373:
enough material for analysis. Another approach, single cell proteomics by mass spectrometry (SCoPE-MS) has quantified thousands of proteins in mammalian cells with typical cell sizes (diameter of 10-15 μm) by combining carrier-cells and single-cell barcoding. The second generation, SCoPE2, increased the throughput by automated and miniaturized sample preparation; It also improved quantitative reliability and proteome coverage by data-driven optimization of LC-MS/MS and peptide identification. The sensitivity and consistency of these methods have been further improved by prioritization, and massively parallel sample preparation in nanoliter size droplets. Another direction for single-cell protein analysis is based on a scalable framework of multiplexed data-independent acquisition (plexDIA) enables time saving by parallel analysis of both peptide ions and protein samples, thereby realizing multiplicative gains in throughput.
345:. Since different colored quantum dots or unique fluorophores are attached to each antibody it is possible to identify multiple different proteins in a single cell. Quantum dots can be washed off of the antibodies without damaging the sample, making it possible to do multiple rounds of protein quantification using this method on the same sample. For the methods based on organic fluorophores, the fluorescent tags are attached by a reversible linkage such as a DNA-hybrid (that can be melted/dissociated under low-salt conditions) or chemically inactivated, allowing multiple cycles of analysis, with 3-5 targets quantified per cycle. These approaches have been used for quantifying protein abundance in patient biopsy samples (e.g. cancer) to map variable protein expression in tissues and/or tumors, and to measure changes in protein expression and cell signaling in response to cancer treatment.
66:(FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome and transcriptome of single cells, as well as to quantify their proteome and
133:
acoustic. There is a need to explore the insights of SCA in the cell's natural state and development of these techniques is highly essential for that study. Researchers have highlighted the vast potential field that needs to be explored to develop biochip devices to suit market/researcher demands. Hydrodynamic microfluidics facilitates the development of passive lab-on-chip applications. A latest review gives an account of the recent advances in this field, along with their mechanisms, methods and applications.
564:
method utilizing capillary microsampling combined with mass spectrometry with ion mobility separation has been demonstrated to enhance the molecular coverage and ion separation for single cell metabolomics. Researchers are trying to develop a technique that can fulfil what current techniques are lacking: high throughput, higher sensitivity for metabolites that have a lower abundance or that have low ionization efficiencies, good replicability and that allow quantification of metabolites.
161:
uses oil-filled channels to hold separated aqueous droplets. This allows the single cell to be contained and isolated from the inside the oil based channels. Pneumatic membrane valves use the manipulation of air pressure, to isolate individual cells by membrane deflection. The manipulation of the pressure source allows the opening or closing of channels in a microfluidic network. Typically, the system requires an operator and is limited in throughput.
364:
antibodies are then modified to have single stranded DNA connected to them that are complementary. When the two antibodies bind to a protein the complementary strands will anneal and produce a double stranded segment of DNA that can then be amplified using PCR. Each pair of antibodies designed for one protein is tagged with a different DNA sequence. The DNA amplified from PCR can then be sequenced, and the protein levels quantified.
176:
acoustic. There is a need to explore the insights of SCA in the cell's natural state, and development of these techniques is highly essential for that study. Researchers have highlighted the need to develop biochip devices to suit market and researcher demands. Hydrodynamic microfluidics facilitate the development of passive lab-on-chip applications.
238:(MALBAC). Bias in this system is reduced by only copying off the original DNA strand instead of making copies of copies. The main draw backs to using MALBA, is it has reduced accuracy compared to DOP-PCR and MDA due to the enzyme used to copy the DNA. Once amplified using any of the above techniques, the DNA can be sequenced using Sanger or
234:, to accomplish the amplification of larger fragments and greater genome coverage than DOP-PCR. Despite these improvement MDA still has a sequence dependent bias (certain parts of the genome are amplified more than others because of their sequence). The method shown to largely avoid the bias seen in DOP-PCR and MDA is
251:
cell sequencing. The second major application is to study the genetic evolution of cancer. Since cancer cells are constantly mutating it is of great interest to see how cancers evolve at the genetic level. These patterns of somatic mutations and copy number aberration can be observed using single cell sequencing.
210:
The
Adapter Linker PCR WGA is reported in many comparative studies to be best performing for diploid single cell mutation analysis, thanks to its very low Allelic Dropout effect, and for copy number variation profiling due to its low noise, both with aCGH and with NGS low Pass Sequencing. This method
175:
The development of hydrodynamic-based microfluidic biochips has been increasing over the years. In this technique, the cells are trapped in a particular region for single cell analysis (SCA). This usually occurs without any application of external force fields such as optical, electrical, magnetic or
160:
Microfluidics allows for the isolation of individual cells for further analyses. The following principles outline the various microfluidic processes for single-cell separation: droplet-in-oil based isolation, pneumatic membrane valving, and hydrodynamic cell traps. Droplet-in-oil based microfluidics
149:
The Laser
Capture Microdissection technique utilizes a laser to dissect and separate individual cells, or sections, from tissue samples of interest. The methods involve the observation of a cell under a microscope, so that a section for analysis can be identified and labeled so that the laser can cut
141:
Dielectrophoretic digital sorting method utilizes a semiconductor controlled array of electrodes in a microfluidic chip to trap single cells in
Dielectrophoretic (DEP) cages. Cell identification is ensured by the combination of fluorescent markers with image observation. Precision delivery is ensured
132:
The development of hydrodynamic-based microfluidic biochips has been increasing over the years. In this technique, the cells or particles are trapped in a particular region for single cell analysis (SCA) usually without any application of external force fields such as optical, electrical, magnetic or
61:
at the single cell level. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell in a bulk population of cells at a particular condition using optical or electronic
372:
In mass spectroscopy based proteomics there are three major steps needed for peptide identification: sample preparation, separation of peptides, and identification of peptides. Several groups have focused on oocytes or very early cleavage-stage cells since these cells are unusually large and provide
413:
A huge variety of ionization techniques can be used to analyze single cells. The choice of ionization method is crucial for analyte detection. It can be decisive which type of compounds are ionizable and in which state they appear, e.g., charge and possible fragmentation of the ions. A few examples
581:
Single-cell transcriptomic assays have allowed reconstruction development trajectories. Branching of these trajectories describes cell differentiation. Various methods have been developed for reconstructing branching developmental trajectories from single-cell transcriptomic data. They use various
292:
There are three major reasons gene expression has been studied using this technique: to study gene dynamics, RNA splicing, and cell typing. Gene dynamics are usually studied to determine what changes in gene expression affect different cell characteristics. For example, this type of transcriptomic
250:
There are two major applications to studying the genome at the single cell level. One application is to track the changes that occur in bacterial populations, where phenotypic differences are often seen. These differences are missed by bulk sequencing of a population, but can be observed in single
563:
range. Similar to the methods discussed in proteomics, there has also been success in combining mass spectroscopy with separation techniques such as capillary electrophoresis to quantify metabolites. This method is also capable of detecting metabolites present in femtomole concentrations. Another
554:
biosensors, and mass spectroscopy. The first three methods listed use fluorescence microscopy to detect molecules in a cell. Usually these assays use small fluorescent tags attached to molecules of interest, however this has been shown be too invasive for single cell metabolomics, and alters the
363:
Antibody-DNA quantification: another antibody-based method converts protein levels to DNA levels. The conversion to DNA makes it possible to amplify protein levels and use NGS to quantify proteins. In one such approach, two antibodies are selected for each protein needed to be quantified. The two
120:
The
Dielectrophoretic digital sorting method utilizes a semiconductor controlled array of electrodes in a microfluidic chip to trap single cells in Dielectrophoretic (DEP) cages. Cell identification is ensured by the combination of fluorescent markers with image observation. Precision delivery is
22:
532:
The purpose of studying the proteome is to better understand the activity of cells at the single cells level. Since proteins are responsible for determining how the cell acts, understanding the proteome of single cell gives the best understanding of how a cell operates, and how gene expression
400:
which are used to identify which cell a certain protein came from (proteins coming from each cell have a different label) while others use not labels (quantify cells individually). The mass spectroscopy data is then analyzed by running data through databases that convert the information about
128:
combines microfluidics with MEMS on a CMOS chip to provide individual control over a large number of print nozzles, using the same technology as home inkjet printing. IJP allows for the adjustment of shear force to the sample ejection, greatly improving cell survivability. This approach, when
279:
so that the contents of the cell can be sequenced using NGS methods as was done in genomics. Once converted, there is not enough cDNA to be sequenced so the same DNA amplification techniques discussed in single cell genomics are applied to the cDNA to make sequencing possible. Alternately,
572:
The purpose of single cell metabolomics is to gain a better understanding at the molecular level of major biological topics such as: cancer, stem cells, aging, as well as the development of drug resistance. In general the focus of metabolomics is mostly on understanding how cells deal with
70:. Mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. Recent advances have enabled quantifying thousands of protein across hundreds of single cells, and thus make possible new types of analysis. In situ sequencing and
304:
RNA expression can serve as a proxy for protein abundance. However, protein abundance is governed by the complex interplay between RNA expression and post-transcriptional processes. While more challenging technically, translation can be monitored by ribosome profiling in single cells.
519:
that is capable of absorbing energy from a laser. Similar to SIMS, ionization happens in vacuum. Laser irradiation ablates the matrix material from the surface and results in charged gas phase matrix particles, the analyte molecules are ionized from this charged chemical matrix. Liu
288:
The purpose of single cell transcriptomics is to determine what genes are being expressed in each cell. The transcriptome is often used to quantify the gene expression instead of the proteome because of the difficulty currently associated with amplifying protein levels.
447:
directs the solvent with extracted molecules to the MS inlet. Nano-DESI mass spectrometry (MS) enables sensitive molecular profiling and quantification of endogenous species as small as a few hundred fmol-s in single cells in a higher throughput manner. Lanekoff
129:
combined with optical inspection and AI driven image recognition, not only guarantees single cell dispensing into the wellplate or other medium but also can qualify the cell sample for quality of sample, rejecting defective cells, debris and fragments.
