:vitamin E
10490:vitamin K
10478:vitamin A
10473:Retinoids
10465:vitamin D
10440:vitamin B
10405:vitamin C
10400:Ascorbate
10393:histidine
10380:Shikimate
10359:Ketogenic
10326:Glutamate
10303:Aspartate
10255:glutarate
10170:Pentose-P
10154:Succinate
10145:Propionyl
10138:Tetrose-P
10131:Pentose-P
10027:Glycosyl-
9967:Shikimate
9915:Lipolysis
9857:Oxidative
9814:gluconeo-
9756:oxylation
9737:Gluconeo-
9694:genolysis
9677:oxidation
9653:oxidation
9625:synthesis
9569:phosphate
9359:lipolysis
9166:anabolism
8910:anabolism
8552:Aconitase
7013:CiteSeerX
5225:: 23–39.
4862:CiteSeerX
3541:: 23–65.
3285:in 1939.
3204:ubiquinol
3200:oxidation
3187:Piscicide
3149:Pesticide
3136:uncouples
3111:subunit.
2953:vitamin E
2949:vitamin C
2909:mutations
2862:−
2836:−
2813:_
2810:∙
2788:−
2484:⇀
2477:−
2466:−
2459:↽
2399:phosphate
2395:complex V
2354:anabolism
2315:Succinate
2197:Succinate
2174:Gluconate
2148:2-oxoacid
1790:⟶
1699:magnesium
1697:, one of
1531:⟶
1448:ubiquinol
1300:⟶
1232:parasite
1165:⟶
1155:Succinate
1134:succinate
1087:ubiquinol
988:⟶
906:complex I
655:Succinate
624:catabolic
550:ubiquinol
491:from its
381:exergonic
174:nutrients
161:in which
10850:cofactor
10729:Steroids
10669:Acyl-CoA
10653:Glycero-
10557:Proteins
10520:minerals
10515:Vitamins
10486:Quinones
10422:pigments
10347:Creatine
10340:Arginine
10244:Succinyl
10198:Pyruvate
10117:Glycerol
10110:Triose-P
10103:Hexose-P
10055:multiple
9752:Pyruvate
9543:fixation
9282:Nonhuman
9267:O-linked
9262:N-linked
9054:Specific
8584:Fumarase
8455:Archived
8443:Archived
8431:Archived
8396:Archived
8293:Advanced
8273:(2006).
8200:Archived
8123:Archived
8119:13738472
8068:Archived
8042:55202414
8011:13771349
7960:13111237
7880:: 1–37.
7855:Archived
7851:18116985
7773:26999163
7697:18598358
7689:16283557
7654:10620491
7609:14249115
7574:26620903
7566:15262965
7448:71209231
7374:30713504
7348:: 1941.
7323:10890886
7256:11780125
7213:19409964
7170:11089981
7127:11050436
7092:16978905
7043:11125090
7035:17090411
6969:: 1–31.
6945:24887884
6937:15168615
6850:23763996
6807:Archived
6759:16607397
6710:11893513
6675:30953216
6667:11080505
6626:10836500
6577:14633978
6528:Archived
6524:10600677
6428:11533724
6393:35989618
6301:Archived
6245:39165641
6161:16517654
6071:11803023
5934:10477309
5885:12289639
5877:11536899
5775:Archived
5771:11483615
5730:12206908
5689:10775262
5640:18123642
5632:17322303
5591:46138989
5583:11741580
5534:10393984
5440:15012279
5329:15370881
5239:15725055
5204:16904626
5163:20860573
5077:13942619
5069:12788490
5017:Archived
4926:14977419
4888:Archived
4884:10966481
4832:16055629
4780:Archived
4732:Archived
4687:17050691
4571:17330044
4528:11803022
4487:15078221
4452:29222766
4444:12560550
4401:14527321
4366:20025615
4331:14741580
4290:17157874
4247:20505720
4196:16469879
4144:16828051
4100:15916556
4030:15744302
3987:15582386
3944:10573417
3901:15952888
3863:Archived
3859:10463148
3776:28013583
3768:11771674
3590:14641005
3555:11340051
3529:(2001).
3505:Archived
3501:10600673
3397:28 April
3391:Archived
3325:See also
3170:Poisons
3162:Malonate
3144:Rotenone
3075:Poisons
3016:rotenone
2961:catalase
2947:such as
2945:vitamins
2905:hydroxyl
2868:Peroxide
2826:→
2778:→
2747:peroxide
2698:O + NAD
2644:affinity
2373:isozymes
2311:Fumarate
2193:Fumarate
2126:Pyruvate
2111:Pyruvate
2050:Hydrogen
2015:(Volts)
1964:bacteria
1918:protists
1169:Fumarate
1138:fumarate
920:and the
795:complex
713:(Volts)
640:coenzyme
594:cysteine
553:form (QH
475:Coenzyme
375:such as
313:such as
304:rotation
10866:protein
10846:vitamin
10548:Nucleic
10451:Various
10333:proline
10287:Alanine
10253:α-Keto-
10230:acetate
10221:Citrate
10205:Lactate
10061:glycans
9969:pathway
9889:shuttle
9887:Citrate
9816:genesis
9799:genesis
9754:decarb-
9739:genesis
9709:genesis
9571:pathway
9567:Pentose
9422:Ketosis
9034:Ethanol
8917:General
8756:(CoQ10)
8735:Primary
8591:and ETC
8533:enzymes
8182:4517936
8150:Bibcode
8019:1784050
7991:Bibcode
7968:4153659
7940:Bibcode
7896:1883194
7765:4279328
7645:1220743
7484:8380331
7412:1831660
7365:6346031
7291:Bibcode
7264:4349744
7236:Bibcode
7178:2502238
7150:Bibcode
6983:8660387
6902:8169202
6842:7832594
6803:6214554
6750:1456893
6617:1692760
6485:9620972
6436:3926411
6385:8603713
6365:Bibcode
6342:9242922
6297:8491720
6237:8412675
6202:2856189
6152:1393235
6131:Bibcode
6030:9230919
5986:6387427
5857:Bibcode
5834:6326133
5802:Bibcode
5561:Bibcode
5502:Bibcode
5475:9426242
5405:1883834
5370:9698817
5280:8798503
5155:8638158
5135:Bibcode
5127:Science
5112:7940677
5047:Bibcode
5013:2164001
4969:9651245
4949:Bibcode
4941:Science
4823:1197437
4728:6401712
4678:1637562
4655:Bibcode
4628:3593226
4619:1147643
4579:4421676
4551:Bibcode
4424:Bibcode
4416:Science
4255:4372778
4227:Bibcode
4204:1892332
4174:Bibcode
4166:Science
4038:4401178
4010:Bibcode
3952:4431405
3924:Bibcode
3799:Bibcode
3791:Science
3733:7685958
3725:6317447
3703:Bibcode
3676:3881803
3635:7654171
3626:1135905
3527:Chan SI
3459:4149605
3451:4291593
3431:Bibcode
3232:History
3059:Cyanide
2917:disease
2377:E. coli
2327:E. coli
2254:Ammonia
2250:Nitrite
2235:Nitrite
2231:Nitrate
2178:Glucose
2152:ammonia
2130:Lactate
2103:Acetate
2088:Gly-3-P
2031:Formate
2013:
1995:E. coli
1983:E. coli
1968:archaea
1619:Q cycle
1413:complex
1374:choline
1230:malaria
941:quinone
770:complex
754:or FADH
711:
670:protons
506:soluble
455:glucose
365:coupled
339:inhibit
327:disease
262:protons
205:glucose
171:oxidize
167:enzymes
157:is the
29:in the
10767:Major
10720:&
10685:Glyco-
10625:&
10597:Acetyl
10567:&
10518:&
10494:&
10391:&
10350:&
10331:&
10319:lysine
10317:&
10264:bodies
10262:Ketone
10237:Malate
10228:Oxalo-
10212:Acetyl
10161:Acetyl
10087:&
10070:sugars
10068:Simple
10059:&
10057:sugars