558:
Mass spectroscopy is becoming the most frequently used method for single cell metabolomics. Its advantages are that there is no need to develop fluorescent proteins for all molecules of interest, and is capable of detecting metabolites in the
297:. Single cell transcriptomics has also been used for cell typing, where the genes expressed in a cell are used to identify types of cells. The main goal in cell typing is to find a way to determine the identity of cells that don't have known
3724:
Bourceau P, Geier B, Suerdieck V, Bien T, Soltwisch J, Dreisewerd K, et al. (October 2023). "Visualization of metabolites and microbes at high spatial resolution using MALDI mass spectrometry imaging and in situ fluorescence labeling".
62:
microscope. To date, due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as
2715:"Label-free Quantification of Proteins in Single Embryonic Cells with Neural Fate in the Cleavage-Stage Frog (Xenopus laevis) Embryo using Capillary Electrophoresis Electrospray Ionization High-Resolution Mass Spectrometry (CE-ESI-HRMS)"
145:
Hydrodynamic traps allow for the isolation of an individual cell in a "trap" at a single given time by passive microfluidic transport. The number of isolated cells can be manipulated based on the number of traps in the system.
438:
are set up in a V-shaped form, closing an angle of approx. 85 degrees. The two capillaries are touching therefore a liquid bridge can be formed between them and enable the sampling of surfaces as small as a single cell. The
3530:
Taylor MJ, Liyu A, Vertes A, Anderton CR (September 2021). "Ambient Single-Cell
Analysis and Native Tissue Imaging Using Laser-Ablation Electrospray Ionization Mass Spectrometry with Increased Spatial Resolution".
533:
changes in a cell due to different environmental stimuli. Although transcriptomics has the same purpose as proteomics it is not as accurate at determining gene expression in cells as it does not take into account
3688:
Xie W, Gao D, Jin F, Jiang Y, Liu H (July 2015). "Study of
Phospholipids in Single Cells Using an Integrated Microfluidic Device Combined with Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry".
1987:
Normand E, Qdaisat S, Bi W, Shaw C, Van den Veyver I, Beaudet A, et al. (September 2016). "Comparison of three whole genome amplification methods for detection of genomic aberrations in single cells".
3168:
Huffman RG, Leduc A, Wichmann C, di Gioia M, Borriello F, Specht H, et al. (2022-03-18). "Prioritized single-cell proteomics reveals molecular and functional polarization across primary macrophages".
3449:
326:
The antibody based methods use designed antibodies to bind to proteins of interest, allowing the relative abundance of multiple individual targets to be identified by one of several different techniques.
555:
activity of the metabolites. The current solution to this problem is to use fluorescent proteins which will act as metabolite detectors, fluorescing when ever they bind to a metabolite of interest.
495:. As they hit the surface, molecules are emitted from the surface and ionized. The choice of primary ions determines the size of the beam and also the extent of ionization and fragmentation. Pareek
464:(LAESI) a laser is used to ablate the surface of the sample and the emitted molecules are ionized in the gas phase by charged droplets from electrospray. Similar to DESI the ionization happens in
360:
for simultaneous and sensitive identification of proteins. These techniques can be highly multiplexed for simultaneous quantification of many targets (panels of up to 38 markers) in single cells.
280:
fluorescent compounds attached to RNA hybridization probes are used to identify specific sequences and sequential application of different RNA probes will build up a comprehensive transcriptome.
82:
Many single-cell analysis techniques require the isolation of individual cells. Methods currently used for single cell isolation include: Dielectrophoretic digital sorting, enzymatic digestion,
214:
One widely adopted WGA techniques is called degenerate oligonucleotide–primed polymerase chain reaction (DOP-PCR). This method uses the well established DNA amplification method
537:. Transcriptomics is still important as studying the difference between RNA levels and protein levels could give insight on which genes are post-transcriptionally regulated.
4290:
Ding L, Radfar P, Rezaei M, Warkiani ME (July 2021). "An easy-to-operate method for single-cell isolation and retrieval using a microfluidic static droplet array".
192:
is heavily dependent on increasing the copies of DNA found in the cell so there is enough to be sequenced. This has led to the development of strategies for
318:
There are three major approaches to single-cell proteomics: antibody based methods, fluorescent protein based methods, and mass-spectroscopy based methods.
2771:"Single Cell Proteomics Using Frog (Xenopus laevis) Blastomeres Isolated from Early Stage Embryos, Which Form a Geometric Progression in Protein Content"
3867:"In situ metabolic analysis of single plant cells by capillary microsampling and electrospray ionization mass spectrometry with ion mobility separation"
586:
to principal graphs. Some software libraries for reconstruction and visualization of lineage differentiation trajectories are freely available online.
2879:
Budnik B, Levy E, Slavov N (2017-03-15). "Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation".
2555:
524:
used MALDI-MS to detect eight phospholipids from single A549 cells. MALDI MS imaging can be used for spatial metabolomics and single cell analysis.
550:
There are four major methods used to quantify the metabolome of single cells, they are: fluorescence–based detection, fluorescence biosensors,
4010:
Setty M, et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34, 637–645 (2016).
423:
293:
analysis has often been used to study embryonic development. RNA splicing studies are focused on understanding the regulation of different
1726:
Faria EC, Gardner P (January 2012). "Analysis of Single
Eukaryotic Cells Using Raman Tweezers". In Lindström S, Andersson-Svahn H (eds.).
512:
385:
803:"Automated profiling of individual cell-cell interactions from high-throughput time-lapse imaging microscopy in nanowell grids (TIMING)"
1687:"Energy Charge, Redox State, and Metabolite Turnover in Single Human Hepatocytes Revealed by Capillary Microsampling Mass Spectrometry"
850:
Loo J, Ho H, Kong S, Wang T, Ho Y (September 2019). "Technological
Advances in Multiscale Analysis of Single Cells in Biomedicine".
2620:"Simple Method To Prepare Oligonucleotide-Conjugated Antibodies and Its Application in Multiplex Protein Detection in Single Cells"
105:
Manual single cell picking is a method where cells in a suspension are viewed under a microscope, and individually picked using a
153:
Manual single cell picking is a method where cells in a suspension are viewed under a microscope and individually picked using a
461:
2455:"Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes"
2033:"STR profiling and Copy Number Variation analysis on single, preserved cells using current Whole Genome Amplification methods"
3459:
2181:
1743:
91:
2453:
Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, et al. (July 2018). Chakraborty AK, Raj A, Marr C, Horváth P (eds.).
551:
356:
rare metal isotopes, not normally found in cells or tissues, can be attached to the individual antibodies and detected by
3423:
583:
223:
503:
and used isotope labeling and SIMS imaging to directly observe hotspots of metabolic activity within frozen HeLa cells.
611:
427:
268:
83:
71:
26:
3343:
Derks J, Slavov N (2022-11-05). "Strategies for increasing the depth and throughput of protein analysis by plexDIA".
2902:"SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation"
2506:"Mass cytometry as a platform for the discovery of cellular biomarkers to guide effective rheumatic disease therapy"
1832:"Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells"
4372:"Microfluidic-Based Technologies for CTC Isolation: A Review of 10 Years of Intense Efforts towards Liquid Biopsy"
4025:"Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming"
472:. used this ionization technique coupled to a Fourier transform mass spectrometer to analyzed 200 single cells of
452:
identified 14 amino acids, 6 metabolites, and several lipid molecules from single cheek cells using nano-DESI MS.
534:
484:
4445:
4440:
573:
environmental stresses at the molecular level, and to give a more dynamic understanding of cellular functions.
4425:
222:. Although simple, this method has been shown to have very low genome coverage. An improvement on DOP-PCR is
87:
2395:"Single-cell barcode analysis provides a rapid readout of cellular signaling pathways in clinical specimens"
1101:"Single-Cell Gene Network Analysis and Transcriptional Landscape of MYCN-Amplified Neuroblastoma Cell Lines"
995:"Miniaturization Technologies for Efficient Single-Cell Library Preparation for Next-Generation Sequencing"
487:(SIMS) is a technique similar to DESI, but while DESI is an ambient ionization technique, SIMS happens in
401:
peptides identified to quantification of protein levels. These methods are very similar to those used to
264:
3451:
Introduction to Mass
Spectrometry: Instrumentation, Applications, and Strategies for Data Interpretation
434:
that enables the sampling of small surfaces, therefore suitable for single-cell analysis. In nano-DESI,
392:(MS/MS). The major difference between quantification methods is some use labels on the peptides such as
1299:"Categorizing Cells on the Basis of their Chemical Profiles: Progress in Single-Cell Mass Spectrometry"
219:
3376:"Global absolute quantification reveals tight regulation of protein expression in single Xenopus eggs"
2661:"Global absolute quantification reveals tight regulation of protein expression in single Xenopus eggs"
2393:
Giedt RJ, Pathania D, Carlson JC, McFarland PJ, Del
Castillo AF, Juric D, et al. (October 2018).
641:
Siuzdak G (September 2023). "Subcellular quantitative imaging of metabolites at the organelle level".
74:(FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.
2031:
Vander Plaetsen AS, Deleye L, Cornelis S, Tilleman L, Van Nieuwerburgh F, Deforce D (December 2017).
1879:
Binder V, Bartenhagen C, Okpanyi V, Gombert M, Moehlendick B, Behrens B, et al. (October 2014).
594:
377:
215:
58:
1792:
1793:"Microfluidic hydrodynamic trapping for single cell analysis: mechanisms, methods and applications"
389:
993:
Mora-Castilla S, To C, Vaezeslami S, Morey R, Srinivasan S, Dumdie JN, et al. (August 2016).
4023:
Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, et al. (February 2019).
616:
342:
3768:
Rappez L, Stadler M, Triana S, Gathungu RM, Ovchinnikova K, Phapale P, et al. (July 2021).
3866:
3633:"Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells"
3374:
Smits AH, Lindeboom RG, Perino M, van Heeringen SJ, Veenstra GJ, Vermeulen M (September 2014).
2659:
Smits AH, Lindeboom RG, Perino M, van Heeringen SJ, Veenstra GJ, Vermeulen M (September 2014).
189:
3822:
Zenobi R (December 2013). "Single-cell metabolomics: analytical and biological perspectives".
2980:
Specht H, Emmott E, Petelski AA, Huffman RG, Perlman DH, Serra M, et al. (January 2021).
4435:
4430:
4136:"Inference of cell state transitions and cell fate plasticity from single-cell with MARGARET"
3865:
Zhang L, Foreman DP, Grant PA, Shrestha B, Moody SA, Villiers F, et al. (October 2014).
2953:"High-throughput single-cell proteomics quantifies the emergence of macrophage heterogeneity"
2549:
1265:
408:
276:
4077:"Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM"
3031:
Specht H, Harmange G, Perlman DH, Emmott E, Niziolek Z, Budnik B, et al. (2018-08-25).
2982:"Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2"
1930:"Comparison of whole genome amplification techniques for human single cell exome sequencing"
231:
4088:
3925:
3878:
3644:
3488:
3122:
2880:
2825:"Identification of Maturation-Specific Proteins by Single-Cell Proteomics of Human Oocytes"
2406:
2245:
2044:
1941:
1830:
Babayan A, Alawi M, Gormley M, Müller V, Wikman H, McMullin RP, et al. (August 2017).
1538:
1055:
755:
388:
is also known as LC-MS). This step gives order to the peptides before quantification using
294:
1099:
Mercatelli D, Balboni N, Palma A, Aleo E, Sanna PP, Perini G, et al. (January 2021).
1042:
Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. (January 2017).
8:
1218:
801:
Merouane A, Rey-Villamizar N, Lu Y, Liadi I, Romain G, Lu J, et al. (October 2015).
560:
4092:
3929:
3882:
3648:
3492:
3126:
2410:
2266:
2249:
2233:
2048:
1945:
1542:
1205:
Wu AR, Wang J, Streets AM, Huang Y (June 2017). "Single-Cell Transcriptional Analysis".
1171:
1154:
1059:
759:
4398:
4371:
4353:
4329:"Single nucleus multi-omics identifies human cortical cell regulatory genome diversity"
4328:
4315:
4273:
4246:
4228:
4201:
4160:
4135:
4111:
4076:
4075:
Chen H, Albergante L, Hsu JY, Lareau CA, Lo Bosco G, Guan J, et al. (April 2019).
4049:
4024:
3993:
3847:
3794:
3769:
3750:
3665:
3632:
3613:
3564:
3400:
3375:
3356:
3320:
3295:
3294:
Derks J, Leduc A, Wallmann G, Huffman RG, Willetts M, Khan S, et al. (July 2022).
3276:
3228:
3201:
3182:
3145:
3110:
3086:
3061:
3008:
2981:
2928:
2901:
2851:
2824:
2795:
2770:
2741:
2714:
2685:
2660:
2595:
2570:
2532:
2505:
2481:
2454:
2427:
2394:
2367:
2342:
2318:
2293:
2209:
2157:
2132:
2113:
2065:
2032:
2013:
1964:
1929:
1910:
1856:
1831:
1659:
1632:
1608:
1583:
1559:
1526:
1499:
1474:
1455:
1442:
1411:
1399:
1380:
1323:
1298:
1274:
1249:
1230:
1184:
1127:
1100:
1076:
1043:
1019:
994:
970:
943:
919:
894:
875:
827:
802:
778:
743:
719:
694:
666:
165:
110:
2343:"Multicolor multicycle molecular profiling with quantum dots for single-cell analysis"
4403:
4358:
4319:
4307:
4278:
4233:
4165:
4116:
4054:
3985:
3943:
3894:
3839:
3799:
3754:
3742:
3706:
3670:
3617:
3605:
3568:
3556:
3548:
3512:
3504:
3455:
3405:
3360:
3325:
3280:
3268:
3233:
3186:
3150:
3091:
3013:
2933:
2856:
2800:
2746:
2690:
2641:
2600:
2537:
2486:
2432:
2372:
2323:
2271:
2214:
2162:
2105:
2070:
2005:
1969:
1902:
1861:
1812:
1749:
1739:
1708:
1664:
1613:
1564:
1504:
1459:
1447:
1429:
1372:
1328:
1279:
1222:
1176:
1155:"Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications"
1132:
1081:
1024:
975:
924:
879:
867:
832:
783:
724:
670:
658:
397:
357:
272:
267:
uses sequencing techniques similar to single cell genomics or direct detection using
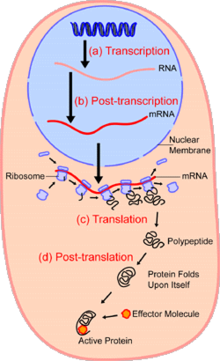
29:, which are all steps researchers are interested to quantify (DNA, RNA, and Protein).
3851:
2636:
2619:
1914:
1234:
1188:
818:
4393:
4383:
4348:
4340:
4299:
4268:
4258:
4223:
4213:
4155:
4147:
4106:
4096:
4044:
4036:
3997:
3977:
3933:
3886:
3831:
3789:
3781:
3734:
3698:
3660:
3652:
3595:
3540:
3496:
3477:"Profiling and quantifying endogenous molecules in single cells using nano-DESI MS"
3395:
3387:
3348:
3315:
3307:
3260:
3223:
3213:
3174:
3140:
3130:
3081:
3073:
3040:
3003:
2993:
2960:
2923:
2913:
2846:
2836:
2790:
2782:
2736:
2726:
2680:
2672:
2631:
2618:
Gong H, Holcomb I, Ooi A, Wang X, Majonis D, Unger MA, et al. (January 2016).
2590:
2582:
2527:
2517:
2476:
2466:
2422:
2414:
2362:
2354:
2313:
2305:
2261:
2253:
2204:
2196:
2182:"RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells"
2152:
2144:
2117:
2097:
2060:
2052:
2017:
1997:
1959:
1949:
1892:
1851:
1843:
1804:
1731:
1698:
1654:
1644:
1603:
1595:
1554:
1546:
1494:
1486:
1437:
1421:
1384:
1362:
1318:
1310:
1269:
1261:
1214:
1166:
1122:
1112:
1071:
1063:
1014:
1006:
965:
955:
914:
906:
859:
822:
814:
773:
763:
744:"Computation and measurement of cell decision making errors using single cell data"
714:
706:
650:
169:
114:
95:
34:
3077:
1490:
710:
3702:
3600:
3583:
3135:
2786:
2769:
Sun L, Dubiak KM, Peuchen EH, Zhang Z, Zhu G, Huber PW, et al. (July 2016).
1954:
1735:
1703:
1686:
1599:
768:
393:
125:
99:
4327:
Luo C, Liu H, Xie F, Armand EJ, Siletti K, Bakken TE, et al. (March 2022).
3962:
1631:
Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P (July 2015).
443:
delivers the solvent to the sample surface where the extraction happens and the
4344:
4303:
4263:
4101:
4040:
3785:
3738:
3311:
3264:
3218:
2998:
2586:
2504:
Nair N, Mei HE, Chen SY, Hale M, Nolan GP, Maecker HT, et al. (May 2015).
2418:
2257:
2232:
Ozadam H, Tonn T, Han CM, Segura A, Hoskins I, Rao S, et al. (June 2023).
2056:
1425:
910:
742:
Habibi I, Cheong R, Lipniacki T, Levchenko A, Emamian ES, Abdi A (April 2017).
654:
381:
348:
298:
239:
63:
3352:
3178:
2918:
2522:
1847:
1730:. Methods in Molecular Biology. Vol. 853. Humana Press. pp. 151–67.
121:
ensured by the semiconductor controlled motion of DEP cages in the flow cell.
4419:
3508:
2234:"Single-cell quantification of ribosome occupancy in early mouse development"
1816:
1433:
1010:
960:
46:
4218:
4202:"Single-Cell Analysis of Circulating Tumor Cells: Why Heterogeneity Matters"
3835:
3656:
2841:
2731:
2200:
1550:
1351:"Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity"
4407:
4362:
4311:
4282:
4237:
4169:
4120:
4058:
3989:
3947:
3898:
3843:
3803:
3746:
3710:
3674:
3609:
3560:
3516:
3409:
3329:
3272:
3237:
3154:
3095:
3017:
2937:
2860:
2804:
2750:
2694:
2645:
2604:
2541:
2490:
2436:
2376:
2358:
2327:
2275:
2218:
2166:
2109:
2074:
2009:
1973:
1906:
1865:
1753:
1712:
1668:
1617:
1568:
1508:
1451:
1376:
1367:
1350:
1332:
1283:
1226:
1180:
1136:
1085:
1028:
979:
928:
871:
863:
836:
787:
728:
662:
606:
54:
4245:
Zhou WM, Yan YY, Guo QR, Ji H, Wang H, Xu TT, et al. (October 2021).
4151:
3544:
2101:
1649:
409:
Ionization techniques used in mass spectrometry-based single-cell analysis
376:
The separation of differently sized proteins can be accomplished by using
4388:
3391:
3202:"Exploring functional protein covariation across single cells using nPOP"
3033:"Automated sample preparation for high-throughput single-cell proteomics"
2676:
1314:
1117:
338:
334:
2471:
2309:
1067:
4247:"Microfluidics applications for high-throughput single cell sequencing"
3981:
3938:
3913:
3890:
3500:
3476:
2148:
1897:
1880:
1808:
500:
402:
271:. The first step in quantifying the transcriptome is to convert RNA to
67:
50:
3961:
Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ (October 2016).
3552:
1520:
1518:
1044:"Massively parallel digital transcriptional profiling of single cells"
196:(WGA). Currently WGA strategies can be grouped into three categories:
142:
by the semiconductor controlled motion of DEP cages in the flow cell.
2030:
1790:
2001:
491:. The solid sample surface is bombarded by a highly focused beam of
3045:
3032:
2965:
2952:
2885:
1791:
Narayanamurthy V, Nagarajan S, Samsuri F, Sridhar TM (2017-06-30).
1515:
1416:
621:
499:
performed metabolomics to trace how purines are synthesized within
422:
One of the possible ways to measure the content of single cells is
42:
21:
3251:"Framework for multiplicative scaling of single-cell proteomics".
3200:
Leduc A, Huffman RG, Cantlon J, Khan S, Slavov N (December 2022).
2180:
Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X (April 2015).