10051:Double
9722:Glyco-
9707:Glyco-
9692:Glyco-
9582:Citric
9554:Photo-
9541:Carbon
8944:Energy
8825:COQ10B
8820:COQ10A
8364:
8345:
8326:
8307:
8281:
8271:Lane N
8259:
8240:
8180:
8173:427120
8170:
8117:
8040:
8017:
8009:
7983:Nature
7966:
7958:
7932:Nature
7894:
7849:
7806:
7771:
7763:
7695:
7687:
7652:
7642:
7607:
7572:
7564:
7525:156853
7523:
7513:
7482:
7446:
7436:
7410:
7372:
7362:
7321:
7311:
7262:
7254:
7228:Nature
7211:
7176:
7168:
7142:Nature
7125:
7090:
7041:
7033:
7015:
6981:
6943:
6935:
6900:
6893:205391
6890:
6848:
6840:
6801:
6757:
6747:
6708:
6673:
6665:
6624:
6614:
6575:
6568:291849
6565:
6522:
6483:
6476:107823
6473:
6434:
6426:
6391:
6383:
6340:
6295:
6288:204621
6285:
6243:
6235:
6200:
6159:
6149:
6069:
6028:
5984:
5977:373010
5974:
5932:
5925:103747
5922:
5883:
5875:
5832:
5825:345118
5822:
5769:
5728:
5687:
5680:302020
5677:
5638:
5630:
5589:
5581:
5532:
5522:
5473:
5438:
5403:
5368:
5327:
5288:893754
5286:
5278:
5237:
5202:
5161:
5153:
5110:
5075:
5067:
5011:
4967:
4924:
4882:
4864:
4830:
4820:
4776:925004
4774:
4726:
4685:
4675:
4626:
4616:
4577:
4569:
4543:Nature
4526:
4485:
4450:
4442:
4399:
4364:
4329:
4288:
4253:
4245:
4219:Nature
4202:
4194:
4142:
4098:
4036:
4028:
4002:Nature
3985:
3950:
3942:
3916:Nature
3899:
3857:
3819:388618
3817:
3774:
3766:
3731:
3723:
3674:
3633:
3623:
3588:
3553:
3499:
3457:
3449:
3423:Nature
3034:, and
3020:amytal
3002:toxins
2977:oxygen
2963:, and
2899:These
2620:stator
2527:matrix
2319:+0.03
2300:+0.13
2277:+0.16
2258:+0.36
2239:+0.42
2220:+0.82
2212:Oxygen
2201:+0.03
2182:−0.14
2134:−0.19
2092:−0.19
2073:−0.32
2054:−0.42
2046:Proton
2035:−0.43
1910:plants
1896:plants
1780:matrix
1695:copper
1521:matrix
1427:: one
1263:flavin
978:matrix
910:enzyme
867:+0.82
852:+0.29
832:+0.22
810:+0.12
786:+0.06
760:−0.20
750:/ FMNH
733:−0.32
634:, and
630:, the
604:10 m.
586:sulfur
578:flavin
377:oxygen
361:energy
10917:Redox
10736:Endo-
10701:Waxes
10678:acids
10676:Fatty
10550:acids
10429:Hemes
10413:δ-ALA
10389:acids
10361:&
10328:group
10314:group
10305:group
10273:chain
10044:acids
10042:Sugar
10029:ation
9797:Keto-
9784:lysis
9782:Keto-
9724:lysis
9636:Fatty
9621:Fatty
9610:cycle
9597:cycle
9440:Other
9400:Other
9175:Human
9056:paths
8844:Other
8835:PDSS2
8830:PDSS1
8540:Cycle
8071:(PDF)
8060:(PDF)
8015:S2CID
7964:S2CID
7769:S2CID
7693:S2CID
7570:S2CID
7314:27006
7260:S2CID
7174:S2CID
7050:(PDF)
7039:S2CID
7001:(PDF)
6941:S2CID
6846:S2CID
6671:S2CID
6432:S2CID
6389:S2CID
6304:(PDF)
6263:(PDF)
6241:S2CID
5881:S2CID
5636:S2CID
5587:S2CID
5525:22224
5284:S2CID
5159:S2CID
5073:S2CID
5020:(PDF)
4987:(PDF)
4891:(PDF)
4850:(PDF)
4783:(PDF)
4750:(PDF)
4575:S2CID
4448:S2CID
4251:S2CID
4200:S2CID
4034:S2CID
3948:S2CID
3866:(PDF)
3837:(PDF)
3772:S2CID
3729:S2CID
3455:S2CID
3342:Notes
3283:]
3067:Azide
2998:drugs
2921:aging
2690:1/2 O
2563:This
2216:Water
1914:fungi
1421:dimer
864:/ HO
622:Many
537:redox
529:lipid
461:of a
331:aging
163:cells
149:) or
99:, US
10864:and
10848:and
10599:-CoA
10534:PRPP
10420:Bile
10246:-CoA
10214:-CoA
10163:-CoA
10147:-CoA
9675:beta
9651:Beta
9638:acid
9623:acid
9608:Urea
9164:and
8815:COQ9
8810:COQ7
8805:COQ6
8800:COQ5
8795:COQ4
8790:COQ3
8785:COQ2
8641:and
8578:SDHA
8362:ISBN
8343:ISBN
8324:ISBN
8305:ISBN
8279:ISBN
8257:ISBN
8238:ISBN
8178:PMID
8115:PMID
8038:OCLC
8007:PMID
7956:PMID
7892:PMID
7847:PMID
7804:ISBN
7761:PMID
7685:PMID
7650:PMID
7605:PMID
7562:PMID
7521:PMID
7511:ISBN
7480:PMID
7444:OCLC
7434:ISBN
7408:PMID
7396:1067
7370:PMID
7319:PMID
7252:PMID
7209:PMID
7197:1800
7166:PMID
7123:PMID
7088:PMID
7031:PMID
6979:PMID
6933:PMID
6898:PMID
6838:PMID
6799:PMID
6755:PMID
6706:PMID
6663:PMID
6622:PMID
6573:PMID
6520:PMID
6481:PMID
6424:PMID
6381:PMID
6338:PMID
6293:PMID
6233:PMID
6198:PMID
6157:PMID
6067:PMID
6053:1553
6026:PMID
6012:1320
5982:PMID
5930:PMID
5873:PMID
5830:PMID
5767:PMID
5726:PMID
5712:1555
5685:PMID
5628:PMID
5579:PMID
5530:PMID
5471:PMID
5455:Gene
5436:PMID
5401:PMID
5389:1059
5366:PMID
5325:PMID
5276:PMID
5235:PMID
5200:PMID
5186:1757
5151:PMID
5108:PMID
5065:PMID
5009:PMID
4965:PMID
4922:PMID
4880:PMID
4828:PMID
4772:PMID
4724:PMID
4683:PMID
4624:PMID
4567:PMID
4524:PMID
4510:1553
4483:PMID
4440:PMID
4397:PMID
4362:PMID
4327:PMID
4313:1608
4286:PMID
4243:PMID
4192:PMID
4140:PMID
4126:1757
4096:PMID
4026:PMID
3983:PMID
3940:PMID
3897:PMID
3855:PMID
3815:PMID
3764:PMID
3721:PMID
3672:PMID
3631:PMID
3586:PMID
3551:PMID
3497:PMID
3447:PMID
3399:2018
3240:and
3224:for
3164:and
3117:CCCP
3047:Use
3000:and
2951:and
2730:and
2616:axle
2334:and
2292:TMAO
2269:DMSO
2084:DHAP
2069:NADH
1966:and
1703:zinc
1441:heme
1372:and
1130:heme
957:NADH
929:NADH
729:NADH
643:NADH
616:and
518:heme
514:iron
477:and
369:NADH
353:and
333:and
317:and
220:FADH
218:and
216:NADH
165:use
52:(UK
31:cell
10771:in
10590:MVA
10583:MEP
9512:map
9188:⇄
9029:ABE
8691:to
8672:to
8653:to
8608:to
8422:PDB
8168:PMC
8158:doi
8105:doi
8101:235
7999:doi
7987:191
7948:doi
7936:172
7882:doi
7837:doi
7833:178
7796:doi
7753:doi
7724:doi
7677:doi
7640:PMC
7632:doi
7597:doi
7552:doi
7548:279
7503:doi
7472:doi
7400:doi
7360:PMC
7350:doi
7309:PMC
7299:doi
7244:doi
7232:415
7201:doi
7158:doi
7146:408
7115:doi
7080:doi
7023:doi
6971:doi
6925:doi
6888:PMC
6880:doi
6876:176
6830:doi
6789:doi
6785:257
6745:PMC
6737:doi
6698:doi
6653:doi
6649:276
6612:PMC
6604:doi
6600:355
6563:PMC
6555:doi
6512:doi
6508:203
6471:PMC
6463:doi
6459:180
6416:doi
6373:doi
6361:379
6330:doi
6283:PMC
6275:doi
6271:175
6225:doi
6188:doi
6147:PMC
6139:doi
6098:doi