1881:"A new workflow for whole-genome sequencing of single human cells"
405:, with modifications to accommodate the very small sample volume.
3373:
2822:
2658:
2131:
Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L (April 2014).
1928:
Borgström E, Paterlini M, Mold JE, Frisen J, Lundeberg J (2017).
200:
Controlled priming and PCR Amplification: Adapter-Linker PCR WGA
154:
106:
4183:
3584:"Single-cell metabolomics: where are we and where are we going?"
1767:
16:
Testbg biochemical processes and reactions in an individual cell
2823:
Virant-Klun I, Leicht S, Hughes C, Krijgsveld J (August 2016).
2133:"Single-cell in situ RNA profiling by sequential hybridization"
1878:
488:
235:
227:
3963:"Diffusion pseudotime robustly reconstructs lineage branching"
3914:"Advances in mass spectrometry based single-cell metabolomics"
3296:"Increasing the throughput of sensitive proteomics by plexDIA"
3062:"DO-MS: Data-Driven Optimization of Mass Spectrometry Methods"
1630:
992:
741:
4184:
Single-Cell Trajectory Reconstruction Exploration and Mapping
2392:
800:
3960:
3167:
2712:
2130:
1927:
576:
3767:
3030:
2979:
2713:
Lombard-Banek C, Reddy S, Moody SA, Nemes P (August 2016).
895:"Cellular heterogeneity: do differences make a difference?"
211:
is only applicable to human cells, both fixed and unfixed.
4022:
3723:
3630:
2088:
Kalisky T, Quake SR (April 2011). "Single-cell genomics".
1098:
226:(MDA), which uses random primers and a high fidelity
218:
to try and amplify the entire genome using a large set of
3864:
3424:"SCoPE-MS -- We can finally do single cell proteomics!!!"
3293:
1829:
1475:"Transformative Opportunities for Single-Cell Proteomics"
1041:
333:
Antibodies can be bound to fluorescent molecules such as
236:
Multiple Annealing and Looping–Based Amplification Cycles
117:, which uses a laser beam to trap, and manipulate cells.
4289:
3631:
Pareek V, Tian H, Winograd N, Benkovic SJ (April 2020).
3529:
3454:(4th ed.). John Wiley & Sons, 2007. p. 8.
2950:
2899:
2179:
597:
are characterized by stable and transient interactions.
515:
and ionization (MALDI), the sample is incorporated in a
150:
the cell. Then, the cell can be extracted for analysis.
4074:
3199:
3059:
2900:
Budnik B, Levy E, Harmange G, Slavov N (October 2018).
1297:
Comi TJ, Do TD, Rubakhin SS, Sweedler JV (March 2017).
426:(nanospray desorption electrospray ionization). Unlike
172:, which use a laser beam to trap and manipulate cells.
3911:
3109:
Chen AT, Franks A, Slavov N (July 2019). Cox J (ed.).
1296:
1254:
Annual Review of Chemical and Biomolecular Engineering
1247:
3581:
3533:
Journal of the American Society for Mass Spectrometry
2951:
Specht H, Emmott E, Koller T, Slavov N (2019-07-09).
2768:
414:
of ionization are mentioned in the paragraphs below.
203:
Random priming and PCR Amplification: DOP-PCR, MALBAC
3060:
Huffman RG, Chen A, Specht H, Slavov N (June 2019).
1986:
4369:
2231:
1400:"Single-cell protein analysis by mass spectrometry"
695:"Single cell analysis: the new frontier in 'omics'"
4018:
4016:
3912:Duncan KD, Fyrestam J, Lanekoff I (January 2019).
2617:
2340:
2294:"Single cell protein analysis for systems biology"
1204:
430:, which is a desorption technique, nano-DESI is a
367:
3770:"SpaceM reveals metabolic states of single cells"
3111:"DART-ID increases single-cell proteome coverage"
1584:"De Novo Gene Expression Reconstruction in Space"
4417:
4370:Descamps L, Le Roy D, Deman AL (February 2022).
3108:
2878:
1248:Tsioris K, Torres AJ, Douce TB, Love JC (2014).
1152:
206:Random priming and isothermal amplification: MDA
4326:
4013:
3474:
2503:
2452:
1153:Huang L, Ma F, Chapman A, Lu S, Xie XS (2015).
3687:
3582:Lanekoff I, Sharma VV, Marques C (June 2022).
3447:
3416:
2341:Zrazhevskiy P, True LD, Gao X (October 2013).
941:
164:The technique Raman tweezers combines the use
2571:"Mass Cytometry: Single Cells, Many Features"
2554:: CS1 maint: DOI inactive as of March 2024 (
849:
39:single-cell analysis and subcellular analysis
4244:
2568:
2225:
2087:
1725:
1472:
1159:Annual Review of Genomics and Human Genetics
892:
692:
386:liquid chromatography with mass spectroscopy
109:. Raman tweezers is a technique where
4376:International Journal of Molecular Sciences
4133:
4070:
4068:
3342:
3024:
2874:
2872:
2870:
1684:
1637:International Journal of Molecular Sciences
1348:
948:Frontiers in Cell and Developmental Biology
942:Hu P, Zhang W, Xin H, Deng G (2016-10-25).
4127:
2291:
1250:"A new toolbox for assessing single cells"
136:
25:This single cell shows the process of the
4397:
4387:
4352:
4272:
4262:
4227:
4217:
4199:
4159:
4110:
4100:
4048:
3937:
3793:
3664:
3599:
3399:
3319:
3227:
3217:
3144:
3134:
3085:
3044:
3007:
2997:
2964:
2927:
2917:
2884:
2850:
2840:
2794:
2740:
2730:
2684:
2635:
2594:
2531:
2521:
2480:
2470:
2426:
2366:
2317:
2265:
2208:
2156:
2064:
1963:
1953:
1896:
1855:
1702:
1658:
1648:
1607:
1558:
1498:
1441:
1415:
1366:
1322:
1273:
1170:
1126:
1116:
1075:
1018:
969:
959:
918:
826:
777:
767:
718:
577:Reconstructing developmental trajectories
321:
4065:
2867:
1633:"Technologies for Single-Cell Isolation"
1527:"Unpicking the proteome in single cells"
1303:Journal of the American Chemical Society
1266:10.1146/annurev-chembioeng-060713-035958
589:
476:(red onion) in high spatial resolution.
77:
20:
4200:Lim SB, Lim CT, Lim WT (October 2019).
3448:Watson JT, Sparkman OD (October 2007).
640:
4418:
3821:
1680:
1678:
1524:
1397:
1344:
1342:
462:Laser ablation electrospray ionization
3817:
3815:
3813:
2818:
2816:
2814:
2764:
2762:
2760:
2708:
2706:
2704:
2448:
2446:
2388:
2386:
2287:
2285:
1207:Annual Review of Analytical Chemistry
1200:
1198:
1148:
1146:
1219:10.1146/annurev-anchem-061516-045228
944:"Single Cell Isolation and Analysis"
688:
686:
684:
682:
680:
582:advanced mathematical concepts from
3858:
2829:Molecular & Cellular Proteomics
2719:Molecular & Cellular Proteomics
1675:
1581:
1404:Current Opinion in Chemical Biology
1339:
1290:
1172:10.1146/annurev-genom-090413-025352
403:quantify the proteome of bulk cells
224:Multiple displacement amplification
64:fluorescence-activated cell sorting
13:
4192:
3810:
2811:
2757:
2701:
2443:
2383:
2282:
1685:Zhang L, Vertes A (October 2015).
1473:Specht H, Slavov N (August 2018).
1195:
1143:
612:Micro-arrays for mass spectrometry
428:desorption electrospray ionization
269:fluorescence in situ hybridization
254:
72:fluorescence in situ hybridization
27:central dogma of molecular biology
14:
4457:
4134:Pandey K, Zafar H (August 2022).
2569:Spitzer MH, Nolan GP (May 2016).
2292:Levy E, Slavov N (October 2018).
893:Altschuler SJ, Wu LF (May 2010).
677:
3588:Current Opinion in Biotechnology
2510:Arthritis Research & Therapy
1349:Zhang L, Vertes A (April 2018).
999:Journal of Laboratory Automation
693:Wang D, Bodovitz S (June 2010).
513:matrix-assisted laser desorption
4176:
4004:
3954:
3905:
3761:
3717:
3681:
3624:
3575:
3523:
3475:Bergman HM, Lanekoff I (2017).
3468:
3441:
3367:
3336:
3287:
3244:
3193:
3161:
3102:
3053:
2973:
2944:
2893:
2652:
2637:10.1021/acs.bioconjchem.5b00613
2611:
2562:
2497:
2334:
2173:
2124:
2081:
2024:
1980:
1921:
1872:
1823:
1784:
1760:
1719:
1624:
1575:
1466:
1391:
1241:
1092:
545:
540:
535:post-transcriptional regulation
485:Secondary ion mass spectrometry
368:Mass spectrometry–based methods
313:
259:
1035:
986:
935:
886:
843:
794:
735:
634:
1:
3078:10.1021/acs.jproteome.9b00039
1491:10.1021/acs.jproteome.8b00257
819:10.1093/bioinformatics/btv355
711:10.1016/j.tibtech.2010.03.002
627:
308:
184:
88:laser capture microdissection
4251:Journal of Nanobiotechnology
3703:10.1021/acs.analchem.5b00010
3601:10.1016/j.copbio.2022.102693
3136:10.1371/journal.pcbi.1007082
3066:Journal of Proteome Research
2787:10.1021/acs.analchem.6b01921
1955:10.1371/journal.pone.0171566
1736:10.1007/978-1-61779-567-1_12
1704:10.1021/acs.analchem.5b02502
1600:10.1016/j.molmed.2017.05.004
1588:Trends in Molecular Medicine
1479:Journal of Proteome Research
769:10.1371/journal.pcbi.1005436
567:
527:
436:two fused silica capillaries
417:
283:
7:
3428:News in Proteomics Research
600:
432:liquid extraction technique
265:Single-cell transcriptomics
179:
10:
4462:
4345:10.1016/j.xgen.2022.100107
4304:10.1007/s00604-021-04897-9
4264:10.1186/s12951-021-01045-6
4102:10.1038/s41467-019-09670-4
4041:10.1016/j.cell.2019.01.006
3786:10.1038/s41592-021-01198-0
3739:10.1038/s41596-023-00864-1
3312:10.1038/s41587-022-01389-w
3265:10.1038/s41587-022-01411-1
3219:10.1186/s13059-022-02817-5
3115:PLOS Computational Biology
2999:10.1186/s13059-021-02267-5
2587:10.1016/j.cell.2016.04.019
2419:10.1038/s41467-018-07002-6
2258:10.1038/s41586-023-06228-9
2057:10.1038/s41598-017-17525-5
1426:10.1016/j.cbpa.2020.04.018
911:10.1016/j.cell.2010.04.033
748:PLOS Computational Biology
655:10.1038/s42255-023-00882-z
245:
240:next-generation sequencing
194:whole genome amplification
94:, Inkjet Printing or IJP,
3353:10.1101/2022.11.05.515287
3179:10.1101/2022.03.16.484655
2919:10.1186/s13059-018-1547-5
2523:10.1186/s13075-015-0644-z
1848:10.18632/oncotarget.10701
1525:Slavov N (January 2020).