6057:doi
6016:doi
5972:PMC
5964:doi
5920:PMC
5912:doi
5865:doi
5820:PMC
5810:doi
5757:doi
5753:276
5716:doi
5675:PMC
5667:doi
5618:doi
5614:282
5569:doi
5557:509
5520:PMC
5510:doi
5463:doi
5459:203
5428:doi
5393:doi
5356:doi
5315:doi
5266:doi
5262:271
5227:doi
5190:doi
5143:doi
5131:272
5100:doi
5055:doi
5043:545
4999:doi
4995:265
4957:doi
4945:281
4914:doi
4872:doi
4818:PMC
4810:doi
4762:doi
4758:252
4714:doi
4710:258
4673:PMC
4663:doi
4651:103
4614:PMC
4606:doi
4602:241
4559:doi
4547:446
4514:doi
4475:doi
4432:doi
4420:299
4389:doi
4354:doi
4350:425
4317:doi
4278:doi
4274:366
4235:doi
4223:465
4182:doi
4170:311
4130:doi
4088:doi
4018:doi
4006:434
3975:doi
3932:doi
3920:402
3889:doi
3845:doi
3807:doi
3795:206
3756:doi
3711:doi
3699:164
3662:doi
3621:PMC
3613:doi
3578:doi
3543:doi
3489:doi
3485:203
3439:doi
3427:213
3202:of
2975:As
2913:DNA
2911:in
2745:or
2683:/ H
2493:ATP
2419:ADP
2296:TMA
2273:DMS
2065:NAD
1925:ATP
1797:Cyt
1740:red
1731:Cyt
1555:red
1546:Cyt
1490:Cyt
1364:of
1304:ETF
1284:red
1280:ETF
1136:to
1120:or
1093:).
1089:(QH
992:NAD
935:or
931:by
904:or
848:red
828:red
806:red
782:red
748:FAD
746:or
744:FMN
725:NAD
660:In
371:to
241:In
226:to
169:to
153:or
10898::
10633:)
10502:)
10480:)
10467:)
10445:)
10442:12
10407:)
9361:,
9205:⇄
9022:→
8982:+
8973:→
8969:→
8965:→
8908:,
8904:,
8780:10
8754:10
8630:E3
8628:,
8626:E2
8624:,
8622:E1
8528::
8469::
8176:.
8166:.
8156:.
8146:70
8144:.
8140:.
8121:.
8113:.
8099:.
8095:.
8062:.
8036:.
8013:.
8005:.
7997:.
7985:.
7962:.
7954:.
7946:.
7934:.
7915:.
7890:.
7878:60
7876:.
7872:.
7853:.
7845:.
7831:.
7827:.
7802:.
7790:.
7767:.
7759:.
7747:.
7718:.
7714:.
7691:.
7683:.
7673:25
7671:.
7648:.
7638:.
7626:.
7603:.
7593:92
7591:.
7568:.
7560:.
7546:.
7542:.
7519:.
7509:.
7478:.
7468:32
7466:.
7442:.
7420:^
7406:.
7394:.
7382:^
7368:.
7358:.
7344:.
7340:.
7317:.
7307:.
7297:.
7287:97
7285:.
7281:.
7258:.
7250:.
7242:.
7230:.
7207:.
7195:.
7172:.
7164:.
7156:.
7144:.
7121:.
7111:25
7109:.
7086:.
7076:39
7074:.
7037:.
7029:.
7021:.
7009:40
7007:.
7003:.
6977:.
6967:61
6965:.
6953:^
6939:.
6931:.
6921:36
6919:.
6896:.
6886:.
6874:.
6870:.
6858:^
6844:.
6836:.
6826:65
6824:.
6805:.
6797:.
6783:.
6779:.
6767:^
6753:.
6743:.
6731:.
6727:.
6704:.
6694:27
6692:.
6669:.
6661:.
6647:.
6643:.
6620:.
6610:.
6598:.
6594:.
6571:.
6561:.
6551:22
6549:.
6545:.
6526:.
6518:.
6506:.
6502:.
6479:.
6469:.
6457:.
6453:.
6430:.
6422:.
6410:.
6387:.
6379:.
6371:.
6359:.
6336:.
6326:66
6324:.
6312:^
6299:.
6291:.
6281:.
6269:.
6265:.
6239:.
6231:.
6219:.
6196:.
6184:54
6182:.
6178:.
6155:.
6145:.
6137:.
6127:72
6125:.
6121:.
6094:66
6092:.
6088:.
6065:.
6051:.
6047:.
6024:.
6010:.
6006:.
5994:^
5980:.
5970:.
5960:48
5958:.
5954:.
5942:^
5928:.
5918:.
5908:63
5906:.
5902:.
5879:.
5871:.
5863:.
5853:29
5851:.
5828:.
5818:.
5808:.
5798:81
5796:.
5792:.
5773:.
5765:.
5751:.
5747:.
5724:.
5710:.
5706:.
5683:.
5673:.
5663:19
5661:.
5657:.
5634:.
5626:.
5612:.
5608:.
5585:.
5577:.
5567:.
5555:.
5551:.
5528:.
5518:.
5508:.
5498:96
5496:.
5492:.
5469:.
5457:.
5434:.
5424:48
5422:.
5399:.
5387:.
5364:.
5352:31
5350:.
5346:.
5323:.
5311:56
5309:.
5305:.
5282:.
5274:.
5260:.
5256:.
5233:.
5223:55
5221:.
5198:.
5184:.
5180:.
5157:.
5149:.
5141:.
5129:.
5106:.
5096:19
5094:.
5071:.
5063:.
5053:.
5041:.
5037:.
5015:.
5007:.
4993:.
4989:.
4963:.
4955:.
4943:.
4920:.
4910:66
4908:.
4886:.
4878:.
4870:.
4858:69
4856:.
4852:.
4826:.
4816:.
4806:17
4804:.
4800:.
4778:.
4770:.
4756:.
4752:.
4730:.
4722:.
4708:.
4704:.
4681:.
4671:.
4661:.
4649:.
4645:.
4622:.
4612:.
4600:.
4596:.
4573:.
4565:.
4557:.
4545:.
4522:.
4508:.
4504:.
4481:.
4469:.
4446:.
4438:.
4430:.
4418:.
4395:.
4385:72
4383:.
4360:.
4348:.
4325:.
4311:.
4307:.
4284:.
4272:.
4249:.
4241:.
4233:.
4221:.
4198:.
4190:.
4180:.
4168:.
4164:.
4152:^
4138:.
4124:.
4120:.
4108:^
4094:.
4084:33
4082:.
4070:^
4046:^
4032:.
4024:.
4016:.
4004:.
3981:.
3971:14
3969:.
3946:.
3938:.
3930:.
3918:.
3895:.
3885:74
3883:.
3861:.
3853:.
3839:.
3813:.
3805:.
3793:.
3770:.
3762:.
3752:20
3750:.
3727:.
3719:.
3709:.
3697:.
3693:.
3670:.
3658:45
3656:.
3652:.
3629:.
3619:.
3607:.
3584:.
3574:31
3572:.
3549:.
3539:30
3537:.
3533:.
3513:^
3503:.
3495:.
3483:.
3479:.
3467:^
3453:.
3445:.
3437:.
3425:.
3389:.
3385:.
3281:uk
3206:.
3087:Cu
3083:Fe
2959:,
2923:.
2705:.
2401:(P
2313:/
2294:/
2271:/
2252:/
2233:/
2214:/
2195:/
2176:/
2163:?
2154:/
2150:+
2128:/
2115:?
2109:/
2105:+
2086:/
2067:/
2048:/
2029:/
1997:.
1935:.
1916:,
1806:ox
1705:.
1499:ox
1469:QH
1407:,
1322:QH
1308:ox
1177:QH
1005:QH
844:ox
824:ox
802:ox
778:ox
727:/
691:.
653:.
524:.
499:).
449:.
404:pH
280:pH
200:.
130:eɪ
109:ɑː
10880:.
10836:.
10824:.
10812:.
10800:.
10788:.
10629:(
10498:(
10492:)
10488:(
10476:(
10463:(
10438:(
10403:(
10053:/
9502:e
9495:t
9488:v
9387:)
9383:(
9365:)
9357:(
9168:)
9159:(
8987:)
8977:(
8894:e
8887:t
8880:v
8724:/
8645:)
8632:)
8620:(
8580:)
8576:(
8518:e
8511:t
8504:v
8370:.
8351:.
8332:.
8313:.
8287:.
8265:.
8246:.
8209:.
8184:.
8160::
8152::
8107::
8080:.
8044:.
8021:.
8001::
7993::
7970:.
7950::
7942::
7917:4
7898:.
7884::
7839::
7812:.
7798::
7792:1
7775:.
7755::
7749:5
7732:.
7726::
7720:B
7699:.
7679::
7656:.
7634::
7611:.
7599::
7576:.
7554::
7527:.
7505::
7486:.
7474::
7450:.
7414:.