378:capillary electrophoresis
1011:10.1177/2211068216630741
961:10.3389/fcell.2016.00116
506:
455:
390:tandem mass-spectroscopy
4219:10.3390/cancers11101595
3836:10.1126/science.1243259
3657:10.1126/science.aaz6465
3259:(1): 23–24. July 2022.
2842:10.1074/mcp.M115.056887
2732:10.1074/mcp.M115.057760
2526:(inactive 2024-03-30).
2201:10.1126/science.aaa6090
1551:10.1126/science.aaz6695
699:Trends in Biotechnology
617:Single-cell variability
479:
343:fluorescence microscopy
337:or tagged with organic
137:Associated Technologies
4140:Nucleic Acids Research
3380:Nucleic Acids Research
2665:Nucleic Acids Research
2624:Bioconjugate Chemistry
2359:10.1038/nprot.2013.112
2298:Essays in Biochemistry
1398:Slavov N (June 2020).
1368:10.1002/anie.201709719
864:10.1002/adbi.201900138
595:Cell–cell interactions
584:optimal transportation
322:Antibody–based methods
102:, and Raman tweezers.
86:, hydrodynamic traps,
59:cell–cell interactions
30:
4446:Scientific techniques
4441:Laboratory techniques
4081:Nature Communications
3545:10.1021/jasms.1c00149
3347:: 2022.11.05.515287.
3173:: 2022.03.16.484655.
2399:Nature Communications
2102:10.1038/nmeth0411-311
1650:10.3390/ijms160816897
1048:Nature Communications
590:Cell–cell interaction
382:liquid chromatography
277:reverse transcriptase
78:Single-cell isolation
33:In the field of
24:
4426:Analytical chemistry
4389:10.3390/ijms23041981
3691:Analytical Chemistry
3300:Nature Biotechnology
3253:Nature Biotechnology
2775:Analytical Chemistry
1728:Single-Cell Analysis
1691:Analytical Chemistry
1582:Lee JH (July 2017).
1315:10.1021/jacs.6b12822
1118:10.3390/biom11020177
190:Single-cell genomics
4152:10.1093/nar/gkac412
4093:2019NatCo..10.1903C
3930:2019Ana...144..782D
3883:2014Ana...139.5079Z
3649:2020Sci...368..283P
3493:2017Ana...142.3639B
3127:2019PLSCB..15E7082C
2472:10.7554/eLife.31657
2411:2018NatCo...9.4550G
2310:10.1042/EBC20180014
2250:2023Natur.618.1057O
2244:(7967): 1057–1064.
2049:2017NatSR...717189V
1946:2017PLoSO..1271566B
1842:(34): 56066–56080.
1543:2020Sci...367..512S
1068:10.1038/ncomms14049
1060:2017NatCo...814049Z
852:Advanced Biosystems
760:2017PLSCB..13E5436H
445:secondary capillary
295:transcript isoforms
4035:(4): 928–943.e22.
3982:10.1038/nmeth.3971
3939:10.1039/C8AN01581C
3891:10.1039/C4AN01018C
3501:10.1039/C7AN00885F
3392:10.1093/nar/gku661
2677:10.1093/nar/gku661
2149:10.1038/nmeth.2892
2037:Scientific Reports
1990:Prenatal Diagnosis
1898:10.1002/humu.22625
1809:10.1039/C7AY00656J
1797:Analytical Methods
466:ambient conditions
232:Φ29 DNA polymerase
166:Raman spectroscopy
111:Raman spectroscopy
90:, manual picking,
31:
4292:Mikrochimica Acta
3830:(6163): 1243259.
3733:(10): 3050–3079.
3697:(14): 7052–7059.
3643:(6488): 283–290.
3487:(19): 3639–3647.
3461:978-0-470-51634-8
2781:(13): 6653–6657.
2671:(15): 9880–9891.
2195:(6233): aaa6090.
1803:(25): 3751–3772.
1745:978-1-61779-566-4
1697:(20): 10397–405.
1537:(6477): 512–513.
1361:(17): 4466–4477.
1355:Angewandte Chemie
1309:(11): 3920–3929.
643:Nature Metabolism
441:primary capillary
358:mass spectrometry
341:for detection by
113:is combined with
96:micromanipulation
4453:
4411:
4401:
4391:
4366:
4356:
4323:
4286:
4276:
4266:
4241:
4231:
4221:
4186:
4180:
4174:
4173:
4163:
4131:
4125:
4124:
4114:
4104:
4072:
4063:
4062:
4052:
4020:
4011:
4008:
4002:
4001:
3967:
3958:
3952:
3951:
3941:
3909:
3903:
3902:
3862:
3856:
3855:
3819:
3808:
3807:
3797:
3765:
3759:
3758:
3727:Nature Protocols
3721:
3715:
3714:
3685:
3679:
3678:
3668:
3628:
3622:
3621:
3603:
3579:
3573:
3572:
3539:(9): 2490–2494.
3527:
3521:
3520:
3472:
3466:
3465:
3445:
3439:
3438:
3436:
3435:
3420:
3414:
3413:
3403:
3371:
3365:
3364:
3340:
3334:
3333:
3323:
3291:
3285:
3284:
3248:
3242:
3241:
3231:
3221:
3197:
3191:
3190:
3165:
3159:
3158:
3148:
3138:
3106:
3100:
3099:
3089:
3072:(6): 2493–2500.
3057:
3051:
3050:
3048:
3028:
3022:
3021:
3011:
3001:
2977:
2971:
2970:
2968:
2948:
2942:
2941:
2931:
2921:
2897:
2891:
2890:
2888:
2876:
2865:
2864:
2854:
2844:
2835:(8): 2616–2627.
2820:
2809:
2808:
2798:
2766:
2755:
2754:
2744:
2734:
2725:(8): 2756–2768.
2710:
2699:
2698:
2688:
2656:
2650:
2649:
2639:
2615:
2609:
2608:
2598:
2566:
2560:
2559:
2553:
2545:
2535:
2525:
2501:
2495:
2494:
2484:
2474:
2450:
2441:
2440:
2430:
2390:
2381:
2380:
2370:
2347:Nature Protocols
2338:
2332:
2331:
2321:
2289:
2280:
2279:
2269:
2229:
2223:
2222:
2212:
2186:
2177:
2171:
2170:
2160:
2128:
2122:
2121:
2085:
2079:
2078:
2068:
2028:
2022:
2021:
1984:
1978:
1977:
1967:
1957:
1925:
1919:
1918:
1900:
1876:
1870:
1869:
1859:
1827:
1821:
1820:
1788:
1782:
1781:
1779:
1778:
1764:
1758:
1757:
1723:
1717:
1716:
1706:
1682:
1673:
1672:
1662:
1652:
1643:(8): 16897–919.
1628:
1622:
1621:
1611:
1579:
1573:
1572:
1562:
1522:
1513:
1512:
1502:
1485:(8): 2565–2571.
1470:
1464:
1463:
1445:
1419:
1395:
1389:
1388:
1370:
1346:
1337:
1336:
1326:
1294:
1288:
1287:
1277:
1245:
1239:
1238:
1202:
1193:
1192:
1174:
1150:
1141:
1140:
1130:
1120:
1096:
1090:
1089:
1079:
1039:
1033:
1032:
1022:
990:
984:
983:
973:
963:
939:
933:
932:
922:
890:
884:
883:
858:(11): e1900138.
847:
841:
840:
830:
798:
792:
791:
781:
771:
739:
733:
732:
722:
690:
675:
674:
649:(9): 1446–1448.
638:
394:tandem mass tags
170:optical tweezers
115:optical tweezers
41:is the study of
35:cellular biology
4461:
4460:
4456:
4455:
4454:
4452:
4451:
4450:
4416:
4415:
4414:
4195:
4193:Further reading
4190:
4189:
4181:
4177:
4132:
4128:
4073:
4066:
4021:
4014:
4009:
4005:
3965:
3959:
3955:
3910:
3906:
3877:(20): 5079–85.
3863:
3859:
3820:
3811:
3766:
3762:
3722:
3718:
3686:
3682:
3629:
3625:
3580:
3576:
3528:
3524:
3473:
3469:
3462:
3446:
3442:
3433:
3431:
3422:
3421:
3417:
3386:(15): 9880–91.
3372:
3368:
3341:
3337:
3292:
3288:
3250:
3249:
3245:
3198:
3194:
3166:
3162:
3121:(7): e1007082.
3107:
3103:
3058:
3054:
3029:
3025:
2978:
2974:
2949:
2945:
2898:
2894:
2877:
2868:
2821:
2812:
2767:
2758:
2711:
2702:
2657:
2653:
2616:
2612:
2567:
2563:
2547:
2546:
2502:
2498:
2451:
2444:
2391:
2384:
2353:(10): 1852–69.
2339:
2335:
2290:
2283:
2230:
2226:
2184:
2178:
2174:
2129:
2125:
2086:
2082:
2029:
2025:
2002:10.1002/pd.4866
1985:
1981:
1940:(2): e0171566.