7402::
7376:.
7352::
7346:9
7325:.
7301::
7293::
7266:.
7246::
7238::
7215:.
7203::
7180:.
7160::
7152::
7129:.
7117::
7094:.
7082::
7059:.
7025::
6985:.
6973::
6947:.
6927::
6904:.
6882::
6852:.
6832::
6791::
6761:.
6739::
6733:7
6712:.
6700::
6677:.
6655::
6628:.
6606::
6579:.
6557::
6514::
6487:.
6465::
6438:.
6418::
6412:2
6395:.
6375::
6367::
6344:.
6332::
6277::
6247:.
6227::
6221:9
6204:.
6190::
6163:.
6141::
6133::
6106:.
6100::
6073:.
6059::
6032:.
6018::
5988:.
5966::
5936:.
5914::
5887:.
5867::
5859::
5836:.
5812::
5804::
5759::
5732:.
5718::
5691:.
5669::
5642:.
5620::
5593:.
5571::
5563::
5536:.
5512::
5504::
5477:.
5465::
5442:.
5430::
5407:.
5395::
5372:.
5358::
5331:.
5317::
5290:.
5268::
5241:.
5229::
5206:.
5192::
5165:.
5145::
5137::
5114:.
5102::
5079:.
5057::
5049::
5001::
4971:.
4959::
4951::
4928:.
4916::
4874::
4834:.
4812::
4764::
4716::
4689:.
4665::
4657::
4630:.
4608::
4581:.
4561::
4553::
4530:.
4516::
4489:.
4477::
4471:5
4454:.
4434::
4426::
4403:.
4391::
4368:.
4356::
4333:.
4319::
4292:.
4280::
4257:.
4237::
4229::
4206:.
4184::
4176::
4146:.
4132::
4102:.
4090::
4040:.
4020::
4012::
3989:.
3977::
3954:.
3934::
3926::
3903:.
3891::
3847::
3821:.
3809::
3801::
3778:.
3758::
3735:.
3713::
3705::
3678:.
3664::
3637:.
3615::
3592:.
3580::
3557:.
3545::
3491::
3461:.
3441::
3433::
3401:.
3109:o
3085:–
2981:2
2892:)
2890:7
2888:(
2857:2
2852:2
2848:O
2832:e
2804:2
2800:O
2784:e
2768:2
2764:O
2703:2
2696:2
2692:2
2685:2
2681:2
2669:o
2667:A
2665:1
2661:o
2659:A
2657:1
2604:O
2597:1
2592:1
2588:1
2584:O
2556:)
2554:6
2552:(
2532:+
2523:H
2518:4
2515:+
2512:O
2504:2
2500:H
2496:+
2448:+
2439:H
2434:4
2431:+
2428:i
2425:P
2422:+
2403:i
2285:N
2157:D
2141:D
1944:"
1882:)
1880:5
1878:(
1858:+
1849:H
1844:4
1841:+
1836:O
1828:2
1824:H
1819:2
1816:+
1802:c
1793:4
1785:+
1776:H
1771:8
1768:+
1758:2
1754:O
1750:+
1736:c
1727:4
1682:.
1658:2
1650:2
1646:2
1642:2
1631:2
1627:2
1623:2
1615:2
1606:)
1604:4
1602:(
1582:+
1573:H
1568:4
1565:+
1551:c
1542:2
1539:+
1534:Q
1526:+
1517:H
1512:2
1509:+
1495:c
1486:2
1483:+
1473:2
1433:1
1431:c
1411:1
1353:)
1351:3
1349:(
1326:2
1318:+
1297:Q
1294:+
1208:)
1206:2
1204:(
1181:2
1173:+
1162:Q
1159:+
1109:.
1091:2
1075:2
1058:)
1056:1
1054:(
1034:+
1025:H
1020:4
1017:+
1009:2
1001:+
996:+
983:+
974:H
969:5
966:+
963:Q
960:+
862:2
860:O
793:1
768:1
756:2
752:2
666:2
602:×
567:2
559:2
555:2
497:2
440:O
224:2
145:/
142:v
139:ɪ
136:t
133:.
127:d
124:ˌ
121:ɪ
118:s
115:.
112:k
106:ˈ
103:/
95:/
92:v
89:ɪ
86:t
83:.
80:ə
77:.
74:d
71:ɪ
68:s
65:ˈ
62:k
59:ɒ
56:/
45:.
39:2
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.