1926:
1922:
1891:(10): 1260–70.
1877:
1873:
1828:
1824:
1789:
1785:
1776:
1774:
1766:
1765:
1761:
1746:
1724:
1720:
1683:
1676:
1629:
1625:
1580:
1576:
1523:
1516:
1471:
1467:
1396:
1392:
1347:
1340:
1295:
1291:
1246:
1242:
1203:
1196:
1151:
1144:
1097:
1093:
1040:
1036:
991:
987:
940:
936:
891:
887:
848:
844:
813:(19): 3189–97.
799:
795:
754:(4): e1005436.
740:
736:
691:
678:
639:
635:
630:
603:
592:
579:
570:
548:
543:
530:
517:chemical matrix
509:
482:
458:
420:
411:
398:dimethyl labels
370:
324:
316:
311:
299:genetic markers
286:
262:
257:
255:Transcriptomics
248:
230:, usually
187:
182:
139:
100:serial dilution
80:
47:transcriptomics
17:
12:
11:
5:
4459:
4449:
4448:
4443:
4438:
4433:
4428:
4413:
4412:
4367:
4324:
4287:
4242:
4196:
4194:
4191:
4188:
4187:
4175:
4126:
4064:
4012:
4003:
3970:Nature Methods
3953:
3924:(3): 782–793.
3904:
3857:
3809:
3780:(7): 799–805.
3774:Nature Methods
3760:
3716:
3680:
3623:
3574:
3522:
3467:
3460:
3440:
3415:
3366:
3335:
3286:
3243:
3206:Genome Biology
3192:
3160:
3101:
3052:
3046:10.1101/399774
3023:
2986:Genome Biology
2972:
2966:10.1101/665307
2943:
2906:Genome Biology
2892:
2886:10.1101/102681
2866:
2810:
2756:
2700:
2651:
2610:
2561:
2496:
2442:
2382:
2333:
2304:(4): 595–605.
2281:
2224:
2172:
2137:Nature Methods
2123:
2090:Nature Methods
2080:
2023:
1979:
1920:
1885:Human Mutation
1871:
1822:
1783:
1759:
1744:
1718:
1674:
1623:
1594:(7): 583–593.
1574:
1514:
1465:
1390:
1338:
1289:
1240:
1213:(1): 439–462.
1194:
1142:
1091:
1034:
985:
934:
885:
842:
807:Bioinformatics
793:
734:
676:
632:
631:
629:
626:
625:
624:
619:
614:
609:
602:
599:
591:
588:
578:
575:
569:
566:
547:
544:
542:
539:
529:
526:
508:
505:
481:
478:
457:
454:
419:
416:
410:
407:
369:
366:
350:Mass Cytometry
323:
320:
315:
312:
310:
307:
285:
282:
261:
258:
256:
253:
247:
244:
208:
207:
204:
201:
186:
183:
181:
178:
138:
135:
126:nkjet Printing
79:
76:
15:
9:
6:
4:
3:
2:
4458:
4447:
4444:
4442:
4439:
4437:
4434:
4432:
4429:
4427:
4424:
4423:
4421:
4409:
4405:
4400:
4395:
4390:
4385:
4381:
4377:
4373:
4368:
4364:
4360:
4355:
4350:
4346:
4342:
4339:(3): 100107.
4338:
4334:
4333:Cell Genomics
4330:
4325:
4321:
4317:
4313:
4309:
4305:
4301:
4297:
4293:
4288:
4284:
4280:
4275:
4270:
4265:
4260:
4256:
4252:
4248:
4243:
4239:
4235:
4230:
4225:
4220:
4215:
4211:
4207:
4203:
4198:
4197:
4185:
4182:Pinello Lab.
4179:
4171:
4167:
4162:
4157:
4153:
4149:
4145:
4141:
4137:
4130:
4122:
4118:
4113:
4108:
4103:
4098:
4094:
4090:
4086:
4082:
4078:
4071:
4069:
4060:
4056:
4051:
4046:
4042:
4038:
4034:
4030:
4026:
4019:
4017:
4007:
3999:
3995:
3991:
3987:
3983:
3979:
3976:(10): 845–8.
3975:
3971:
3964:
3957:
3949:
3945:
3940:
3935:
3931:
3927:
3923:
3919:
3915:
3908:
3900:
3896:
3892:
3888:
3884:
3880:
3876:
3872:
3868:
3861:
3853:
3849:
3845:
3841:
3837:
3833:
3829:
3825:
3818:
3816:
3814:
3805:
3801:
3796:
3791:
3787:
3783:
3779:
3775:
3771:
3764:
3756:
3752:
3748:
3744:
3740:
3736:
3732:
3728:
3720:
3712:
3708:
3704:
3700:
3696:
3692:
3684:
3676:
3672:
3667:
3662:
3658:
3654:
3650:
3646:
3642:
3638:
3634:
3627:
3619:
3615:
3611:
3607:
3602:
3597:
3593:
3589:
3585:
3578:
3570:
3566:
3562:
3558:
3554:
3550:
3546:
3542:
3538:
3534:
3526:
3518:
3514:
3510:
3506:
3502:
3498:
3494:
3490:
3486:
3482:
3478:
3471:
3463:
3457:
3453:
3452:
3444:
3429:
3425:
3419:
3411:
3407:
3402:
3397:
3393:
3389:
3385:
3381:
3377:
3370:
3362:
3358:
3354:
3350:
3346:
3339:
3331:
3327:
3322:
3317:
3313:
3309:
3305:
3301:
3297:
3290:
3282:
3278:
3274:
3270:
3266:
3262:
3258:
3254:
3247:
3239:
3235:
3230:
3225:
3220:
3215:
3211:
3207:
3203:
3196:
3188:
3184:
3180:
3176:
3172:
3164:
3156:
3152:
3147:
3142:
3137:
3132:
3128:
3124:
3120:
3116:
3112:
3105:
3097:
3093:
3088:
3083:
3079:
3075:
3071:
3067:
3063:
3056:
3047:
3042:
3038:
3034:
3027:
3019:
3015:
3010:
3005:
3000:
2995:
2991:
2987:
2983:
2976:
2967:
2962:
2958:
2954:
2947:
2939:
2935:
2930:
2925:
2920:
2915:
2911:
2907:
2903:
2896:
2887:
2882:
2875:
2873:
2871:
2862:
2858:
2853:
2848:
2843:
2838:
2834:
2830:
2826:
2819:
2817:
2815:
2806:
2802:
2797:
2792:
2788:
2784:
2780:
2776:
2772:
2765:
2763:
2761:
2752:
2748:
2743:
2738:
2733:
2728:
2724:
2720:
2716:
2709:
2707:
2705:
2696:
2692:
2687:
2682:
2678:
2674:
2670:
2666:
2662:
2655:
2647:
2643:
2638:
2633:
2630:(1): 217–25.
2629:
2625:
2621:
2614:
2606:
2602:
2597:
2592:
2588:
2584:
2581:(4): 780–91.
2580:
2576:
2572:
2565:
2557:
2551:
2543:
2539:
2534:
2529:
2524:
2519:
2515:
2511:
2507:
2500:
2492:
2488:
2483:
2478:
2473:
2468:
2464:
2460:
2456:
2449:
2447:
2438:
2434:
2429:
2424:
2420:
2416:
2412:
2408:
2404:
2400:
2396:
2389:
2387:
2378:
2374:
2369:
2364:
2360:
2356:
2352:
2348:
2344:
2337:
2329:
2325:
2320:
2315:
2311:
2307:
2303:
2299:
2295:
2288:
2286:
2277:
2273:
2268:
2263:
2259:
2255:
2251:
2247:
2243:
2239:
2235:
2228:
2220:
2216:
2211:
2206:
2202:
2198:
2194:
2190:
2183:
2176:
2168:
2164:
2159:
2154:
2150:
2146:
2142:
2138:
2134:
2127:
2119:
2115:
2111:
2107:
2103:
2099:
2095:
2091:
2084:
2076:
2072:
2067:
2062:
2058:
2054:
2050:
2046:
2042:
2038:
2034:
2027:
2019:
2015:
2011:
2007:
2003:
1999:
1996:(9): 823–30.
1995:
1991:
1983:
1975:
1971:
1966:
1961:
1956:
1951:
1947:
1943:
1939:
1935:
1931:
1924:
1916:
1912:
1908:
1904:
1899:
1894:
1890:
1886:
1882:
1875:
1867:
1863:
1858:
1853:
1849:
1845:
1841:
1837:
1833:
1826:
1818:
1814:
1810:
1806:
1802:
1798:
1794:
1787:
1773:
1769:
1763:
1755:
1751:
1747:
1741:
1737:
1733:
1729:
1722:
1714:
1710:
1705:
1700:
1696:
1692:
1688:
1681:
1679:
1670:
1666:
1661:
1656:
1651:
1646:
1642:
1638:
1634:
1627:
1619:
1615:
1610:
1605:
1601:
1597:
1593:
1589:
1585:
1578:
1570:
1566:
1561:
1556:
1552:
1548:
1544:
1540:
1536:
1532:
1528:
1521:
1519:
1510:
1506:
1501:
1496:
1492:
1488:
1484:
1480:
1476:
1469:
1461:
1457:
1453:
1449:
1444:
1439:
1435:
1431:
1427:
1423:
1418:
1413:
1409:
1405:
1401:
1394:
1386:
1382:
1378:
1374:
1369:
1364:
1360:
1356:
1352:
1345:
1343:
1334:
1330:
1325:
1320:
1316:
1312:
1308:
1304:
1300:
1293:
1285:
1281:
1276:
1271:
1267:
1263:
1259:
1255:
1251:
1244:
1236:
1232:
1228:
1224:
1220:
1216:
1212:
1208:
1201:
1199:
1190:
1186:
1182:
1178:
1173:
1168:
1165:(1): 79–102.
1164:
1160:
1156:
1149:
1147:
1138:
1134:
1129:
1124:
1119:
1114:
1110:
1106:
1102:
1095:
1087:
1083:
1078:
1073:
1069:
1065:
1061:
1057:
1053:
1049:
1045:
1038:
1030:
1026:
1021:
1016:
1012:
1008:
1005:(4): 557–67.
1004:
1000:
996:
989:
981:
977:
972:
967:
962:
957:
953:
949:
945:
938:
930:
926:
921:
916:
912:
908:
905:(4): 559–63.
904:
900:
896:
889:
881:
877:
873:
869:
865:
861:
857:
853:
846:
838:
834:
829:
824:
820:
816:
812:
808:
804:
797:
789:
785:
780:
775:
770:
765:
761:
757:
753:
749:
745:
738:
730:
726:
721:
716:
712:
708:
705:(6): 281–90.
704:
700:
696:
689:
687:
685:
683:
681:
672:
668:
664:
660:
656:
652:
648:
644:
637:
633:
623:
620:
618:
615:
613:
610:
608:
605:
604:
598:
596:
587:
585:
574:
565:
562:
556:
553:
538:
536:
525:
523:
518:
514:
504:
502:
498:
494:
490:
486:
477:
475:
471:
467:
463:
453:
451:
446:
442:
437:
433:
429:
425:
415:
406:
404:
399:
395:
391:
387:
383:
379:
374:
365:
361:
359:
355:
352:
351:
346:
344:
340:
336:
332:
328:
319:
306:
302:
300:
296:
290:
281:
278:
274:
270:
266:
252:
243:
241:
237:
233:
229:
225:
221:
217:
212:
205:
202:
199:
198:
197:
195:
191:
177:
173:
171:
167:
162:
158:
156:
151:
147:
143:
134:
130:
127:
122:
118:
116:
112:
108:
103:
101:
97:
93:
92:microfluidics
89:
85:
75:
73:
69:
65:
60:
56:
52:
48:
44:
40:
36:
28:
23:
19:
4436:Cell biology
4431:Biochemistry
4379:
4375:
4336:
4332:
4295:
4291:
4254:
4250:
4212:(10): 1595.
4209:
4205:
4178:
4143:
4139:
4129:
4084:
4080:
4032:
4028:
4006:
3973:
3969:
3956:
3921:
3917:
3907:
3874:
3870:
3860:
3827:
3823:
3777:
3773:
3763:
3730:
3726:
3719:
3694:
3690:
3683:
3640:
3636:
3626:
3591:
3587:
3577:
3536:
3532:
3525:
3484:
3480:
3470:
3450:
3443:
3432:. Retrieved
3430:. 2017-03-09
3427:
3418:
3383:
3379:
3369:
3344:
3338:
3306:(1): 50–59.
3303:
3299:
3289:
3256:
3252:
3246:
3209:
3205:
3195:
3170:
3163:
3118:
3114:
3104:
3069:
3065:
3055:
3036:
3026:
2989:
2985:
2975:
2956:
2946:
2909:
2905:
2895:
2832:
2828:
2778:
2774:
2722:
2718:
2668:
2664:
2654:
2627:
2623:
2613:
2578:
2574:
2564:
2550:cite journal
2513:
2509:
2499:
2462:
2458:
2402:
2398:
2350:
2346:
2336:
2301:
2297:
2241:
2237:
2227:
2192:
2188:
2175:
2143:(4): 360–1.
2140:
2136:
2126:
2096:(4): 311–4.
2093:
2089:
2083:
2043:(1): 17189.
2040:
2036:
2026:
1993:
1989:
1982:
1937:
1933:
1923:
1888:
1884:
1874:
1839:
1835:
1825:
1800:
1796:
1786:
1775:. Retrieved
1771:
1762:
1727:
1721:
1694:
1690:
1640:
1636:
1626:
1591:
1587:
1577:
1534:
1530:
1482:
1478:
1468:
1407:
1403:
1393:
1358:
1354:
1306:
1302:
1292:
1257:
1253:
1243:
1210:
1206:
1162:
1158:
1108:
1105:Biomolecules
1104:
1094:
1051:
1047:
1037:
1002:
998:
988:
951:
947:
937:
902:
898:
888:
855:
851:
845:
810:
806:
796:
751:
747:
737:
702:
698:
646:
642:
636:
607:Cell sorting
593:
580:
571:
557:
549:
541:Metabolomics
531:
521:
516:
510:
496:
493:primary ions
492:
483:
473:
469:
465:
459:
449:
444:
440:
435:
431:
421:
412:
384:(LC) (using
375:
371:
362:
353:
349:
347:
339:fluorophores
335:quantum dots
330:
329:
325:
317:
303:
291:
287:
263:
249:
213:
209:
193:
188:
174:
163:
159:
155:micropipette
152:
148:
144:
140:
131:
123:
119:
107:micropipette
104:
81:
55:metabolomics
38:
32:
18:
4382:(4): 1981.
4146:(15): e86.
4087:(1): 1903.
3918:The Analyst
3871:The Analyst
3481:The Analyst
2405:(1): 4550.
501:purinosomes
474:Allium cepa
468:. Anderton
4420:Categories
4298:(8): 242.
4257:(1): 312.
3594:: 102693.
3434:2017-06-28
3212:(1): 261.
3039:: 399774.
2912:(1): 161.
2516:(1): 127.
2465:: e31657.
1836:Oncotarget
1777:2024-05-15
1417:2004.02069
1260:: 455–77.
1111:(2): 177.
628:References
546:Techniques
314:Techniques
309:Proteomics
260:Techniques
185:Techniques
68:metabolome
51:proteomics
4320:235738076
3755:261580460
3618:246773056
3569:236968123
3509:0003-2654
3361:253399025
3281:250642572
3187:247599981
2992:(1): 50.
1817:1759-9679
1460:219966629
1434:1367-5931
1054:: 14049.
880:203101696
671:261607846
561:femtomole
424:nano-DESI
418:Nano-DESI
396:(TMT) or
380:(CE) or
4408:35216097
4363:35419551
4312:34226955
4283:34635104
4238:31635038
4170:35639499
4121:31015418
4059:30712874
3990:27571553
3948:30426983
3899:25109271
3852:21381091
3844:24311695
3804:34226721
3747:37674095
3711:26110742
3675:32299949
3610:35151979
3561:34374553
3517:28835951
3410:25056316
3330:35835881
3273:35851377
3238:36527135
3155:31260443
3096:31081635
3018:33504367
2938:30343672
2861:27215607
2805:27314579
2751:27317400
2695:25056316
2646:26689321
2605:27153492
2542:25981462
2491:29993362
2437:30382095
2377:24008381
2328:30072488
2276:37344592
2267:10307641
2219:25858977
2167:24681720
2110:21451520
2075:29215049
2010:27368744
1974:28207771
1934:PLOS ONE
1915:27392899
1907:25066732
1866:28915574
1754:22323146
1713:26398405
1669:26213926
1618:28571832
1569:32001644
1509:29945450
1452:32599342
1377:29218763
1333:28135079
1284:24910919
1235:40069109
1227:28301747
1189:12987987
1181:26077818
1137:33525507
1086:28091601
1029:26891732
980:27826548
929:20478246
872:32648696
837:26059718
788:28379950
729:20434785
663:37679555
622:CITE-Seq
601:See also
331:Imaging:
180:Genomics
43:genomics
4399:8875744
4354:9004682
4274:8507141
4229:6826423
4206:Cancers
4161:9410915
4112:6478907
4089:Bibcode
4050:6402800
3998:3594049
3926:Bibcode
3879:Bibcode
3824:Science
3795:7611214
3666:7494208
3645:Bibcode
3637:Science
3553:1824325
3489:Bibcode
3401:4150773
3345:bioRxiv
3321:9839897
3229:9756690
3171:bioRxiv
3146:6625733
3123:Bibcode
3087:6737531
3037:bioRxiv
3009:7839219
2957:bioRxiv
2929:6196420
2881:bioRxiv
2852:4974340
2796:4940028
2742:4974349
2686:4150773
2596:4860251
2533:4436107
2482:6075866
2428:6208406
2407:Bibcode
2368:4108347
2319:6204083
2246:Bibcode
2210:4662681
2189:Science
2158:4085791
2118:5601612
2066:5719346
2045:Bibcode
2018:5537482
1965:5313163
1942:Bibcode
1857:5593545
1660:4581176
1609:5514424
1560:7029782
1539:Bibcode
1531:Science
1500:6089608
1443:7767890
1410:: 1–9.
1385:4928231
1324:5364434
1275:4309009
1128:7912277
1077:5241818
1056:Bibcode
1020:4948133
971:5078503
954:: 116.
920:2918286
828:4693004
779:5397092
756:Bibcode
720:2876223
568:Purpose
528:Purpose
284:Purpose
246:Purpose
242:(NGS).
220:primers
37:,
4406:
4396:
4361:
4351:
4318:
4310:
4281:
4271:
4236:
4226:
4168:
4158:
4119:
4109:
4057:
4047:
3996:
3988:
3946:
3897:
3850:
3842:
3802:
3792:
3753:
3745:
3709:
3673:
3663:
3616:
3608:
3567:
3559:
3551:
3515:
3507:
3458:
3408:
3398:
3359:
3328:
3318:
3279:
3271:
3236:
3226:
3185:
3153:
3143:
3094:
3084:
3016:
3006:
2936:
2926:
2883:
2859:
2849:
2803:
2793:
2749:
2739:
2693:
2683:
2644:
2603:
2593:
2540:
2530:
2489:
2479:
2435:
2425:
2375:
2365:
2326:
2316:
2274:
2264:
2238:Nature
2217:
2207:
2165:
2155:
2116:
2108:
2073:
2063:
2016:
2008:
1972:
1962:
1913:
1905:
1864:
1854:
1815:
1752:
1742:
1711:
1667:
1657:
1616:
1606:
1567:
1557:
1507:
1497:
1458:
1450:
1440:
1432:
1383:
1375:
1331:
1321:
1282:
1272:
1233:
1225:
1187:
1179:
1135:
1125:
1084:
1074:
1027:
1017:
978:
968:
927:
917:
878:
870:
835:
825:
786:
776:
727:
717:
669:
661:
522:et al.
497:et al.
489:vacuum
450:et al.
275:using
228:enzyme
4316:S2CID
3994:S2CID
3966:(PDF)
3848:S2CID
3751:S2CID
3614:S2CID
3565:S2CID
3357:S2CID
3277:S2CID
3183:S2CID
2459:eLife
2185:(PDF)
2114:S2CID
2014:S2CID
1911:S2CID
1768:"MMS"
1456:S2CID
1412:arXiv
1381:S2CID
1231:S2CID
1185:S2CID
876:S2CID
667:S2CID
507:MALDI
470:et al
456:LAESI
4404:PMID
4359:PMID
4308:PMID
4279:PMID
4234:PMID
4166:PMID
4117:PMID
4055:PMID
4029:Cell
3986:PMID
3944:PMID
3895:PMID
3840:PMID
3800:PMID
3743:PMID
3707:PMID
3671:PMID
3606:PMID
3557:PMID
3549:OSTI
3513:PMID
3505:ISSN
3456:ISBN
3406:PMID
3326:PMID
3269:PMID
3234:PMID
3151:PMID
3092:PMID
3014:PMID
2934:PMID
2857:PMID
2801:PMID
2747:PMID
2691:PMID
2642:PMID
2601:PMID
2575:Cell
2556:link
2538:PMID
2487:PMID
2433:PMID
2373:PMID
2324:PMID
2272:PMID
2215:PMID
2163:PMID
2106:PMID
2071:PMID
2006:PMID
1970:PMID
1903:PMID
1862:PMID
1813:ISSN
1750:PMID
1740:ISBN
1709:PMID
1665:PMID
1614:PMID
1565:PMID
1505:PMID
1448:PMID
1430:ISSN
1373:PMID
1329:PMID
1280:PMID
1223:PMID
1177:PMID
1133:PMID
1082:PMID
1025:PMID
976:PMID
925:PMID
899:Cell
868:PMID
833:PMID
784:PMID
725:PMID
659:PMID
552:FRET
480:SIMS
273:cDNA
168:and
84:FACS
57:and
4394:PMC
4384:doi
4349:PMC
4341:doi
4300:doi
4296:188
4269:PMC
4259:doi
4224:PMC
4214:doi
4156:PMC
4148:doi
4107:PMC
4097:doi
4045:PMC
4037:doi
4033:176
3978:doi
3934:doi
3922:144
3887:doi
3875:139
3832:doi
3828:342
3790:PMC
3782:doi
3735:doi
3699:doi
3661:PMC
3653:doi
3641:368
3596:doi
3541:doi
3497:doi
3485:142
3396:PMC
3388:doi
3349:doi
3316:PMC
3308:doi
3261:doi
3224:PMC
3214:doi
3175:doi
3141:PMC
3131:doi
3082:PMC
3074:doi
3041:doi
3004:PMC
2994:doi
2961:doi
2924:PMC
2914:doi
2847:PMC
2837:doi
2791:PMC
2783:doi
2737:PMC
2727:doi
2681:PMC
2673:doi
2632:doi
2591:PMC
2583:doi
2579:165
2528:PMC
2518:doi
2477:PMC
2467:doi
2423:PMC
2415:doi
2363:PMC
2355:doi
2314:PMC
2306:doi
2262:PMC
2254:doi
2242:618
2205:PMC
2197:doi
2193:348
2153:PMC
2145:doi
2098:doi
2061:PMC
2053:doi
1998:doi
1960:PMC
1950:doi
1893:doi
1852:PMC
1844:doi
1805:doi
1772:MMS
1732:doi
1699:doi
1655:PMC
1645:doi
1604:PMC
1596:doi
1555:PMC
1547:doi
1535:367
1495:PMC
1487:doi
1438:PMC
1422:doi
1363:doi
1319:PMC
1311:doi
1307:139
1270:PMC
1262:doi
1215:doi
1167:doi
1123:PMC
1113:doi
1072:PMC
1064:doi
1015:PMC
1007:doi
966:PMC
956:doi
915:PMC
907:doi
903:141
860:doi
823:PMC
815:doi
774:PMC
764:doi
715:PMC
707:doi
651:doi
511:In
460:In
216:PCR
4422::
4402:.
4392:.
4380:23
4378:.
4374:.
4357:.
4347:.
4335:.
4331:.
4314:.
4306:.
4294:.
4277:.
4267:.
4255:19
4253:.
4249:.
4232:.
4222:.
4210:11
4208:.
4204:.
4164:.
4154:.
4144:50
4142:.
4138:.
4115:.
4105:.
4095:.
4085:10
4083:.
4079:.
4067:^
4053:.
4043:.
4031:.
4027:.
4015:^
3992:.
3984:.
3974:13
3972:.
3968:.
3942:.
3932:.
3920:.
3916:.
3893:.
3885:.
3873:.
3869:.
3846:.
3838:.
3826:.
3812:^
3798:.
3788:.
3778:18
3776:.
3772:.
3749:.
3741:.
3731:18
3729:.
3705:.
3695:87
3693:.
3669:.
3659:.
3651:.
3639:.
3635:.
3612:.
3604:.
3592:75
3590:.
3586:.
3563:.
3555:.
3547:.
3537:32
3535:.
3511:.
3503:.
3495:.
3483:.
3479:.
3426:.
3404:.
3394:.
3384:42
3382:.
3378:.
3355:.
3324:.
3314:.
3304:41
3302:.
3298:.
3275:.
3267:.
3257:41
3255:.
3232:.
3222:.
3210:23
3208:.
3204:.
3181:.
3149:.
3139:.
3129:.
3119:15
3117:.
3113:.
3090:.
3080:.
3070:18
3068:.
3064:.
3035:.
3012:.
3002:.
2990:22
2988:.
2984:.
2959:.
2955:.
2932:.
2922:.
2910:19
2908:.
2904:.
2869:^
2855:.
2845:.
2833:15
2831:.
2827:.
2813:^
2799:.
2789:.
2779:88
2777:.
2773:.
2759:^
2745:.
2735:.
2723:15
2721:.
2717:.
2703:^
2689:.
2679:.
2669:42
2667:.
2663:.
2640:.
2628:27
2626:.
2622:.
2599:.
2589:.
2577:.
2573:.
2552:}}
2548:{{
2536:.
2514:17
2512:.
2508:.
2485:.
2475:.
2461:.
2457:.
2445:^
2431:.
2421:.
2413:.
2401:.
2397:.
2385:^
2371:.
2361:.
2349:.
2345:.
2322:.
2312:.
2302:62
2300:.
2296:.
2284:^
2270:.
2260:.
2252:.
2240:.
2236:.
2213:.
2203:.
2191:.
2187:.
2161:.
2151:.
2141:11
2139:.
2135:.
2112:.
2104:.
2092:.
2069:.
2059:.
2051:.
2039:.
2035:.
2012:.
2004:.
1994:36
1992:.
1968:.
1958:.
1948:.
1938:12
1936:.
1932:.
1909:.
1901:.
1889:35
1887:.
1883:.
1860:.
1850:.
1838:.
1834:.
1811:.
1799:.
1795:.
1770:.
1748:.
1738:.
1707:.
1695:87
1693:.
1689:.
1677:^
1663:.
1653:.
1641:16
1639:.
1635:.
1612:.
1602:.
1592:23
1590:.
1586:.
1563:.
1553:.
1545:.
1533:.
1529:.
1517:^
1503:.
1493:.
1483:17
1481:.
1477:.
1454:.
1446:.
1436:.
1428:.
1420:.
1408:60
1406:.
1402:.
1379:.
1371:.
1359:57
1357:.
1353:.
1341:^
1327:.
1317:.
1305:.
1301:.
1278:.
1268:.
1256:.
1252:.
1229:.
1221:.
1211:10
1209:.
1197:^
1183:.
1175:.
1163:16
1161:.
1157:.
1145:^
1131:.
1121:.
1109:11
1107:.
1103:.
1080:.
1070:.
1062:.
1050:.
1046:.
1023:.
1013:.
1003:21
1001:.
997:.
974:.
964:.
950:.
946:.
923:.
913:.
901:.
897:.
874:.
866:.
854:.
831:.
821:.
811:31
809:.
805:.
782:.
772:.
762:.
752:13
750:.
746:.
723:.
713:.
703:28
701:.
697:.
679:^
665:.
657:.
645:.
301:.
157:.
98:,
53:,
49:,
45:,
4410:.
4386::
4365:.
4343::
4337:2
4322:.
4302::
4285:.
4261::
4240:.
4216::
4172:.
4150::
4123:.
4099::
4091::
4061:.
4039::
4000:.
3980::
3950:.
3936::
3928::
3901:.
3889::
3881::
3854:.
3834::
3806:.
3784::
3757:.
3737::
3713:.
3701::
3677:.
3655::
3647::
3620:.
3598::
3571:.
3543::
3519:.
3499::
3491::
3464:.
3437:.
3412:.
3390::
3363:.
3351::
3332:.
3310::
3283:.
3263::
3240:.
3216::
3189:.
3177::
3157:.
3133::
3125::
3098:.
3076::
3049:.
3043::
3020:.
2996::
2969:.
2963::
2940:.
2916::
2889:.
2863:.
2839::
2807:.
2785::
2753:.
2729::
2697:.
2675::
2648:.
2634::
2607:.
2585::
2558:)
2544:.
2520::
2493:.
2469::
2463:7
2439:.
2417::
2409::
2403:9
2379:.
2357::
2351:8
2330:.
2308::
2278:.
2256::
2248::
2221:.
2199::
2169:.
2147::
2120:.
2100::
2094:8
2077:.
2055::
2047::
2041:7
2020:.
2000::
1976:.
1952::
1944::
1917:.
1895::
1868:.
1846::
1840:8
1819:.
1807::
1801:9
1780:.
1756:.
1734::
1715:.
1701::
1671:.
1647::
1620:.
1598::
1571:.
1549::
1541::
1511:.
1489::
1462:.
1424::
1414::
1387:.
1365::
1335:.
1313::
1286:.
1264::
1258:5
1237:.
1217::
1191:.
1169::
1139:.
1115::
1088:.
1066::
1058::
1052:8
1031:.
1009::
982:.
958::
952:4
931:.
909::
882:.
862::
856:3
839:.
817::
790:.
766::
758::
731:.
709::
673:.
653::
647:5
354::
124:I
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.